Biomolecular gradients in cell culture systems
Thomas M.
Keenan
and
Albert
Folch
*
Department of Bioengineering, University of Washington, Washington 98195, USA. E-mail: afolch@u.washington.edu
First published on 6th December 2007
Abstract
Biomolecule gradients have been shown to play roles in a wide range of biological processes including development, inflammation, wound healing, and cancer metastasis. Elucidation of these phenomena requires the ability to expose cells to biomolecule gradients that are quantifiable, controllable, and mimic those that are present in vivo. Here we review the major biological phenomena in which biomolecule gradients are employed, traditional in vitro gradient-generating methods developed over the past 50 years, and new microfluidic devices for generating gradients. Microfluidic gradient generators offer greater levels of precision, quantitation, and spatiotemporal gradient control than traditional methods, and may greatly enhance our understanding of many biological phenomena. For each method, we outline the salient features, capabilities, and applications.
 Thomas M. Keenan | Tom Keenan is a postdoctoral fellow in the Stem Cell Training Program at the University of Wisconsin, Madison working with Drs David Beebe and Clive Svendsen. He received his doctoral training from Dr Albert Folch at the University of Washington, Seattle and graduated in 2006. His current research interests are to use engineered microenvironments to study central nervous system development and develop new strategies for deriving relevant cell types from human stem cells. |
 Albert Folch | Albert Folch received his BSc in physics from the University of Barcelona (UB), Spain, in 1989. In 1994, he received his PhD in surface science and nanotechnology from the UB’s Physics Dept. During 1990–91 he was also a visiting scientist at the Lawrence Berkeley Lab working on AFM under Dr Miquel Salmeron. From 1994–1996, he was a postdoc at MIT developing MEMS under the advice of Martin Schmidt (EECS) and Mark Wrighton (Chemistry). In 1997, he joined the laboratory of Mehmet Toner as a postdoct at Harvard’s Center for Engineering in Medicine to work on BioMEMS. He has been at UW BioE since June 2000 where he is an Associate Professor. His lab works at the interface between neurobiology and microfluidics. In 2001 he received a NSF Career Award. He has two children, Jordi and Tàlia. |
Introduction
Biomolecular gradients are an important, evolutionarily-conserved signaling mechanism for guiding the growth, migration, and differentiation of cells within the dynamic, three-dimensional environment of living tissue. Gradients play essential roles in many phenomena including development, inflammation, wound healing, and cancer. Interest in elucidating these phenomena has led to the development of numerous in vitro methods for exposing cells to chemical gradients. In combination with in vivo studies, these methods have revealed gradient signaling to be an intricate, highly-regulated process, in which the ultimate cellular response is determined by the unique complement, concentration, and spatiotemporal characteristics of the gradients to which cells are exposed.Traditional in vitro gradient-generating methods have been instrumental in shaping our current understanding of gradient signaling, but they are not ideal for examining the quantitative or combinatorial nature of gradient signaling due to their inability to produce precise, user-defined gradients with tailored spatial and temporal profiles. The chemical gradients generated by traditional methods often evolve unpredictably or uncontrollably over space and time, and can be difficult to characterize quantitatively. The gradients form and dissipate within a few hours, greatly limiting the cell types and questions that can be studied. Elucidation of the complexities of gradient signaling requires more detailed knowledge and control over the spatiotemporal distribution of chemical species in the extracellular environment and the ability to directly visualize cells within that environment.
Because molecules diffuse isotropically and unrestricted in free solution, maintaining one or more gradients requires constant supply and removal of molecules at precise locations within well-defined liquid volumes. Microfluidic technology, whereby fluids can be routed precisely and with significant levels of automation at micrometer dimensions, provides an appealing strategy to control the fluid flow necessary to create gradients on a scale suitable for cellular studies. Recently, several microfluidic devices have been developed for generating gradients that are predictable, reproducible, and easily quantified. Many devices offer significant control over the shape and temporal characteristics of the gradient. In this review, we discuss the major biological phenomena that employ chemical gradients, traditional in vitro methods used to generate biomolecular gradients, and the new gradient-generating methods that exploit microfluidic technology.
Biomolecule gradients in biology
Development
The development of a fertilized egg into a fully-formed animal requires a delicately orchestrated sequence of cell proliferation, differentiation, and migration events.1 At developmentally-conserved time points, select groups of cells within the embryo secrete signaling proteins into the extracellular environment. These secreted signaling proteins diffuse away from source cells, forming chemical gradients that induce proliferation,2 differentiation,3 or migration1 in other cells. The extracellular space within the developing embryo is thus a highly dynamic chemical environment composed of multiple signaling protein gradients with overlapping spatial and temporal expression profiles. One classic example is the development of the mammalian pituitary gland. The pituitary gland is a small pouch of endocrine tissue at the base of the brain that plays a critical role in maintaining homeostasis, inducing growth, regulating reproduction, and stimulating lactation.4 These essential functions result from the highly-regulated activity of 6 distinct endocrine cell types that comprise the pituitary gland. The differentiation of primordial pituitary cells into these cell types and the cells' juxtaposition relative to one another are determined by overlapping gradients of bone morphogenic proteins (BMPs), fibroblast growth factors (FGFs), and molecules from the Wnt and sonic hedgehog (Shh) families.4 Gradients emanating from adjacent tissues induce primordial pituitary cells to secrete proteins that either modulate the effects of existing gradients or generate new gradients to further define the cell types and tissue architecture of the pituitary gland.4In addition to their role in cell differentiation, gradients of diffusible signaling proteins also provide directional cues to guide cell migration and growth during development.1 One of the most prominent examples is in the developing nervous system, when young neurons send out membrane protrusions called axons to innervate target cells that can reside at significant distances from where the neurons are born. For example, commissural neurons are specialized spinal neurons whose role is to relay sensory information to the brain. The ventral extension of commissural axons through the spinal cord is guided by a gradient of a protein called netrin-1, which is secreted by cells at the ventral midline forming a high-ventral to low-dorsal gradient.5,6 Once the axons reach the ventral midline they cross it and extend towards the brain (rostrally). Netrin-1 signaling is thought to sensitize the axons to a protein called Slit, which is also secreted by cells at the ventral midline. Upregulation of the Slit receptor Robo in the commissural axon growth cone allows the growth cone to sense the Slit gradient and respond in a repulsive manner to extend beyond the ventral midline.7,8 Studies in frogs have shown that spinal neuron contact with laminin can convert Netrin-1 attraction into repulsion, indicating that contact guidance cues may also play a role.9–13 Other molecules such as semaphorins14 and ephrins15 may also help prevent midline recrossing by commissural axons. Once the axon crosses the ventral midline, it turns rostrally in response to attractive gradients of signaling proteins including Wnt4.16
Because of the high fidelity with which developmental processes need to occur for proper biological function, the signaling protein gradients used to guide the growth, differentiation, and migration of developing cells are highly spatially and temporally regulated. Rarely do these gradients act independently8,12,13 as evidenced by the fact that many gradient-induced signaling cascades share or have interacting intracellular second messengers, which facilitate cross talk and signaling pathway modulation.17–19 The specific differentiation or migration response of a cell is thus determined not by a single protein gradient, but by the unique complement of gradients, their concentrations, and the spatiotemporal gradient characteristics to which a cell is exposed.1
Immune response
Mitigating the harmful effects of pathogenic microorganisms that have infected the body depends entirely on the ability of host immune cells to detect and locate invading microorganisms and recruit other immune cells to the site of infection. Biomolecule gradients play a critical role in this process by providing immune cells with the directional cues they need to rapidly migrate to the infection site. When a bacteria or virus enters the body, it is often first recognized by phagocytic immune cells called macrophages that reside within the tissue.20Macrophages that come in contact with bacteria or the proteins they produce begin secreting signaling proteins called cytokines. These molecules diffuse into the tissue space forming gradients originating at the site of infection. As part of the inflammatory response, cytokine-stimulated endothelial cells lining nearby blood vessels begin expressing cell adhesion molecules (e.g. ICAM-1), and adhesive proteoglycans (e.g.P-selectin and E-selectin). Immune cells called neutrophils, which circulate benignly in the blood stream, are ensnared and activated by the adhesive molecules on the endothelial surface. The neutrophils crawl between adjacent endothelial cells into the tissue space. Multiple gradients of cytokines (e.g. CXCL-8, TNF-α), complement protein fragments (e.g. C5a), and bacterial peptides (i.e. formylated peptides) present in the tissue space induce the neutrophil to adopt a polarized morphology, with the side of the cell nearest the gradient source forming a large lamellopod and the other side forming a thin, bulbous pseudopod called a uropod. Differential ligand–receptor binding across the cell keeps the cell in this polarized morphology and through its effects on cytoskeletal dynamics, produces directed migration towards the gradient source (i.e. site of infection).17,21 Upon reaching the infection site, neutrophils and macrophages phagocytose and destroy resident bacteria.Cancer
Biomolecule gradients have been implicated in playing a critical role in the development of metastatic cancer.22 In order for tumors to metastasize, the cells must escape the original tumor, invade new tissues, and recruit endothelial cells to create blood vessels to feed the new tumor site. Many proteins known to form gradients in other systems have been shown to increase cell motility and chemotaxis in cancer cell lines22–25 and promote angiogenesis.26 In particular, the biomolecular gradients used by immune cells to cross tissue boundaries and find sites of infection with the tissue space, can also be used by metastatic cancer cells to exit the bloodstream, take up residence in locations distant from the original tumor, and recruit endothelial cells to begin building blood vessels.27,28 In two recent studies, CXCL-8, the same molecule used to recruit neutrophils to the site of bacterial infection, was found to induce cancer cell proliferation.29,30 Tissues that are common sites of metastasis, such as the lymph nodes, lungs, liver, and bone marrow have been found to express high levels of certain cytokines.22 In fact, a large percentage of solid tumors are filled with non-cancerous chemotaxing cell types such as immune cells and fibroblasts.28Traditional in vitro gradient-generating methods
The importance of biomolecular gradients in directing the growth, differentiation, and migration of various cell types in vivo has motivated researchers to develop numerous methods for generating chemical gradients in vitro. Below we review some of the most common techniques traditionally used to expose cells to biomolecule gradients:Biological hydrogels
Biological hydrogels made from collagen, fibrin, or agarose (Fig. 1) are commonly used to establish biomolecule gradients around cellsin vitro.18,31–33 Dissociated cells or whole-tissue explants are either seeded on a cell culture surface and overlaid with gel, or are homogeneously mixed with the liquid hydrogel solution prior to gelation. A biomolecule gradient is traditionally formed within the gel in one of two ways. The first method is to co-culture the experimental tissue or cells with another cellular source, such as a tissue explant or transfected cells that release the chosen biomolecule.31 The second method is to mold or excise voids within the gel and fill them with the soluble form of the biomolecule at a known concentration.32 More recently biological hydrogel gradients have been created by directly depositing arrays of spaced droplets or lines of biomolecule on the surface of gels and allowing the droplets to diffuse into the hydrogel matrix.34 Defined droplets can be dispensed with a computer-controlled pump34 or through inkjet printing.35,36 For all methods the biomolecule diffuses away from its source into the gel forming a concentration gradient over the cells that evolves in both space and time (Fig. 1c).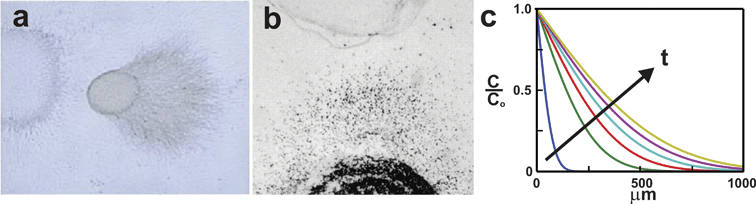 | ||
| Fig. 1 Biological hydrogels. Gels can be used to expose cells to biomolecule gradients. (a) A neural tissue explant co-cultured with semaphorin-expressing COS cells in a collagen gel (adapted from ref. 31, copyright Elsevier, 1998). (b) Neutrophils (black dots) are exposed to opposing gradients of Il-8 and leukotriene B4 in an under-agarose assay (reproduced from ref. 32, copyright 1997 The Rockefeller University Press). (c) The gradient produced in biological hydrogels varies in space and time as shown in this error-function solution of the concentration profiles generated at 5 min intervals for a molecule diffusing away from a constant concentration source using an assumed diffusivity of D = 6.4 × 10–7 cm2 s–1. | ||
The biological hydrogel method has several unique features that have made it popular for exposing cells to biomolecule gradients. First, the gels are easy to make and provide cells with an environment that is, in theory, more similar to in vivo tissue than conventional two-dimensional culture substrates. Second, the high network density of the gels allows the movement of chemical species to occur only via free diffusion, unaffected by bulk fluid movement around the gel. Third, the biological hydrogel method offers significant control over the positions of the biomolecule sources, enabling the generation of gradients with specific orientations, or combinatorial gradients of multiple factors over distances as small as 2 mm.18,32
Unfortunately, the biological hydrogel method offers little control over the spatiotemporal evolution of the gradient and generates gradients with poor reproducibility. The use of cell-based biomolecule sources (i.e. explants, transfected cell lines) results in gradients that are highly variable and difficult to quantitatively characterize due to the unknown level of biomolecule secreted by the cells. Direct quantification of the biomolecule concentration by engineering the cells to express fluorescent fusion proteins is not practical due to the technical challenge of detecting the nanomolar concentrations to which receptive cells typically respond.31–33 Further lack of spatial and temporal gradient control is caused by the fact that the diffusion of chemical species in the biological hydrogel is determined strictly by the chemistry of the polymer chains and the network porosity of the gel. Once the biomolecule has been loaded into the source compartment, the biomolecule concentration, gradient shape, and rate at which the gradient evolves in both space and time cannot be modified. The use of biological hydrogels is also subject to experiment-to-experiment variability due to the manual placement of cell and biomolecule source compartments. Manual placement can at best attain millimeter precision, while cells are sensitive to changes in their environment that occur over distances 1000 times smaller.37 The inability to determine or regulate the nature of the gradient each cell is exposed to at a given time, makes it difficult to correlate an observed cell response with the specific concentrations and spatiotemporal profiles of the applied gradient.
Another disadvantage of biological hydrogels is the difficulty of observing single cell responses due to the 3D architecture of the culture system and the optical properties of some biological hydrogels. In contrast to 2D culture architectures where cells can be imaged with a conventional light microscope at a single focal plane, 3D cultures require cells to be tracked in 3 dimensions using z-series image stacks (e.g.confocal microscopy) and non-trivial (or expensive) 3D tracking algorithms. The optical properties of some gels also make it difficult to distinguish the cell from the hydrogel background. For example, collagen gels have ordered fibrils that refract incident light and interfere with phase contrast microscopy.
In summary, biological matrix gels provide a qualitative and easy method for exposing cells to one or more time-evolving gradients in a 3D matrix with no complex equipment. However, the method is not suitable for quantitatively understanding how cells respond to gradients with specific characteristics, or the integration of multiple gradient signals due to an inability to maintain reproducible gradients for long periods of time, control the spatial and temporal evolution of the gradient, and reliably observe single-cell responses.
Micropipette-generated gradients
Another method for exposing cells to biomolecule gradients is through the use of drawn glass micropipettes. One variation of the micropipette method (Fig. 2) was originally developed by Gundersen and Barrett38 and is now most commonly associated with the work of Mu-Ming Poo's group.9–12,39–41 In this method a glass capillary is heated and pulled axially, forming a fine glass tip with an internal diameter of approximately 1 µm. This micropipette is filled with a biomolecule solution and its tip is positioned at a set distance from cells using a mechanical manipulator and microscope. The biomolecule solution is pneumatically ejected out of the pipette into the extracellular environment. The frequency of the puffs and the volume ejected with each puff are determined by the pneumatic pump frequency and driving pressure. The puffs combine to form a diffusive gradient that emanates radially from the micropipette tip. The second variation of the micropipette method is identical to the first except that instead of being pneumatically ejected, the biomolecule solution is simply allowed to passively diffuse from the micropipette tip.42–45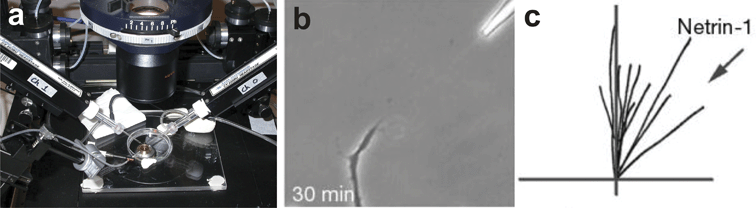 | ||
| Fig. 2 Micropipette gradient generation. (a) Electrophysiology micropipettes loaded with soluble signaling molecules are mounted in micromanipulators arranged around the cell culture dish (courtesy of Dr Donglin Bai, University of Western Ontario, London, Ontario). (b) The micropipette is brought within a defined distance to the cell and the biomolecule is pneumatically ejected from the pipette generating a gradient. Here, a Xenopus spinal neuron is turning in response to a netrin-1 gradient (adapted with permission from Macmillan Publishers Ltd: ref. 41, copyright 2002). (c) The response of the neuron can be quantified by tracing the resulting growth trajectories (adapted with permission from Macmillan Publishers Ltd: ref. 41, copyright 2002). | ||
The micropipette method readily generates gradients and elicits responses from a variety of cell types including neutrophils42–45 and primary neurons.9–12,39–41 Unlike biological hydrogels, the micropipette method is well suited for characterizing single cell responses since cells are cultured on a surface at a single focal plane and in a medium that does not interfere with the ability to discern the cell from the background. The gradient can be oriented at a specific angle and distance relative to the cell to provide a more quantitative assessment of the response of the cell to the gradient. Multiple micropipettes can be placed around the cell46 and in principle could be used to generate multifactor combinatorial gradients.
Like the biological hydrogel method, the micropipette method does not generate highly reproducible or controllable gradients. Because the biomolecule gradient is formed in free solution small vibrations, thermal imbalances, or evaporation can cause convective currents that distort the gradient during the course of an experiment. Variability also arises from differences in the shape of the pulled micropipette tips. The gradient generated by pneumatic ejection depends greatly upon the hydrodynamic properties of the micropipette, which is determined by the shape of the micropipette tip. The shape of the micropipette tip depends on how fast and hot the parent capillary was heated, and the rate at which the capillary was pulled. Because micropipettes pulled even on the best commercial micropipette pullers do not heat and break in precisely the same manner, significant geometric differences between micropipettes can result. The resulting geometric differences can produce different gradients even when the micropipettes are pulled and operated under the same conditions. Direct quantification of the gradient is complicated by the fact that the gradient is created by a point source suspended above the cell culture surface. Gradients are generated in all three dimensions, necessitating the use of a confocal microscope for indirect gradient quantification using fluorimetric dyes.
Although it is theoretically possible to tune the shape and spatiotemporal evolution of the gradient by modifying the driving pressure and puffing frequency, only a single set of parameters has been historically used to generate gradients with micropipettes: 3 psig of pressure is applied for 10–20 ms at 2 Hz to a micropipette ∼1 µm in diameter.10–12,39–41,47,48 The gradient commonly assumed to be generated under these conditions is based on a single study9 which used the diameter of aqueous droplets ejected into oil to estimate the volume of aqueous fluid ejected into an aqueous environment for each pulse. No correction for the water–oil surface tension was incorporated, nor was there any examination of the reproducibility of the gradient when different micropipettes pulled under the same conditions were used.
Another significant limitation of the micropipette method is its reliance on bulky micromanipulators to position the micropipette tips with respect to the cells. The micromanipulators restrict the number and positions of the micropipettes that can be placed around the cells, and make it difficult to utilize microscope stage incubators to control temperature, pH, and humidity. The challenge of on-stage environmental control makes this method most suitable for non-mammalian cell types9–12,39–41,47,48 or mammalian cell types that respond to the biomolecule gradient within ∼1 h.9–12,39–45,47,48
In summary, generating gradients with micropipettes enables quantitative characterization of single cell responses. However, the distortion of gradients by convective currents and the undemonstrated ability to generate controllable, reproducible gradients makes the method unsuitable for examining cell responses to gradients with specific characteristics (i.e. slope, concentration range, etc.) or the integration of multiple biomolecule gradient signals.
Boyden Chamber/Transwell Assay
In 1962, Stephen Boyden developed a gradient-generating method that along with its modern form, the Transwell Assay, has greatly advanced our understanding of chemotaxis.49 The method begins by placing a chemoattractant solution in the lower compartment of a culture well (Fig. 3). Another culture well with a porous membrane bottom is seeded with cells, and placed in the lower compartment causing the chemoattractant contained therein to diffuse across the membrane into the upper compartment. The resulting gradient induces cells seeded on the top side of the membrane to migrate through the transmembrane holes to the lower compartment. The cells that migrate to the bottom of the membrane are then fixed, stained and counted to quantify the degree of chemotaxis induced by the gradient.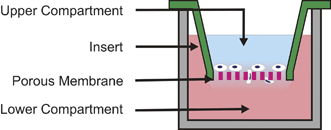 | ||
| Fig. 3 Transwell Assay. This assay is based on the Boyden Chamber method. Cells seeded on a porous membrane are placed in a well containing a chemoattractant solution. The chemoattractant in the lower compartment diffuses into the upper compartment forming a gradient across the membrane. Cells respond by migrating through the membrane to the bottom surface where they can be subsequently fixed, stained, and counted. | ||
The advantages of the Boyden Chamber/Transwell Assay are that it is easy to perform, readily elicits chemotactic responses from cells, and provides a quantitative measure of the level of transmembrane migration induced by chemotaxis. Multiple assays can be run in parallel to increase the statistical strength of observed findings and expand the number of different conditions examined during each experiment.
However, even under ideal conditions the gradient that forms across the membrane varies over space and time and cannot be controlled. Gradient instability is created by fluid level imbalances between the upper and lower compartments that generate gradient-distorting flows across the membrane. Fluid level imbalances are common due to pipetting error when loading the compartments and the requirement that the insert, with its liquid-porous bottom, be dropped into the lower compartment. Complex multi-factor combinatorial gradients (i.e. unique gradient shapes, concentrations, and orientations) cannot be generated in the Boyden Chamber/Transwell Assay due to geometric limitations, which at most allow two opposed gradients to form along a single axis perpendicular to the surface of the membrane. Gradient quantification is complicated by the presence of the membrane (optically translucent, but not transparent) and the geometry of the setup, which requires confocal microscopy for indirect gradient quantification using fluorimetric dyes. The membrane also makes it difficult to visualize single cells, which in combination with the inability to control and quantify the gradient, precludes correlation of observed cell responses with specific gradient characteristics. As a result, the Boyden Chamber/Transwell Assay is best suited as a population-based, migrational assay as it was originally designed.
In summary, the Boyden Chamber/Transwell Assay is a very effective means for quantifying the migrational response of cells under various conditions with high statistical rigor. However, the inability to directly visualize cells, and quantify or control the biomolecule gradient makes the method unsuitable for correlating specific cell responses with particular gradient characteristics (i.e. slope, concentration, temporal evolution, etc.). The inability to generate multiple independent gradients prevents the use of the Boyden Chamber/Transwell Assay for studying multi-gradient signal integration.
Zigmond Chamber
In 1977, Sally Zigmond developed a device for characterizing the migration of individual neutrophils in response to well-defined gradients of various chemotactic factors.50 The Zigmond Chamber, shown in Fig. 4a, was the first to allow direct visualization of cell behavior in the presence of a biomolecule gradient. The device consists of two parallel channels etched into a glass slide. Between the etched channels is a glass ridge that lies below the top surface of the slide. Cells are seeded on a glass coverslip and the coverslip is inverted over the etched channels forming a thin gap 3–10 µm tall and 1 mm wide between the coverslip and the glass ridge. The addition of 100 µL of culture medium in the sink channel and 100 µL of the biomolecule solution in the source channel causes a gradient to form in the thin gap over the glass ridge. Changes in cell growth, differentiation, or migration of individual cells can be viewed with a microscope in the glass ridge region.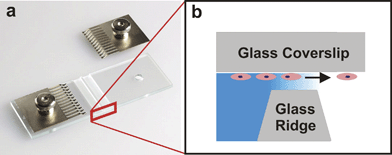 | ||
| Fig. 4 Zigmond Chamber. (a) The device consists of two etched channels separated by a glass ridge. The metal tines are used to clamp an inverted glass coverslip seeded with cells to the device (used with permission from Neuroprobe, Inc., Gaithersburg, MD). (b) A cross section schematic of the device shows cells on the inverted coverslip migrating in response to the gradient established between the coverslip and the glass ridge. | ||
The geometry and precise dimensions of the Zigmond Chamber produces a gradient that is reproducible, mathematically predictable, and can be indirectly estimated using fluorimetric dyes. Although the gradient evolves over time, during approximately the first hour of operation the small volume of the gap region (where diffusive mass transport occurs) and the comparatively large volumes of the source and sink channels approximates an “infinite source-infinite sink” system51 causing a near steady-state gradient to form. In addition, unlike the aforementioned methods, the symmetry and precise dimensions of the Zigmond Chamber creates a uniform 1-D gradient that can be calculated analytically for all time points and biomolecule concentrations loaded into the source or sink channels. The ability of the Zigmond Chamber to allow direct visualization of cell behavior and to create a quantifiable, pseudo-steady-state gradient enables the correlation of single-cell behavior with specific gradient characteristics.
The main limitations of the Zigmond Chamber are that the gradients it creates have life spans of approximately 1 h and the gradients cannot be modified to achieve specific spatial and temporal evolution profiles once the source and sink channels have been filled. Different gradients can be achieved by loading fluids with different biomolecule concentrations into the source and sink channels, but the characteristics of the gradient (i.e. slope, position, spatial and temporal evolution) are determined exclusively by the geometry of the chamber and the diffusion coefficient of the biomolecule. There are no means by which the user can dynamically modify the gradient during the course of an experiment. The rapid overall evolution of the gradient limits the use of the Zigmond Chamber to fast-responding cell types such as neutrophils,52–61 and sperm.62–64 Like the Boyden Chamber/Transwell Assay, the Zigmond Chamber cannot generate complex multi-factor combinatorial gradients. Only two-factor combinatorial gradients can be created along a single axis. In addition, the design of the Zigmond Chamber's source and sink channels makes the generated gradient extremely sensitive to evaporation. Evaporation adversely affects the gradient by concentrating dissolved species in each chamber, inducing fluid flow across the gap region, and reducing the length of the device where an effective gradient can be maintained. Over several hours evaporative losses can destroy the gradient and harm the viability of the cells.
In summary, the Zigmond Chamber is a very effective method for exposing cells to reproducible, predictable, and quantifiable biomolecule gradients over time scales of up to 1 h. The gradient is nearly steady-state for much of the time and can be indirectly estimated using fluorimetric dyes, allowing correlation of observed cell responses with specific gradient characteristics. However, its inability to maintain steady gradients over longer periods of time, susceptibility to evaporation, and inability to generate complex multi-factor gradients prevents its use in understanding cell responses to specific gradient characteristics and elucidating how cells integrate multiple biomolecule gradient signals to bring about a particular biological response.
Dunn Chamber
In 1991, Graham Dunn developed a device that is very similar to the Zigmond Chamber, but is much less susceptible to evaporation.65 In the Dunn Chemotaxis Chamber (Fig. 5) the source and sink chambers are arranged as concentric rings that can be filled with the appropriate solutions prior to affixing the glass coverslip seeded with cells. When the coverslip is inverted and clamped, it seals both chambers and eliminates the air–liquid interfaces that contribute to evaporative losses.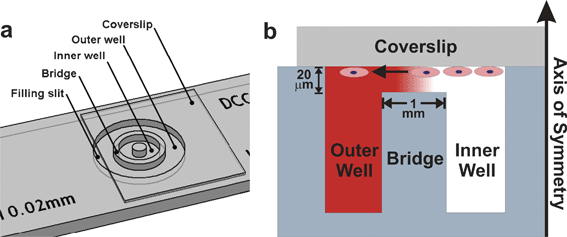 | ||
| Fig. 5 Dunn Chamber. The Dunn Chamber is similar to the Zigmond Chamber but much less susceptible to evaporation. (a) The device consists of two wells arranged as concentric rings, and separated by a glass bridge (image courtesy of Hawksley Medical and Laboratory Equipment, Lancing, Sussex, UK). (b) A gradient forms in the 20 µm gap between the cell-seeded inverted coverslip and the glass bridge. Cell responses can be directly visualized in the bridge region. | ||
Although the gradient generated in the Dunn Chamber is less susceptible to evaporation, the advantages and disadvantages of the Dunn Chamber vary little from those of the Zigmond Chamber. The gradient is pseudo-steady state for periods of time up to 1–2 h, and can be calculated for all time points and biomolecule concentrations. Like the Zigmond Chamber, the Dunn Chamber also has a ridge region that allows cell responses to be imaged directly and correlated with specific gradient characteristics. The gradients are short-lived and time-evolving, and cannot be modified once solutions are loaded and the coverslip is secured. Like the Boyden Chamber/Transwell Assay and the Zigmond Chamber, the Dunn Chamber is only capable of generating multi-factor combinatorial gradients along a single axis. The only major difference in the gradients generated by the Zigmond and Dunn Chambers is their geometric symmetries. From a practical point of view, evaluating the cell response from a radially symmetric design is slightly more difficult since cells must be tracked using polar instead of Cartesian coordinates. Tracking cells along a single axis, as in the Zigmond Chamber, is more straightforward.
Questions that cannot be addressed with traditional methods
The in vitro methods we have described have been invaluable in identifying new biomolecules that elicit gradient-dependent cell responses and elucidating aspects of the mechanisms by which these responses are exacted. They have revealed gradient signaling to be an intricate, highly-regulated process, dependent upon a complex signaling network that integrates the intracellular signals generated by all the different biomolecule gradients to which a cell is exposed.1,66Unfortunately, further elucidation of gradient-dependent phenomena is hindered by our lack of quantitative knowledge about the biomolecule gradients actually present in vivo. For embryonic development and axon guidance, immunocytochemistry and in situ hybridization have helped determine when certain biomolecules are expressed and their relative distribution in the tissue at key developmental time points. For example, Isbister and colleagues67 used in situ hybridization to reveal that a chemorepellent in the grasshopper limb is expressed in overlapping high dorsal-low ventral and high distal-low proximal gradients. The gradients work sequentially to guide the axons arising from a specific set of neurons along highly-conserved pathways through the tissue to their developmental targets. However, immunocytochemistry and in situ hybridization have not been able to provide sufficient quantitative information to establish the precise biomolecule concentrations and gradient profiles needed to elicit specific cellular responses. Similarly, for immune and wound healing responses, trying to measure and map the levels of various bacterial peptides and chemokines in and around an infected or damaged tissue site is extremely challenging. In vitro experiments using traditional gradient generating methods have helped determine the biomolecules capable of eliciting chemotaxis as well as the prioritization hierarchy used by cells to find infected or damaged tissue sites most efficiently.32,68–75 However, little is known about the specific biomolecule concentrations and gradient profiles that elicit the most efficient responses, how these gradient characteristics influence the cell's prioritization hierarchy, and how they influence the cell's sensitivity to other biomolecule gradients.68,71,74 Complicating matters further is the fact that the in vivo concentration and profile of a biomolecule gradient are not simply a function of the biomolecule's diffusive properties, but also of its binding kinetics to other species in a dynamic physicochemical microenvironment. A wide variety of feedback and control mechanisms are used to regulate the concentration and distribution of biomolecule in living tissue.66 Deciphering the true characteristics of physiologically-relevant gradients and how these highly-dynamic gradients are integrated to produce specific cell responses requires a way to expose cells to one or more biomolecule gradients, each with user-defined spatial and temporal distributions.
Recent advances in microfluidic gradient generators provide a way to create predictable, reproducible, and easily-quantified biomolecule gradients in vitro. Many of these devices offer increased spatial and temporal gradient control that can be used to systematically characterize how specific gradient features such as concentration and gradient profile influence cell response. For fast responding cell types such as those involved in immune response, microfluidic gradient generators have already begun providing great insight. As microfluidic gradient generating technology continues to evolve and the performance of devices extends to longer time scales, the same detailed and quantitative understanding shown here for immune cells can be constructed for slower responding cell types such as those involved in embryonic development and axon guidance. Microfluidic gradient generators also have the potential to create multiple biomolecule gradients each with its own user-defined spatiotemporal distribution. The ability to create complex, user-defined gradient environments would enable quantitative elucidation of multi-gradient signal integration and provide the specific recipes for engineering the growth, migration, and differentiation of a variety of cell types.
Microfluidic methods
The rapid evolution of the microelectronics industry during the past 30 years has produced numerous tools and methods for fabricating micrometer-scale features in and on various substrates with very high precision and at relatively low cost. Realization that these advances in microfabrication technology could be used to control the micrometer-scale environment, or microenvironment, of cultured cells has led to the development and rapid expansion of two closely related fields: Biomedical Electromechanical Systems (BioMEMS) and Microfluidics (µF). BioMEMS and µF research have provided a plethora of ways to explore how cells respond to micrometer-scale modifications of their physical and chemical environments.76–81 Because microfabrication allows creation of cell culture environments that are well-defined with micrometer precision, quantitative characterization and experimental reproducibility are greatly enhanced.Recently, µF researchers have focused on the need for better gradient-generating methods. Microfluidic cell culture environments are uniquely suited to achieving the level of quantification and gradient control required to correlate observed cell responses with specific gradient characteristics, and elucidate how cells integrate different biomolecule gradients to achieve particular biological responses. The precise dimensions of µF devices combined with our detailed understanding of the behavior of fluids at the micrometer scale confers unique advantages to µF gradient-generators over traditional methods. Fluids flowing in micrometer-scale conduits, or microchannels, are dominated by the viscous properties of the fluid at the expense of the inertial forces generated by the fluid. This specialized flow regime, called laminar flow, is well-understood both conceptually and mathematically82 allowing the movement of momentum, heat, and chemical species inside a microfluidic device to be calculated with great accuracy.83 When used with commercially-available, finite element modeling software packages, the defining equations developed for laminar flow can be used to characterize an existing device or help prototype new devices to create specific, engineered cell culture environments. Virtual prototyping streamlines the fabrication process, ensuring that the appropriate cell culture environment will be created in the actual device.
Microfluidic devices have practical advantages that increase the throughput and reduce the cost of gradient experiments when compared to traditional gradient-generating methods. The parallel nature of microfabrication methods, or scalability, and the small physical footprint of most microfluidic devices allow multiple gradient-generating cell culture environments to be constructed and placed in the same physical space typically occupied by a single traditional set up. The greater experimental throughput reduces the time and monetary cost of each experiment. In addition, the small volumes of microfluidic devices require vastly smaller amounts of precious reagents. This reduces the cost of each experiment and enables the creation of biomolecule gradients that would be too expensive to create using traditional gradient-generating methods.
Microfluidic gradient generators have been used to infect cells with graded concentrations of virus,84 generate gradients of mechanical stiffness85 and cell-adhesion molecules in synthetic extracellular matrices,86 create adsorbed gradients of extracellular matrix proteins,87–90 induce proliferation and differentiation in neural stem cells,91 and examine the effects of various biomolecule gradients on the chemotaxis of neutrophils,92–95 bacteria,96,97sperm,98 breast cancer cells,99,100 and intestinal cells.90 Here we focus our review on the major microfluidic gradient generators that are capable of forming reproducible, well-defined gradients suitable for cell culture studies.
Substrate-bound biomolecule gradients
One of the easiest ways to create gradients using microfabricated tools is to selectively adsorb or tether the biomolecule of interest to the cell culture surface. Surface-attached gradients can be very stable allowing correlation of observed cell response with specific gradient characteristics. However, gradients may have limited biological functionality due to partial protein denaturation or steric hindrance that can result when the biomolecule is adsorbed or tethered to a solid surface. Gradient stability depends on the biomolecule's binding or reaction kinetics as well as components in the cell culture medium that can degrade or replace the adsorbed biomolecule on the surface.Depletion gradients
One microfluidic gradient-generating method capitalizes on the high surface area-to-volume ratio of microfluidic channels to create adsorbed chemical gradients on cell culture substrates. The method developed by Delamarche and colleagues101 and modified by Fosser and Nuzzo88 employs a microfluidic channel with a reservoir at one or both ends (Fig. 6). A biomolecule solution is dispensed into the reservoir at one end of an empty or liquid-filled channel. The solution is drawn into an empty channelvia capillarity,101 or diffuses into a pre-filled channel88 and adsorbs non-specifically to the internal surfaces. Because of the large surface area-to-volume ratio of microfluidic channels, the level of biomolecule adsorption significantly depletes the amount of biomolecule in free solution. A gradient of adsorbed biomolecule is formed within the channel with the highest concentrations nearest the biomolecule reservoir and the lowest at the distal end of the channel. For empty channels, the hydrophilic nature of the channel surfaces controls the rate at which the biomolecule solution will be drawn into the channels. If the channels are not sufficiently hydrophilic, they may fill over time scales that allow biomolecules in the reservoir to readily replenish those that adsorb to the internal surfaces of the microchannel. The result would be very steep, short gradients101 or no gradients at all. For pre-filled, non-adsorbing microchannels, gradient formation is much more straightforward and only depends upon the biomolecule diffusion coefficient and the incubation time; gradients become broader as incubation time increases. The characteristics of the gradient (i.e. slope, concentration, spatiotemporal evolution) are entirely dependent upon the biomolecule diffusion coefficient, the interaction of the diffusing biomolecule with the microchannel material, and the length of time over which the gradients are allowed to form.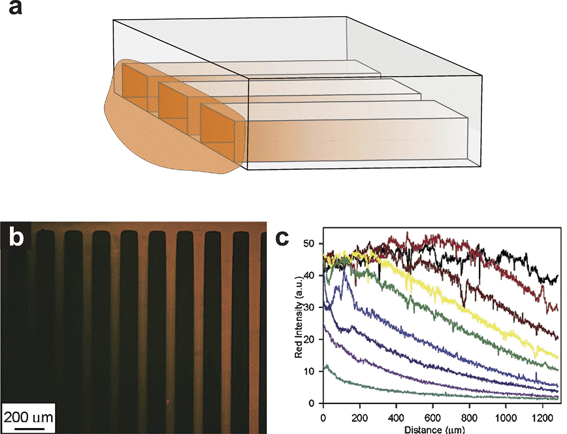 | ||
| Fig. 6 Depletion gradient. (a) Microfluidic channels have high surface area to volume ratios that can deplete the concentration of chemicals inside the microchannel if those chemicals bind to internal surfaces. Chemical solutions applied at one end of a microchannel can be used to form adsorbed chemical gradients. (b) Adsorbed depletion gradients of BSA-TRITC (reprinted with permission from ref. 88, copyright 2003 American Chemical Society). (c) Intensity profiles of each adsorbed gradient shown in (b) (reprinted with permission from ref. 88, copyright 2003 American Chemical Society). | ||
The advantages of using depletion gradients are that they are easy to establish and require no complex equipment or labor-intensive fabrication steps. The adsorbed gradients can be quite stable, allowing correlation of observed biological responses with specific gradient characteristics. Although the method has only been used to generate adsorbed biomolecule gradients, it could theoretically be used to generate soluble gradients as well. Functionalizing the internal microchannel surfaces with different chemistries might also provide the user with some control over the shape and evolution of the gradient.
However, it is difficult to create depletion gradients with user-defined concentration profiles due to the reliance on adsorption kinetics, which is influenced by a variety of factors including number of surface functional sites, molecular access to those sites, temperature, local concentration, concentration of secondary solutes, etc. Because many of these factors cannot be controlled, the types of gradients that can be generated are limited. Also, adsorbed gradients provide no way to dynamically alter the gradient in a prescribed manner. Another potential concern is the change in functionality that may occur when biomolecules are adsorbed or chemically linked to a surface. Proteins partially denature when adsorbed to a surface; depending on where and how rigidly the biomolecule is attached to the surface cell access to binding epitopes may be limited. The adsorbed biomolecule may also interact with or be replaced by other proteins or compounds present in the cell culture medium, reducing the longevity and stability of the gradient.
Micropatterned gradients
Adsorbed biomolecule gradients can also be generated using microstamping102,103 or by flowing biomolecule solutions of different concentrations through microfluidic channels. The method developed by von Philipsborn and colleagues104 used a microfabricated elastomeric stamp to print proteins on a cell culture substrate. By controlling the spacing and density of high-resolution patterns they were able to closely approximate continuous gradients with different slopes and concentration ranges (Fig. 7). They then used the patterns to study the response of chick temporal retinal axons to gradients of ephrin A5.105 Because the patterns were of subcellular resolution, cells presumably detected the discrete gradients as a continuum and responded in a manner dependent upon gradient slope (Fig. 7d–f).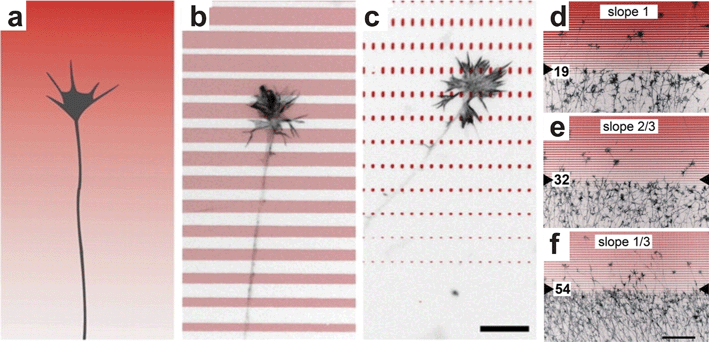 | ||
| Fig. 7 Micropatterned gradients. (a) Schematic of a growth cone navigating a continuous gradient. (b–c) By controlling the spacing and size of printed ephrin A5 (antibody-stained ephrin shown in red) a continuous gradient can be approximated at the cellular level. (d–f) Temporal retinal axons were repelled by the micropatterned gradient in a manner dependent upon the slope (i.e. increasing width) of the ephrin patterns (reproduced from ref. 105 with permission of the Company of Biologists). | ||
The advantages of using micropatterned gradients are that they are relatively easy to make and require no complex equipment to maintain. The user has greater control over the characteristics of the gradient than for depletion gradients since the gradient shape and concentration are determined by the size and spacing of the printed patterns and not by adsorption kinetics.
However, micropatterned gradients are not continuous and like depletion gradients cannot be dynamically altered once formed. Micropatterned gradients are also limited to biomolecules whose functionality does not change significantly when adsorbed or chemically linked to a solid surface. As for all adsorbed gradients, gradient stability is affected by the presence of other proteins or compounds in the cell culture medium that may degrade or supplant the adsorbed biomolecule.
Time-evolving gradient generators
The second class of microfluidic gradient generators consists of devices that employ no active means to control the distribution of chemical species within the device, resulting in gradients that evolve in space and time. Gradients have a limited lifetime, which is determined solely by the diffusion coefficient of the biomolecule and the geometry of the device. Passive gradient generators reliably form gradients, and are easy to mathematically and quantitatively characterize.Nanopore Gradient Generator
One of the major difficulties in generating passive diffusion gradients in microfluidic devices is eliminating uncontrolled convective fluid flow that can distort or destroy the gradient. Microfluidic devices are uniquely sensitive to surface tension- or evaporation-induced fluid flow due to their small size and large surface area-to-volume ratios. Abhyankar and colleagues95 used a novel approach to eliminate convective fluid flow inside a passive diffusion gradient generator. The gradient generator they developed, which we will refer to as the Nanopore Gradient Generator, utilized polyester track etch membranes perforated with nanometer-scale pores. The pores provided sufficient fluid flow resistance to eliminate convection while still providing enough mass transfer area for effective diffusive mass transport. The device, shown in Fig. 8, consists of three layers in a lollipop configuration. The inlet for the biomolecule source solution near the stem of the lollipop is separated from the gradient chamber in the bottom layer by a polyester membrane perforated with 200 nm-diameter holes (Fig. 8a). The cell addition inlet located near the elliptical gradient sink is separated from the gradient-forming region by a similar membrane, but one containing 10 µm diameter holes. Abhyankar and colleagues95 demonstrated that a gradient of an 885 Da fluorescent dye formed within 6 h and lasted more than 24 h. Neutrophils deposited in the cell-seeding inlet settled through the 10 µm holes and attached to the floor of the sink region (Fig. 8b). When the bacterial peptide formyl-methionine-leucine-phenylalanine (fMLF) (438 Da) was added to the source inlet, neutrophils migrated from the sink region towards the source region.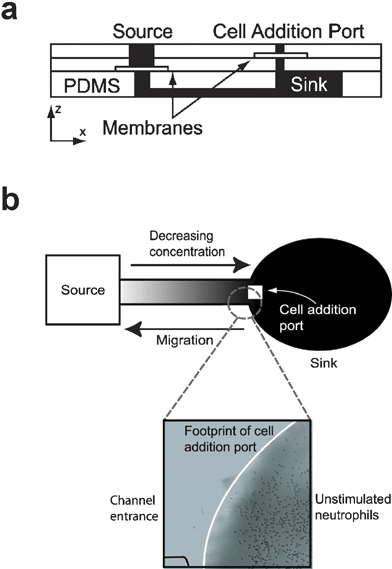 | ||
| Fig. 8 Nanopore Gradient Generator. (a) Cross-section schematic of the device shows the polyester track etch membranes encapsulated in three layers of PDMS, with the gradient/cell culture chamber composing the bottom layer. (b) Cells loaded into the cell addition port attached to the floor of the sink region and migrated towards the source region in response to a gradient of the bacterial peptide f-met-leu-phe (fMLF) (adapted from ref. 95, reproduced by permission of The Royal Society of Chemistry). | ||
The Nanopore Gradient Generator generates quantifiable gradients capable of eliciting chemotactic responses from cells. The fact that gradient formation does not require complex equipment or pumping schemes and utilizes frugal amounts of reagents are clear advantages. In addition, because convective fluid flow is eliminated, essential autocrine and paracrine cell signaling factors are allowed to accumulate. Although not demonstrated by Abhyankar and colleagues, the Nanopore Gradient Generator design could be extrapolated to generate multiple independent gradients with various orientations and positions.
However, the Nanopore Gradient Generator is not suitable for all gradient studies, especially those requiring long-term gradient exposure. Because no active means are used to maintain the source and sink concentration, the device is restricted to forming time-evolving gradients, and has no means of controlling the gradient's spatiotemporal evolution once the gradient fluids have been loaded. As is common with all non-perfused, finite-volume microchannels cells growing within the device become increasingly affected by depleted nutrient levels and increasing concentrations of cell waste products as time in culture progresses. Also, although the transparent properties of the device allow indirect gradient quantification using fluorimetric dyes, full mathematical characterization is more complicated due to required estimation of the mass flux across the track etch membrane.
Microvalve Chemotaxis Device (µVCD)
A method developed by our group106 utilizes an elastomeric valve to provide temporal control over the initiation of a biomolecule gradient. The Microvalve Chemotaxis Device (µVCD), shown in Fig. 9, consists of three layers. The gradient forms in the microchannels in the top, or fluidic, layer. A thin elastomeric membrane layer separates the fluidic layer from pneumatic control microchannels in the control layer. The fluidic and control layers are aligned so that the wall between the microchannels in the fluidic layer acts as a valve seat for the elastomeric membrane overlying the control layer microchannel. When vacuum is pulled in the control layer microchannel, the elastomeric membrane deflects downwards, bringing the liquids in the fluidic layer microchannels into contact. If the fluidic layer microchannels are loaded with chemically dissimilar fluids, a gradient forms when the valve is opened. The device generates a radially-symmetric gradient that evolves over time. Surface tension and evaporation-induced fluid flow are eliminated through the use of additional elastomeric valves that isolate the fluidic layer microchannels from the respective inlets. The device was used to examine the chemotaxis of human neutrophils in response to a quantifiable gradient of the chemokine CXCL-8.106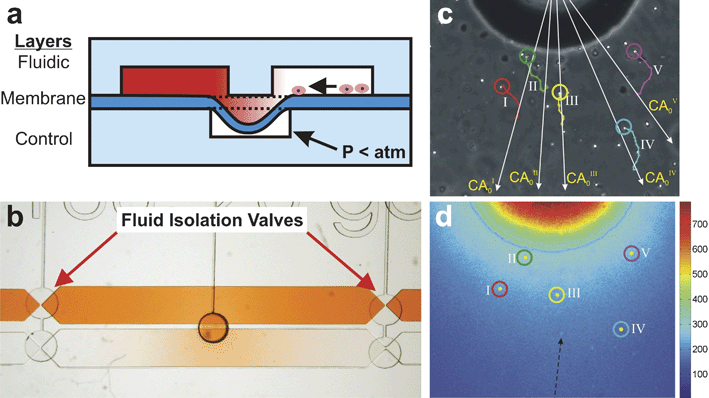 | ||
| Fig. 9 Microvalve Chemotaxis Device (µVCD). (a) Cross-section schematic of the three-layer device. Cells and gradient fluids reside in the two microchannels in the fluidic layer. The microchannels are kept isolated from each other by a thin membrane sealed to the wall separating the microchannels. When vacuum is pulled in the bottom control layer microchannel, the membrane deflects downwards, fluidically connecting the two microchannels. (b) Top-view image of the device shows a gradient of red dye forming in the presumptive cell culture microchannel upon valve opening. Both microchannels are isolated from convective fluid flow contributions from the inlets by the fluid isolation valves at each end of the microchannels. (c) Phase-contrast micrograph of human neutrophils migrating in response to a gradient of CXCL-8. Colored traces indicate the trajectories of 5 cells (adapted from ref. 106, reproduced by permission of The Royal Society of Chemistry). (d) Temperature false-colored image corresponding to (c) showing concentration of FITC-dextran along with positions of the 5 cells shown in (c) (adapted from ref. 106, reproduced by permission of The Royal Society of Chemistry). | ||
The µVCD effectively creates gradients capable of eliciting responses from cells. Its most significant advantage is that it allows the user to control when the gradient is initiated. The device does not require sophisticated equipment or active pumping schemes. It uses low reagent volumes and allows accumulation of autocrine and paracrine cell signals that are essential to many biological processes. Although not demonstrated, the µVCD could be used to generate complex gradient environments containing multiple independent biomolecule gradients.
However, like the Nanopore Gradient Generator, the µVCD is only suited for short-term gradient studies of cells in a time-evolving gradient. The closed microchannel design will ultimately lead to nutrient depletion and environmental toxification if the media are not replaced (which sweeps the gradient away). Although the user can control when the gradient begins to form, subsequent control over the spatiotemporal evolution of the gradient is limited to opening and closing the valve. The gradient diffuses in both space and time, complicating the analysis of the observed cell responses as a function of specific gradient characteristics. Because the generated gradient is radially symmetric, the cell responses must be analyzed in polar coordinates, thus requiring custom-developed analysis software. In principle, arrays of microvalves could be used to create a flat-front (instead of radial) gradient. Like the Nanopore Gradient Generator, the µVCD is best suited for studying fast-responding cells, such as neutrophils or bacteria, over time courses of a few hours.
Microfluidic Multi-Injector (MMI)
A gradient-generating method developed by Chung and colleagues107 replicates the micropipette method developed by Gunderson and Barrett38 in a microfluidic platform. In the MMI gradient generator (Fig. 10) a biomolecule solution is pneumatically ejected out of a round microchannel orifice that is 10 µm in diameter. Pneumatic fluid ejection is controlled by a pneumatically-operated, on-chip diaphragm valve.108 Stacked pulses of biomolecule solution diffuse away from the orifice, forming a gradient in the presumptive cell culture chamber. Stable gradients formed within 10 min and lasted as long as 2–3 h.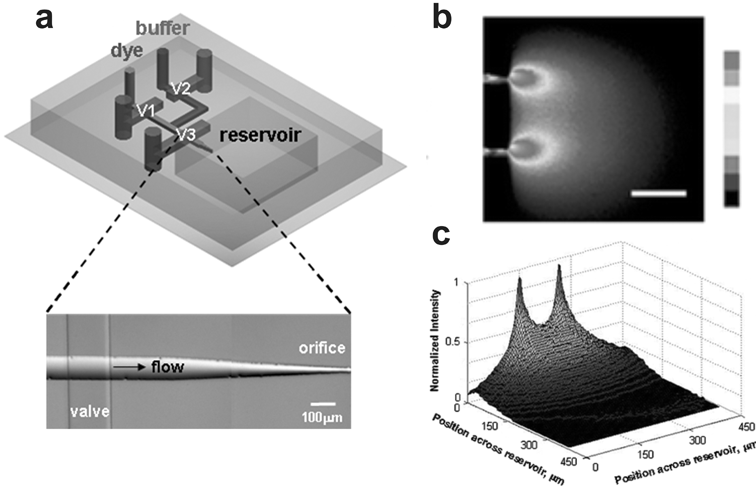 | ||
| Fig. 10 Microfluidic Multi-Injector (MMI). (a) A 3D schematic of the device shows a single orifice injector and the valving necessary to create gradients in the presumptive, enclosed cell culture reservoir. (b) Top view of the device creating gradients of FITC-labeled dextran. (c) 3D intensity plot of data acquired in (b) (adapted from ref. 107, reproduced with permission from the Royal Society of Chemistry). | ||
The primary advantage the MMI device offers over the micropipette method is better reproducibility and quantitation due to the precise dimensions of the device. Like the other microfluidic devices discussed so far, the MMI is also optically transparent allowing observation of single cell responses, indirect gradient quantification using fluorimetric dyes, and the opportunity to establish correlations between the two. Like the µVCD the MMI device provides temporal control over the moment of gradient initiation.
Unfortunately, like the micropipette method, the MMI gradient generator offers little control of the gradient during operation. The precise dimensions and the fixed position of the orifice relative to the cell culture chamber may improve the reproducibility of the generated gradient, but it comes at the expense of lost flexibility in positioning the source relative to the cell. The MMI cell culture chamber is also limited in size due to thermal gradients and other sources of convective flow that adversely affected gradient stability. The chamber size that minimized these convective flows only allowed stable gradient generation for 2–3 h making the MMI best suited for fast responding cell types.
Steady-state gradient generators
Steady-state gradient generators create distinct regions of constant concentration to form time-invariant gradients across a cell culture area. Although the time-invariant gradients they create do not recapitulate the spatially and temporally dynamic gradients cells encounter in vivo, they do provide detailed, quantitative information about how biomolecule concentration or gradient profile influences cell behavior. These concentration and profile dependencies will provide appropriate context for in vitro studies seeking to understand how gradient timing and spatiotemporal dynamics influence cell behavior.Steady-state gradient generators developed to date have employed one of two different strategies. The first group, which we will refer to as Parallel-flow Gradient Generators, exploit the lack of convective mixing that occurs between adjacent fluid streams under laminar flow. Because mixing only occurs via diffusion perpendicular to the direction of fluid flow, chemical gradients form with characteristics (i.e. shape, concentration range, etc.) that depend upon the composition and flow rates of the respective streams and the length of time the streams have been in contact. The resulting gradients are constant as long as the input flow rates and compositions are constant. The major advantage of Parallel-flow Gradient Generators is that they are capable of creating and maintaining a wide variety of gradients for periods of time limited only by the supply of reagents (i.e. size of inlet reservoir, cost, etc.). Rapid formation of steady-state gradients allow correlations to be made between observed cell responses and specific gradient characteristics.92,93 By carefully choosing the concentration of the input fluid streams and the manner in which they are combined, a wide variety of gradient shapes and concentration ranges can be created. Adjustment of the inlet flow rates also offers the user some dynamic control over the gradient formed within the device. The main disadvantage of parallel flow gradient generators is the requirement for constant fluid flow. Flow rates typically have units in µL min–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 93,94,99,109 requiring many milliliters of reagents for a long-term experiment. For some biomolecules, the required liquid volumes are cost-prohibitive. Constant fluid flow also imparts mechanical forces on cells that can introduce experimental bias,109 and may initiate intracellular signaling cascades that can confound experimental results or damage cells. In addition, since the concentration profile smoothens downstream, analysis of cell responses as a function of gradient characteristics must be limited to a short portion of the channel. Another major concern is that any autocrine or paracrine signals released by the cells in response to the applied gradient are quickly removed by constant cell perfusion and accumulated downstream. Since gradients only form perpendicular to the direction of fluid flow, parallel flow gradient generators cannot generate more complex chemical environments containing gradients with various positions and orientations. The Parallel-flow Gradient Generators reviewed here include T-sensor-based devices,84,110–115 the Premixer Gradient Generator,116 and the “Universal” Gradient Generator.117
93,94,99,109 requiring many milliliters of reagents for a long-term experiment. For some biomolecules, the required liquid volumes are cost-prohibitive. Constant fluid flow also imparts mechanical forces on cells that can introduce experimental bias,109 and may initiate intracellular signaling cascades that can confound experimental results or damage cells. In addition, since the concentration profile smoothens downstream, analysis of cell responses as a function of gradient characteristics must be limited to a short portion of the channel. Another major concern is that any autocrine or paracrine signals released by the cells in response to the applied gradient are quickly removed by constant cell perfusion and accumulated downstream. Since gradients only form perpendicular to the direction of fluid flow, parallel flow gradient generators cannot generate more complex chemical environments containing gradients with various positions and orientations. The Parallel-flow Gradient Generators reviewed here include T-sensor-based devices,84,110–115 the Premixer Gradient Generator,116 and the “Universal” Gradient Generator.117
T-sensor
The simplest parallel flow gradient generators are the so-called T-sensors113 (Fig. 11) which consist of two or more microchannels that join into a single microchannel in a T-shaped configuration. T-sensor-based devices have been used to combine dissimilar fluid streams to create combinatorial mixtures of different chemicals,112 perform biological assays,110,111,114,115 study bacterial chemotaxis,118 and infect cells with different viral titrations.84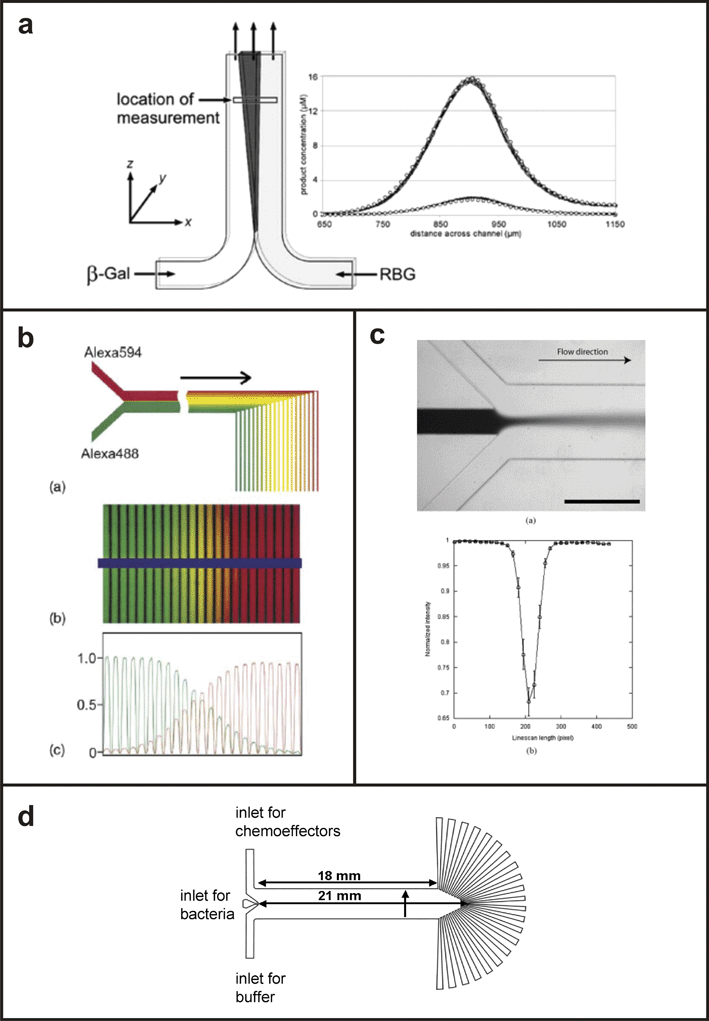 | ||
| Fig. 11 T-sensor based devices. (a) Schematic of the T-sensor with a plot of the concentration profile of a fluorogenic substrate generated by interdiffusion of enzyme (β-gal) and substrate (RBG) solutions (from ref. 115, copyright 2002 American Chemical Society). (b) T-sensor based diffusion diluter developed by ref. 112, validated with the fluorescent dye, Alexa 488 (adapted from ref. 112, copyright Elsevier, 2003). (c) Device used to infect cell populations with a gradient of baculovirus (adapted from ref. 84, copyright Elsevier, 2004). (d) Device used to study bacterial chemotaxis in the presence of various chemoeffectors (from ref. 96, copyright 2003 National Academy of Sciences, USA). | ||
T-sensor-based devices generate steady-state gradients that are reproducible and can be characterized quantitatively. They have simple designs that are easy to fabricate and to describe mathematically. Cells grown within the device can be visualized using standard microscopy and their growth, differentiation, or migration can be correlated with specific gradient characteristics due to the temporal stability of the gradient. Constant perfusion prevents depletion of nutrients and accumulation of cell waste that limits other gradient generators, allowing cells to be maintained for long periods of time in culture. The position and shape of the gradient can be modulated dynamically by adjusting the inlet flow rates and solution concentrations, respectively.
Disadvantages of continuous fluid flow include high reagent consumption, exposure of cells to confounding and potentially damaging mechanical forces, and removal of autocrine/paracrine factors are characteristic of all T-sensor devices. In addition, T-sensor devices can only generate single-solute, sigmoid-shaped gradients or multi-factor gradients in which each biomolecule solution acts as the sink for the other. The gradients are also confined to a single axis perpendicular to the direction of fluid flow, precluding more complex multifactor gradient environments.
Premixer Gradient Generator
Jeon and colleagues116 developed a parallel flow gradient generator (Fig. 12a) that has been used to study the effects of soluble biomolecule gradients on neutrophil migration,92–94 neural stem cell differentiation,91 breast cancer cell chemotaxis,99,100 and rat intestinal cell migration.90 The device has also been used to create substrate-bound biomolecule gradients to direct the growth of hippocampal neurons87 and examine cell cycle progression and exit in intestinal cells.89 The device, which we will refer to as the Premixer Gradient Generator, is based on the T-sensor and generates mathematically predictable, steady-state gradients under constant fluid flow. However, in contrast to the T-sensor, the Premixer Gradient Generator splits and recombines inlet fluids in an upstream microfluidic mixer, which shortens the physical footprint of the device, and allows the formation of more complex gradient profiles. By adding more inlets and reconfiguring the upstream mixer, complex sawtooth and hill gradients can be created (Fig. 12b) in addition to the standard sigmoid gradients observed in T-sensor-based devices. Each type of gradient is generated using a unique mixer configuration.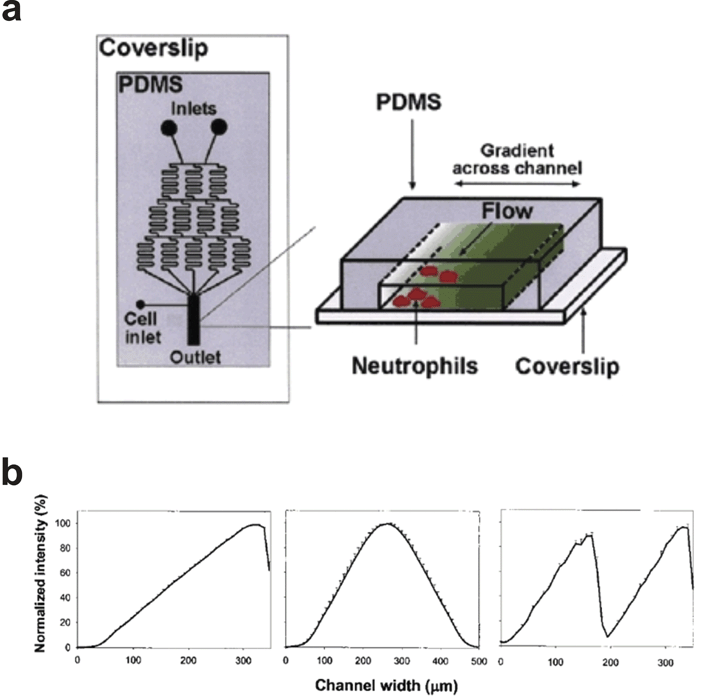 | ||
| Fig. 12 Premixer Gradient Generator. (a) 2D schematic of the device with 3D exploded view of the gradient generated downstream of the microfluidic mixer. (b) By reconfiguring the upstream mixer a variety of complex gradient profiles can be achieved including linear, hill, and sawtooth (adapted with permission from Macmillan Publishers Ltd:92 Copyright 2002). | ||
As with all parallel flow gradient generators, the Premixer Gradient Generator only forms gradients under constant fluid flow, which consumes significant amounts of reagents, flushes away potentially important cell-secreted factors, subjects cells to possibly confounding or damaging shear and drag forces, and causes the gradient upstream to differ from the gradient downstream (i.e. the analyzed region must be small). The device can generate combinatorial gradients with two factors94 and theoretically more, but the gradients can only be arranged along a single axis orthogonal to the direction of flow. The user can dynamically modify the gradient via inlet flow rate control but is constrained by the upstream mixer design. In comparison to T-sensor based devices, the addition of the upstream microfluidic mixer also greatly increases the dead volume of the device, and lengthens the time it takes to realize any dynamic gradient modifications.
“Universal” Gradient Generator
The parallel flow gradient generator (Fig. 13) developed by Irimia and colleagues117 is in essence a T-sensor concept that features a series of physical walls to control the orthogonal diffusion of chemical species between adjacently flowing streams. By controlling the location of the static dividers it was possible to generate gradients with profiles resembling 5th order polynomials, exponential, error, and cubic root functions, and presumably profiles of any other shape.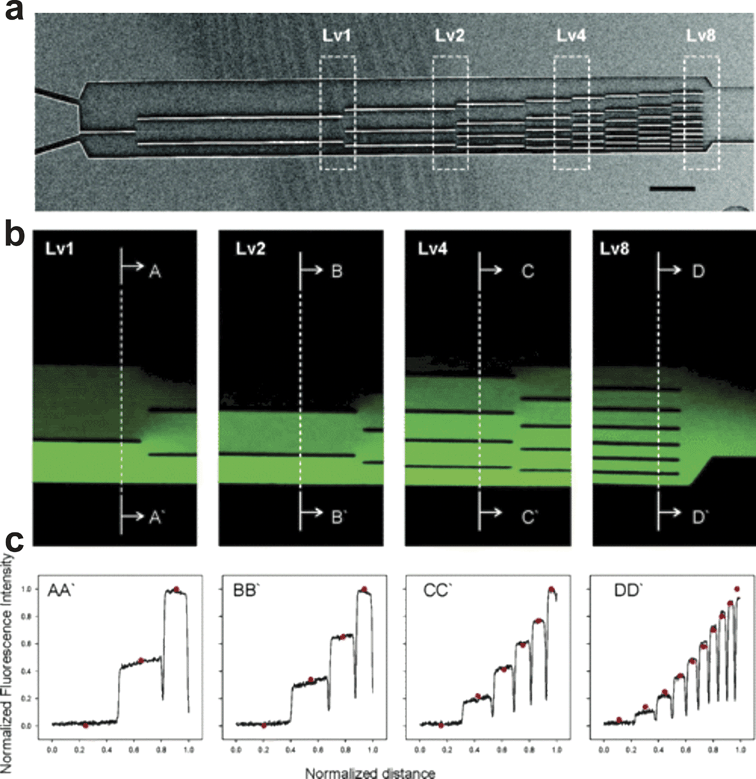 | ||
| Fig. 13 “Universal” Gradient Generator. (a) A scanning electron micrograph of the device shows the position of dividers that restrict the orthogonal diffusion of chemical species. (b) Fluorescence images of the concentration distribution of FITC during at various points within the device shown in (a). (c) Intensity profiles of the images in (b) at the regions indicated by the dotted lines (from ref. 117, copyright 2006 American Chemical Society). | ||
The novel approach used in the “Universal” Gradient Generator greatly enhances the types of gradients that could be applied to cells. It does not have the dead volume associated with the Premixer Gradient Generator and could achieve the same sawtooth, hill, and other complex gradients with the addition of more inlets.
However, because the device relies upon orthogonal diffusion in constantly flowing fluid streams, it suffers from the same disadvantages as the T-sensor and Premixer Gradient Generators. Gradient formation requires significant volumes of potentially expensive reagents. Cells are exposed to fluid flow that flushes away important secreted factors and imparts mechanical forces on the cells that can affect cell behavior and viability. Multifactor biomolecule gradients can only be formed on the axis perpendicular to fluid flow. Like the Premixer device the gradient can be dynamically modified to some degree, but dramatic shifts in gradient position or shape may not be possible depending upon device design.
The second group of steady-state gradient generators, which we will refer to as “Flow-resistive Gradient Generators”, utilize high flow resistance elements between the biomolecule source and sink fluids to minimize or eliminate convection that can prevent gradient formation or destabilize existing gradients. Without fluid flow diffusive mass transport dominates resulting in a stable gradient between the source and sink reservoirs. The two most common strategies for reducing or eliminating convection are to connect the source and sink reservoirs with a hydrogel-filled compartment, or using small slits or microchannels. Hydrogels completely eliminate fluid flow due to the long and torturous fluid paths through the dense polymer network of the hydrogel so that mass transport occurs only via diffusion. Fluid flow still occurs within narrow slits or microchannels when pressure differences between the source and sink reservoirs exist, but the rate of convective mass transport is much less than the diffusive mass transport (i.e. Peclet number is less than 1) allowing a stable gradient to form. The advantages of Flow-resistive Gradient Generators are that they readily form steady-state gradients in cell culture environments devoid of flow (i.e. low shear forces, autocrine/paracrine factor accumulation), do not require complex equipment (e.g. computer-controlled syringe pumps), are easy to design and construct, can create consistent gradients over arbitrarily long cell culture chambers, consume less reagent than Parallel-flow Gradient Generators, and in some cases can be used to create overlapping gradients of different biomolecules with unique orientations, concentration ranges, and gradient profiles. The disadvantages of Fluid-resistive Gradient Generators are that they can require more reagent than traditional methods and often have difficulty creating gradients with complex profiles. The Flow-resistive Gradient Generators reviewed here include the hydrogel-based methods developed by Wu and colleagues,119 Diao and colleagues,97 Saadi and colleagues,120 and the geometric-constriction based methods developed by Keenan and colleagues,83 Paliwal and colleagues,121Li and colleagues,122 and Mosadegh and colleagues.123
Hydrogel-Capped Arbitrary Gradient Generator
A method developed by Wu and colleagues119 utilizes the gradient-stabilizing features of hydrogels to create steady-state gradients of arbitrary profiles in a microfluidic device. The top PDMS layer of the 3-layer device (Fig. 14a,b) consists of two 4 mm long source and sink fluid reservoirs. The sink reservoirs are connected to a cavity 2.8 long × 4 mm wide × 100 µm tall that comprises the second layer. The bottom PDMS layer contains microchannels patterned in a variety of user-defined geometries. To generate the gradient the cavity in layer 2 is filled with a 1% agarose solution and allowed to gel. The reservoirs are then filled with the source and sink fluids causing a gradient to form within the agarose gel. By replenishing the reservoir fluids every 3 h a stable linear gradient is formed. The device is then conformally sealed to the bottom layer and the constituent microchannels filled with liquid. The biomolecule in the agarose diffuses into the microchannels, eventually coming into concentration equilibrium with the overlying gel. If the microchannels are small relative to the dimensions of the gel-filled cavity, the diffusive loss to the microchannels will not significantly alter the overlying gradient. The gradient to which cells growing in the microchannel would be exposed is defined simply by the path the microchannel takes along the bottom of the agarose gel, although complex gradients require turns to which cells may respond differently than in straight paths. Fig. 14c shows a top-view schematic of the various microchannel configurations demonstrated by the authors with the respective gradients shown in Fig. 14d.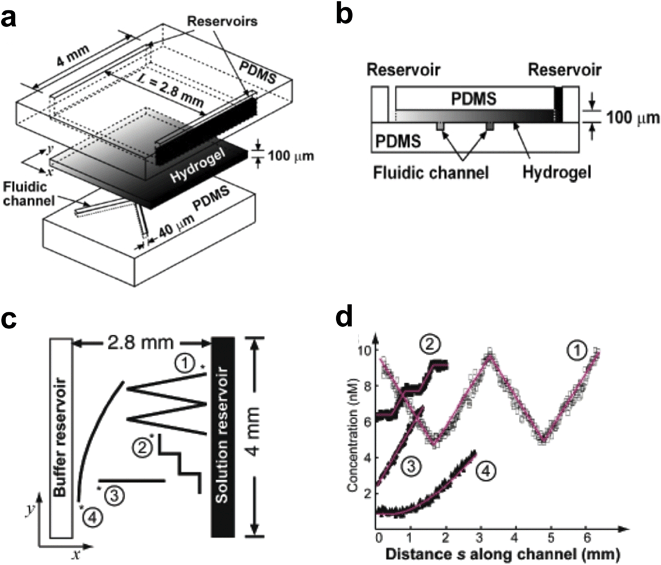 | ||
| Fig. 14 Hydrogel-Capped Arbitrary Gradient Generator. (a) 3D schematic of the device shows the 3-layer architecture. (b) Schematic of the device cross section shows a hydrogel slab separating gradient fluid reservoirs from cell culture microfluidic channels. (c) Top-view schematic of the device showing how microchannels of different configurations and a linear gradient between the buffer and solution reservoirs can be used to generate a wide variety of user-defined gradient profiles (d) (reprinted with permission from ref. 119, copyright 2006 American Chemical Society). | ||
To maintain a constant gradient, the reservoir fluids need to be replaced every 3 h. The device does not offer dynamic control over the shape, position, or concentration of the gradient to explore how temporal gradient dynamics influence cell response. In addition, hydrogels have relatively poor optical transparency compared to glass or PDMS, which hinders phase-contrast microscopy.
Hydrogel Membrane Gradient Generator
The method developed by Diao and colleagues97 creates distinct source, cell culture, and sink reservoirs separated by hydrogel by cutting three parallel microchannels into a nitrocellulose membrane using a CO2 laser (Fig. 15a). The membrane was compressed between an inlet manifold and a piece of glass. The two side channels were filled with a soluble chemotactic agent (L-aspartate or glycerol) and a buffer solution, respectively, and the center channel was filled with E. coli bacteria. The fluids in the side channels were recirculated to maintain constant concentration, but the high flow resistance of the nitrocellulose network resulted in the formation of a purely diffusive gradient within the bacteria-filled channel. E. coli responded by migrating towards L-aspartate and away from glycerol. A similar device made in micromolded agarose (instead of PDMS) has been applied to study chemotaxis of E. coli and a human leukemia cell line, HL-60, to α-methyl-DL-aspartate and formylated bacterial peptide gradients, respectively.124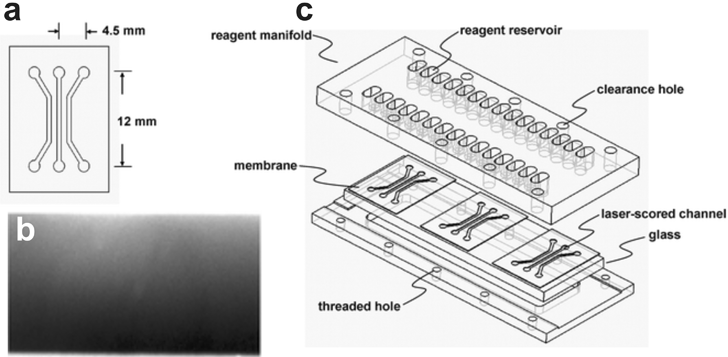 | ||
| Fig. 15 Hydrogel Membrane Gradient Generator. (a) 2D schematic of the device showing the features cut into the nitrocellulose membrane with a CO2 laser. (b) Fluorescence micrograph of a fluorescein gradient generated within the center channel of the device. (c) 3D schematic of the device (adapted from ref. 97 by permission of The Royal Society of Chemistry). | ||
The Hydrogel Membrane Gradient Generator effectively generates steady-state gradients with fixed slopes and concentration ranges. The design and optical transparency of the device allows correlation of specific gradient characteristics with observed biological responses. Cells are not exposed to flow-induced mechanical forces. The linear gradients formed in the device are extremely easy to calculate with formation times dependent solely on the diffusion coefficient of the biomolecule. The design could easily be extended to form multiple stable gradients.
The main disadvantages of the Recirculating Gradient Generator are that it cannot generate the complex gradient profiles possible in Parallel-flow Gradient Generators nor can it provide dynamic gradient tunability. The device lacks the temporal control offered by the µVCD or MMI and the enclosed cell culture chamber makes cells susceptible to nutrient starvation and environmental toxification if the side channels are not continuously perfused with fresh medium.
Microjets Device
A microfluidic gradient generator called the Microjets Device83 (Fig. 16a) generates user-defined, steady-state gradients on open cell culture surfaces without exposing cells to appreciable fluid flow. The Microjets Device generates gradients by pneumatically ejecting fluids out of two opposed arrays of Microjets (small microchannels ∼1.5 µm × 1.5 µm in cross-section) into the cell culture area. Biomolecules are convectively transported in the Microjets, but once they exit the very small amounts of momentum they carry are insufficient to generate appreciable fluid flow in the cell culture area. Convective fluid flow abates and diffusive mass transport dominates, resulting in a purely diffusive gradient. By constantly replenishing the Microjet outlets with fresh gradient fluids, two constant concentration boundaries are established in the cell culture area causing a steady-state gradient to form. The gradient reaches steady-state within 10 min and can be maintained for as long as reagents are available. By adjusting the pneumatic driving pressures delivered to each bank of Microjets, the user has independent control over the slope and position of the gradient in the cell culture area (Fig. 16b–d).83 | ||
| Fig. 16 Microjets Device. (a) 3D schematic of the device showing an open surface gradient created by opposed arrays of small microfluidic channels (i.e. Microjets). (b–d) Top-view confocal fluorescence micrographs and the corresponding 70 kDa dextran surface concentration profiles (solid curves) at equilibrium before and after changes in PL and PR. The dashed white vertical line indicates the position of maximum slope. Yellow-dotted lines mark the buried microchannel and cell culture area boundaries. Comparison of b and c show an increase in gradient slope with no effect on gradient position when equal magnitude pressure increases are applied to the Microjets. Comparison of c and d show a shift in gradient position to the right without a change in slope when equal magnitude pressure offsets are applied (a–d reprinted with permission from ref. 83, copyright 2006 American Institute of Physics). (e) A top-view image of a combination of red, green, and blue dye gradients emanating from Microjets in a T-shaped open cell culture pool. | ||
The Microjets Device has several distinct advantages over other microfluidic gradient generators. First, the high flow resistance of the Microjets creates user-defined, steady-state gradients without exposing cells to confounding or potentially damaging fluid flow. Second, the device can create gradients in open cell culture chambers which facilitates gas/pH equilibration and provides access for other cellular probing methods such as electrophysiological recordings, single cell isolation and analysis, iontophoretic stimulation, etc. Because cells are not exposed to the biomolecule until the Microjet array is pressurized, the open cell culture chamber architecture also allows cells to be cultured for user-defined time periods prior to gradient application. This feature can be used to create in situ controls as well as to precondition cells (e.g. grow, mature, fuse or differentiate cells) prior to examination of gradient-induced effects. Third, the Microjets Device allows the user to dynamically and independently control the characteristics of the gradient to which a cell is exposed. This type of temporal tunability is not offered in other gradient generators. For example, cells migrating in response to a gradient within the Microjets Device can be re-exposed to precisely the same gradient (even in a different area of the cell culture pool) to examine cellular desensitization to a particular biomolecule. As with other gradient generators, cell responses and gradient formation can be tracked easily with an optical microscope due to the optical clarity of the device and the fact that cells are cultured on the same focal plane. By altering the location and orientation of the Microjets complex multi-gradient cell culture environments can also be created (Fig. 16e).
The most significant disadvantage of the Microjets Device is that it is not capable of generating the complex gradients possible with the Premixer, “Universal”, or Gel-stabilized Arbitrary Gradient Generators and is limited to generating only monotonic gradients. The open architecture of the device makes the gradient more susceptible to bulk fluid movement in the overlying medium which can distort or destroy the gradient depending on the cell culture chamber geometry. The small microfluidic channels are also prone to clogging, adversely affecting gradient formation and experimental throughput.
Cross Channel Gradient Generators
Cross Channel Gradient Generators use a series of narrow cross channels to connect parallel microchannels that are continuously perfused with source and sink fluids. In the device developed by Paliwal and colleagues121 (Fig. 17a) the cross channels, or test chambers (Fig. 17b), are 5 µm tall and between 160–600 µm long and are connected to source and sink microchannels that are 25 µm tall. By balancing the hydrostatic pressure delivered to the source and sink microchannels, Paliwal et al. were able to establish gradients in the test chambers (Fig. 17c) and used the gradients to study the chemotactic response of the yeast S. cerevisiae to pheromone (Fig. 17d).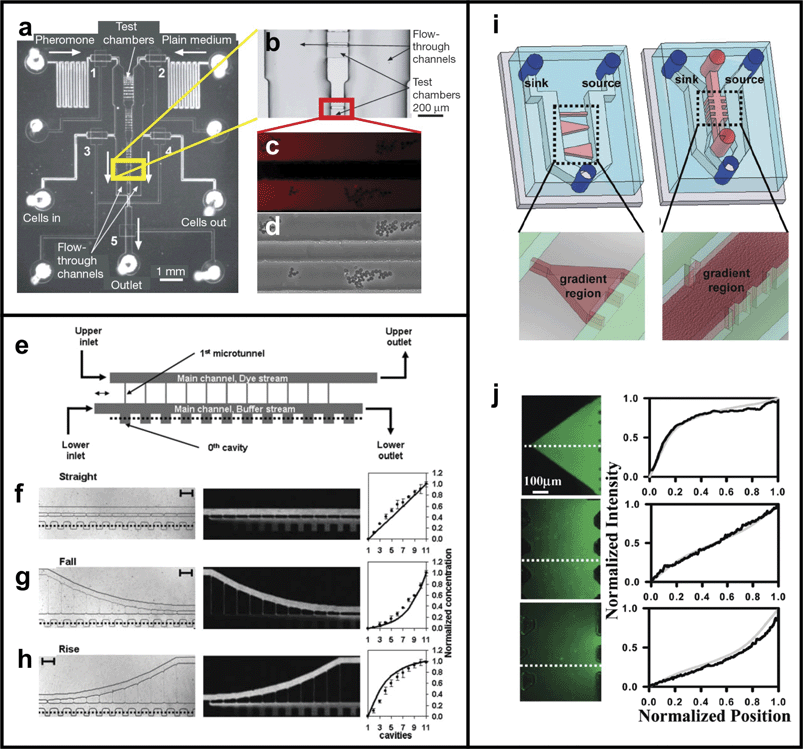 | ||
| Fig. 17 Cross Channel Gradient Generator. (a) Top-view image of the device developed by Paliwal et al..121 (b) Enlarged photo of the test chamber area shows 5 µm tall test chambers connecting source and sink fluid microchannels. (c) By balancing the hydrostatic pressure delivered to each adjacent microchannel gradients of yeast pheromone could be created in the test chambers (visualized by Alexa 555 dye). (d) S. cerevisiae migrating in response to the pheromone gradient (reprinted from ref. 121 by permission of Macmillan Publishers Ltd). (e) Top-view schematic of the device developed by Li and colleagues122 uses constantly perfused main channels and cross channels of varying lengths to create linear (f), exponential (g), and logarithmic (h) gradients (reproduced by permission of The Royal Society of Chemistry). (i) Top-view schematic of the device developed by Mosadegh et al. colleagues.123 (j) By changing the cross sectional dimensions of the cross channels non-linear gradients can be achieved as shown here using fluorescent dyes in Matrigel-filled cross channels (reprinted with permission from ref. 123, copyright 2007 American Chemical Society). | ||
Li and colleagues122 used a slightly different approach to create gradients in a cross-channel device. Fig. 17e shows a top-view schematic of the device. Source and sink fluids are perfused through side microchannels connected by an array of cross channels. Adjacent to the sink fluid channel is an array of dead-volume cavities which come into chemical equilibrium with the fluid in the sink channel. By altering the cross channel length Li and colleagues were able to create linear (Fig. 17f), exponential (Fig. 17g), and logarithmic (Fig. 17h) gradients.
Saadi and colleagues120 used the same strategy as Paliwal and colleagues to generate gradients over 2D culture substrates and in 3D biological hydrogels. Syringe pump-driven fluids were used to maintain constant concentration in the source and sink microchannels allowing steady-state gradients to be established in free solution via 5 µm tall cross channels or in collagen gels via 200 µm wide × 300 µm tall cross channels. Gradients of FITC-dextran (10 kDa) reached steady state in free solution within 18 min and in collagen gels within 30 min. The device has since been modified by Mosadegh and colleagues123 to include cross channels with varying cross section (Fig. 17i). By changing the configuration of the cross channels Mosadegh and colleagues were able to create the same non-linear gradient profiles as Li and colleagues without exposing cells to fluid flow (Fig. 17j).
The advantages of cross-channel gradient generators are that they can generate gradients with user-defined concentration ranges and gradient profiles in both 2D and 3D culture systems, unlike Parallel-flow Gradient Generators can do so uniformly over the entire cell culture chamber, and do not expose cells to appreciable fluid flow that could otherwise induce damage or confound experimental results. Constant perfusion prevents nutrient depletion and accumulation of cell waste that typically restricts cell culture in microchannels. Although not explicitly demonstrated in the aforementioned studies, the principles used to generate single steady-state gradients with different concentration ranges and profiles could be used to generate more complex gradient environments consisting of different biomolecule gradients each with unique user-defined properties.
One of the main limitations of cross-channel gradient generators is the lack of dynamic gradient tunability they afford. In addition, maintaining constant concentration boundaries can require continuous perfusion with costly reagents. Like Parallel-flow Gradient Generators long-term experiments may be prohibitively costly to run. Devices using 5 µm tall chambers could induce cell damage due to the shear forces generated during cell seeding or by restricting access to nutrients and allowing rapid accumulation of waste products because of the small liquid volume in which the cells reside.
Long-term impact
Despite the aforementioned advantages of microfluidic gradient generators, they have yet to reach widespread use in the biological community. A major barrier to widespread use of microfluidic devices is that, in academia, there is little incentive for the labs that have microfluidic “know-how” to produce user-friendly designs. Most microfluidic gradient generators still require a significant level of expertise (e.g. microfabrication) and equipment (e.g. computer controlled syringe pumps) that are not familiar to the greater biological community. Because microfluidic gradient generators are not commercially available and their design is usually question-specific, biological research labs typically need to make their own devices or partner with microfluidics researchers to conduct impactful biological studies. However, more microfluidic labs are now exploring and training students in microfluidics-enabled biological research. The migration of microfluidics-trained personnel into biological labs and the decentralization of microfabrication tools (e.g. spin coaters and aligners) from large user facilities to individual laboratories are helping to lower the “dissemination threshold”. As these biomicrofluidics researchers begin exploring questions that could not be previously answered using conventional methods, the use of microfluidic tools to investigate biological questions will grow.Summary
Biomolecule gradients are an essential component of many biological phenomena and act in concert with one another to bring about cell-, time- and location-specific responses. Traditional methods, although instrumental in developing most of our current understanding of cell behavior under gradients, are ill-equipped to generate the gradients necessary to provide a quantitative understanding of gradient signaling and reveal how different gradient signaling pathways interact. The ability to create complex chemical environments, composed of precisely-engineered gradients with specific spatiotemporal characteristics will greatly advance biological research and help elucidate how developmental programs and other biological processes are affected by gradients of biomolecules. Microfluidic technology has brought about a broad range of methods for exposing cells to engineered gradients, each method having a unique set of attributes and disadvantages (Table 1). Although the complex, multi-factor gradient environments described previously have yet to be fully demonstrated and utilized, we believe the innovations detailed here will be instrumental in shaping the future of chemically-defined cell culture microenvironments.| Gradient type | Exposes cells to flow | Dynamic control of gradient concentration and profile | Could create multiple independent gradients | Biomolecule consumption | Reference | |
|---|---|---|---|---|---|---|
| Traditional methods | ||||||
| Biological hydrogels | Evolving | – | – | + | 20 µL per expt | 32 |
| Micropipette | Evolving | – | – | + | 0.0108 µL h–1 | 9–12 |
| Boyden/Transwell Assay | Evolving | – | – | – | ≥235 µL per expt | |
| Zigmond Chamber | Short-term stable | – | – | – | 100 µL per expt | 50 |
| Dunn Chamber | Short-term stable | – | – | – | 15–30 µL per expt | 65 |
| Microfluidic methods | ||||||
| Substrate-bound | ||||||
| Depletion | Stable | – | – | + | NA | 89,101 |
| Micropatterned | Stable | – | – | + | NA | 102–105 |
| Time-evolving | ||||||
| Nanopore | Evolving | – | – | + | 10 µL per expt | 95 |
| uVCD | Evolving | – | – | + | 0.330 µL per expt | 106 |
| MMI | Short-term stable | – | – | + | 0.160 µL h–1 | 107 |
| Parallel-flow | ||||||
| T-sensor | Stable | + | – | – | 6–300 µL h–1 | 85,110–113 |
| Premixer | Stable | + | – | – | 48–60 µL h–1 | 92–94,99,116 |
| Universal | Stable | + | – | – | 3 µL h–1 | 117 |
| Flow-resistive | ||||||
| Hydrogel Capped Arbitrary | Stable | – | – | – | Not stated | 119 |
| Hydrogel Membrane | Stable | – | – | + | 60 µL h–1 | 97,124 |
| Microjets | Stable | – | + | + | 9 µL h–1 (per jet) | 84 |
| Cross-Channel | Stable | – | – | – | 0.6–6 µL min–1 | 120,122,123 |
References
- S. F. Gilbert, Developmental Biology, Sinauer Associates, Inc. Publishers, Sunderland, MA, 8th edn, 2006, vol. xviii, p. 817 Search PubMed.
- R. Raballo, et al., Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex, J. Neurosci., 2000, 20(13), 5012–5023.
- H. L. Ashe and J. Briscoe, The interpretation of morphogen gradients, Development, 2006, 133(3), 385–394 CrossRef CAS.
- K. M. Scully and M. G. Rosenfeld, Pituitary development: regulatory codes in mammalian organogenesis, Science, 2002, 295(5563), 2231–2235 CrossRef CAS.
- S. A. Colamarino and M. Tessier-Lavigne, The role of the floor plate in axon guidance, Annu. Rev. Neurosci., 1995, 18, 497–529 CrossRef CAS.
- T. Serafini, et al., Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system, Cell, 1996, 87(6), 1001–1014 CrossRef.
- H. Long, et al., Conserved roles for Slit and Robo proteins in midline commissural axon guidance, Neuron, 2004, 42(2), 213–223 CrossRef.
- E. Stein and M. Tessier-Lavigne, Hierarchical organization of guidance receptors: Silencing of netrin attraction by slit through a Robo/DCC receptor complex, Science, 2001, 291(5510), 1928–1938 CrossRef CAS.
- A. M. Lohof, et al., Asymmetric modulation of cytosolic cAMP activity induces growth cone turning, J. Neurosci., 1992, 12(4), 1253–1261 CAS.
- G. L. Ming, et al., cAMP-dependent growth cone guidance by netrin-1, Neuron, 1997, 19(6), 1225–1235 CrossRef.
- H. Song, et al., Conversion of neuronal growth cone responses from repulsion to attraction by cyclic nucleotides, Science, 1998, 281(5382), 1515–1518 CrossRef.
- H. J. Song, G. L. Ming and M. M. Poo, cAMP-induced switching in turning direction of nerve growth cones, Nature, 1997, 388(6639), 275–279 CrossRef CAS.
- V. H. Hopker, et al., Growth-cone attraction to netrin-1 is converted to repulsion by laminin-1, Nature, 1999, 401(6748), 69–73 CrossRef.
- Y. Zou, et al., Squeezing axons out of the gray matter: a role for slit and semaphorin proteins from midline and ventral spinal cord, Cell, 2000, 102(3), 363–375 CrossRef.
- S. R. Kadison, et al., EphB receptors and ephrin-B3 regulate axon guidance at the ventral midline of the embryonic mouse spinal cord, J. Neurosci., 2006, 26(35), 8909–8914 CrossRef.
- A. I. Lyuksyutova, et al., Anterior-posterior guidance of commissural axons by Wnt-frizzled signaling, Science, 2003, 302(5652), 1984–1988 CrossRef.
- R. A. Firtel and C. Y. Chung, The molecular genetics of chemotaxis: sensing and responding to chemoattractant gradients, Bioessays, 2000, 22(7), 603–615 CrossRef CAS.
- B. Heit, et al., An intracellular signaling hierarchy determines direction of migration in opposing chemotactic gradients, J. Cell Biol., 2002, 159(1), 91–102 CrossRef CAS.
- H. J. Song and M. M. Poo, Signal transduction underlying growth cone guidance by diffusible factors, Curr. Opin. Neurobiol., 1999, 9(3), 355–363 CrossRef CAS.
- C. Janeway, Immunobiology: the immune system in health and disease, 2001, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books.
- D. Q. Wu, Signaling mechanisms for regulation of chemotaxis, Cell Res., 2005, 15(1), 52–56 Search PubMed.
- S. A. Eccles, Targeting key steps in metastatic tumour progression, Curr. Opin. Genet. Dev., 2005, 15(1), 77–86 CrossRef CAS.
- Z. Liu and J. Klominek, Chemotaxis and chemokinesis of malignant mesothelioma cells to multiple growth factors, Anticancer Res., 2004, 24(3a), 1625–1630 CAS.
- R. J. Phillips, et al., The stromal derived factor-1/CXCL12-CXC chemokine receptor 4 biological axis in non-small cell lung cancer metastases, Am. J. Respir. Crit. Care Med., 2003, 167(12), 1676–1686 Search PubMed.
- E. Suyama, et al., Cell migration and metastasis as targets of small RNA-based molecular genetic analyses, J. Muscle Res. Cell Motil., 2004, 25(4–5), 303–308 CrossRef.
- R. M. Strieter, et al., CXC chemokines in angiogenesis of cancer, Semin. Cancer Biol., 2004, 14(3), 195–200 CrossRef.
- J. Wyckoff, et al., A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors, Cancer Res., 2004, 64(19), 7022–7029 CrossRef.
- F. Balkwill, Cancer and the chemokine network, Nat. Rev. Cancer, 2004, 4(7), 540–550 CrossRef CAS.
- F. Luppi, et al., Interleukin-8 stimulates cell proliferation in non-small cell lung cancer through epidermal growth factor receptor transactivation, Lung Cancer, 2006 Search PubMed.
- Y. M. Zhu and P. J. Woll, Mitogenic effects of interleukin-8/CXCL8 on cancer cells, Fut. Oncol., 2005, 1(5), 699–704 Search PubMed.
- H. Chen, et al., Semaphorin-neuropilin interactions underlying sympathetic axon responses to class III semaphorins, Neuron, 1998, 21(6), 1283–1290 CrossRef.
- E. F. Foxman, J. J. Campbell and E. C. Butcher, Multistep navigation and the combinatorial control of leukocyte chemotaxis, J. Cell Biol., 1997, 139(5), 1349–1360 CrossRef CAS.
- E. F. Foxman, E. J. Kunkel and E. C. Butcher, Integrating conflicting chemotactic signals. The role of memory in leukocyte navigation, J. Cell Biol., 1999, 147(3), 577–588 CrossRef CAS.
- W. J. Rosoff, et al., A new chemotaxis assay shows the extreme sensitivity of axons to molecular gradients, Nat. Neurosci., 2004, 7(6), 678–682 CrossRef.
- T. Boland, et al., Application of inkjet printing to tissue engineering, Biotechnol. J., 2006, 1(9), 910–917 Search PubMed.
- L. Pardo, W. C. Wilson and T. J. Boland, Characterization of patterned self-assembled monolayers and protein arrays generated by the ink-jet method, Langmuir, 2003, 19(5), 1462–1466 CrossRef CAS.
- G. J. Goodhill and H. Baier, Axon guidance: Stretching gradients to the limit, Neural Comput., 1998, 10(3), 521–527 CrossRef CAS.
- R. W. Gundersen and J. N. Barrett, Neuronal chemotaxis: chick dorsal-root axons turn toward high concentrations of nerve growth factor, Science, 1979, 206(4422), 1079–1080 CrossRef CAS.
- J. R. de la Torre, et al., Turning of retinal growth cones in a netrin-1 gradient mediated by the netrin receptor DCC, Neuron, 1997, 19(6), 1211–1224 CrossRef.
- G. Ming, et al., Electrical activity modulates growth cone guidance by diffusible factors, Neuron, 2001, 29(2), 441–452 CrossRef.
- G. L. Ming, et al., Adaptation in the chemotactic guidance of nerve growth cones, Nature, 2002, 417(6887), 411–418 CrossRef.
- A. Dehghani Zadeh, et al., Chemotactically-induced redistribution of CD43 as related to polarity and locomotion of human polymorphonuclear leucocytes, Biol. Cell, 2003, 95(5), 265–273 CrossRef.
- G. Servant, et al., Dynamics of a chemoattractant receptor in living neutrophils during chemotaxis, Mol. Biol. Cell, 1999, 10(4), 1163–1178 CAS.
- G. Servant, et al., Polarization of chemoattractant receptor signaling during neutrophil chemotaxis, Science, 2000, 287(5455), 1037–1040 CrossRef CAS.
- K. Wong, et al., Neutrophil polarization: spatiotemporal dynamics of RhoA activity support a self-organizing mechanism, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(10), 3639–3644 CrossRef CAS.
- R. M. Fitzsimonds, H. J. Song and M. M. Poo, Propagation of activity-dependent synaptic depression in simple neural networks, Nature, 1997, 388(6641), 439–448 CrossRef CAS.
- J. Q. Zheng, J. J. Wan and M. M. Poo, Essential role of filopodia in chemotropic turning of nerve growth cone induced by a glutamate gradient, J. Neurosci., 1996, 16(3), 1140–1149 CAS.
- J. Q. Zheng, et al., Turning of nerve growth cones induced by neurotransmitters, Nature, 1994, 368(6467), 140–144 CrossRef CAS.
- S. Boyden, The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes, J. Exp. Med., 1962, 115, 453–466 CAS.
- S. H. Zigmond, Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors, J. Cell Biol., 1997, 75(2 Pt 1), 606–616.
- J. Crank, The mathematics of diffusion, Clarendon Press, Oxford, 1975, 2nd edn, vol. viii, p. 414 Search PubMed.
- P. Alstergren, et al., Polarization and directed migration of murine neutrophils is dependent on cell surface expression of CD44, Cell. Immunol., 2004, 231(1–2), 146–157 CrossRef.
- B. D. Bultmann and H. Gruler, Analysis of the directed and nondirected movement of human granulocytes: influence of temperature and ECHO 9 virus on N-formylmethionylleucylphenylalanine-induced chemokinesis and chemotaxis, J. Cell Biol., 1983, 96(6), 1708–1716 CrossRef CAS.
- B. D. Bultmann, et al., Echo 9 virus-induced order-disorder transition of chemotactic response of human polymorphonuclear leucocytes: phenomenology and molecular biology, Blood Cells, 1984, 10(1), 79–106.
- E. S. Hujanen, S. T. Seppa and K. Virtanen, Polymorphonuclear leukocyte chemotaxis induced by zinc, copper and nickel in vitro, Biochim. Biophys. Acta, 1995, 1245(2), 145–152 CrossRef.
- U. Jager, H. Gruler and B. Bultmann, Morphological changes and membrane potential of human granulocytes under influence of chemotactic peptide and/or echo-virus, type 9, Klin. Wochenschr., 1988, 66(10), 434–436 Search PubMed.
- C. J. Kirkpatrick, B. D. Bultmann and H. Gruler, Interaction between enteroviruses and human endothelial cells in vitro. Alterations in the physical properties of endothelial cell plasma membrane and adhesion of human granulocytes, Am. J. Pathol., 1985, 118(1), 15–25 Search PubMed.
- W. A. Marasco, E. L. Becker and J. M. Oliver, The ionic basis of chemotaxis. Separate cation requirements for neutrophil orientation and locomotion in a gradient of chemotactic peptide, Am. J. Pathol., 1980, 98(3), 749–768 Search PubMed.
- C. A. Roobottom, et al., The effects of radiographic contrast media on leucocyte orientation, Br. J. Radiol., 1993, 66(789), 778–780 Search PubMed.
- M. Simon, et al., Glycolipid storage material in Fabry's disease: a study by electron microscopy, freeze-fracture, and digital image analysis, J. Struct. Biol., 1990, 103(1), 40–47 CrossRef.
- K. L. Vollmer, et al., Tumor necrosis factor-alpha decreases neutrophil chemotaxis to N-formyl-1-methionyl-1-leucyl-1-phenylalanine: analysis of single cell movement, J. Leukoc. Biol., 1992, 52(6), 630–636 Search PubMed.
- G. Fabro, et al., Chemotaxis of capacitated rabbit spermatozoa to follicular fluid revealed by a novel directionality-based assay, Biol. Reprod., 2002, 67(5), 1565–1571 Search PubMed.
- L. C. Giojalas and R. A. Rovasio, Mouse spermatozoa modify their motility parameters and chemotactic response to factors from the oocyte microenvironment, Int. J. Androl., 1998, 21(4), 201–206 CAS.
- R. G. Oliveira, et al., Increased velocity and induction of chemotactic response in mouse spermatozoa by follicular and oviductal fluids, J. Reprod. Fertil., 1999, 115(1), 23–27 Search PubMed.
- D. Zicha, G. Dunn and G. Jones, Analyzing chemotaxis using the Dunn direct-viewing chamber, Methods Mol. Biol., 1997, 75, 449–457 Search PubMed.
- A. D. Lander, Morpheus unbound: reimagining the morphogen gradient, Cell, 2007, 128(2), 245–256 CrossRef CAS.
- C. M. Isbister, et al., Gradient steepness influences the pathfinding decisions of neuronal growth cones in vivo, J. Neurosci., 2003, 23(1), 193–202.
- H. Ali, et al., Chemoattractant receptor cross-desensitization, J. Biol. Chem., 1999, 274(10), 6027–6030 CrossRef.
- I. Burgener, et al., Dose-dependent priming or desensitization induced by chemotactic agents in chemiluminescence experiments with canine and human neutrophils, Vet. Immunol. Immunopathol., 1998, 66(1), 11–24 CrossRef.
- J. J. Campbell, E. F. Foxman and E. C. Butcher, Chemoattractant receptor cross talk as a regulatory mechanism in leukocyte adhesion and migration, Eur. J. Immunol., 1997, 27(10), 2571–2578 Search PubMed.
- S. Nourshargh, et al., A comparative study of the neutrophil stimulatory activity in vitro and pro-inflammatory properties in vivo of 72 amino acid and 77 amino acid IL-8, J. Immunol., 1992, 148(1), 106–111.
- A. K. Samanta, J. J. Oppenheim and K. Matsushima, Interleukin 8 (monocyte-derived neutrophil chemotactic factor) dynamically regulates its own receptor expression on human neutrophils, J. Biol. Chem., 1990, 265(1), 183–189 CAS.
- W. B. Smith, et al., Chemotactic desensitization of neutrophils demonstrates interleukin-8 (IL-8)-dependent and IL-8-independent mechanisms of transmigration through cytokine-activated endothelium, Immunology, 1993, 78(3), 491–497.
- E. D. Tomhave, et al., Cross-desensitization of receptors for peptide chemoattractants. Characterization of a new form of leukocyte regulation, J. Immunol., 1994, 153(7), 3267–3275.
- H. U. Zeilhofer and W. Schorr, Role of interleukin-8 in neutrophil signaling, Curr. Opin. Hematol., 2000, 7(3), 178–182 CrossRef CAS.
- R. G. Flemming, et al., Effects of synthetic micro- and nano-structured surfaces on cell behavior, Biomaterials, 1999, 20(6), 573–588 CrossRef CAS.
- A. Folch and M. Toner, Microengineering of cellular interactions, Annu. Rev. Biomed. Eng., 2000, 2, 227–256 CrossRef CAS.
- F. L. Yap and Y. Zhang, Protein and cell micropatterning and its integration with micro/nanoparticles assembly, Biosens. Bioelectron., 2007, 22(6), 775–788 CrossRef CAS.
- D. Falconnet, et al., Surface engineering approaches to micropattern surfaces for cell-based assays, Biomaterials, 2006, 27(16), 3044–3063 CrossRef CAS.
- J. El-Ali, P. K. Sorger and K. F. Jensen, Cells on chips, Nature, 2006, 442(7101), 403–411 CrossRef CAS.
- N. Li, A. Tourovskaia and A. Folch, Biology on a chip: microfabrication for studying the behavior of cultured cells, Crit. Rev. Biomed. Eng., 2003, 31(5–6), 423–488 Search PubMed.
- R. B. Bird, W. E. Stewart and E. N. Lightfoot, Transport phenomena, Wiley, New York, 1960, p. 780 Search PubMed.
- T. M. Keenan, C. H. Hsu and A. Folch, Microfluidic “jets” for generating steady-state gradients of soluble molecules on open surfaces, Appl. Phys. Lett., 2006, 89(11) Search PubMed.
- G. M. Walker, M. S. Ozers and D. J. Beebe, Cell infection within a microfluidic device using virus gradients, Sens. Actuators B: Chem., 2004, 98(2–3), 347–355 CrossRef.
- N. Zaari, et al., Photopolymerization in microfluidic gradient generators: Microscale control of substrate compliance to manipulate cell response, Adv. Mater., 2004, 16(23–24), 2133 CrossRef CAS.
- J. A. Burdick, A. Khademhosseini and R. Langer, Fabrication of gradient hydrogels using a microfluidics/photopolymerization process, Langmuir, 2004, 20(13), 5153–5156 CrossRef.
- S. K. W. Dertinger, et al., Gradients of substrate-bound laminin orient axonal specification of neurons, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(20), 12542–12547 CrossRef CAS.
- K. A. Fosser and R. G. Nuzzo, Fabrication of patterned multicomponent protein gradients and gradient arrays using microfluidic depletion, Anal. Chem., 2003, 75(21), 5775–5782 CrossRef CAS.
- R. C. Gunawan, et al., Regiospecific control of protein expression in cells cultured on two-component counter gradients of extracellular matrix proteins, Langmuir, 2005, 21(7), 3061–3068 CrossRef CAS.
- R. C. Gunawan, et al., Cell migration and polarity on microfabricated gradients of extracellular matrix proteins, Langmuir, 2006, 22(9), 4250–4258 CrossRef.
- B. G. Chung, et al., Human neural stem cell growth and differentiation in a gradient-generating microfluidic device, Lab Chip, 2005, 5(4), 401–406 RSC.
- N. Li Jeon, et al., Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device, Nat. Biotechnol., 2002, 20(8), 826–830 CAS.
- F. Lin, et al., Effective neutrophil chemotaxis is strongly influenced by mean IL-8 concentration, Biochem. Biophys. Res. Commun., 2004, 319(2), 576–581 CrossRef CAS.
- F. Lin, et al., Neutrophil migration in opposing chemoattractant gradients using microfluidic chemotaxis devices, Ann. Biomed. Eng., 2005, 33(4), 475–482 CrossRef.
- V. V. Abhyankar, et al., Characterization of a membrane-based gradient generator for use in cell-signaling studies, Lab Chip, 2006, 6(3), 389–393 RSC.
- H. Mao, P. S. Cremer and M. D. Manson, A sensitive, versatile microfluidic assay for bacterial chemotaxis, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(9), 5449–5454 CrossRef CAS.
- J. Diao, et al., A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis, Lab Chip, 2006, 6(3), 381–388 RSC.
- S. Koyama, et al., Chemotaxis assays of mouse sperm on microfluidic devices, Anal. Chem., 2006, 78(10), 3354–3359 CrossRef CAS.
- W. Saadi, et al., A parallel-gradient microfluidic chamber for quantitative analysis of breast cancer cell chemotaxis, Biomed. Microdev., 2006, 8(2), 109–118 Search PubMed.
- S. J. Wang, et al., Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis, Exp. Cell Res., 2004, 300(1), 180–189 CrossRef CAS.
- E. Delamarche, et al., Microfluidic networks for chemical patterning of substrate: Design and application to bioassays, J. Am. Chem. Soc., 1998, 120(3), 500–508 CrossRef CAS.
- A. Bernard, et al., Microcontact printing of proteins, Adv. Mater., 2000, 12(14), 1067–1070 CrossRef CAS.
- A. P. Quist, E. Pavlovic and S. Oscarsson, Recent advances in microcontact printing, Anal. Bioanal. Chem., 2005, 381(3), 591–600 CrossRef CAS.
- A. C. von Philipsborn, et al., Microcontact printing of axon guidance molecules for generation of graded patterns, Nat. Protoc., 2006, 1(3), 1322–1328 Search PubMed.
- A. C. von Philipsborn, et al., Growth cone navigation in substrate-bound ephrin gradients, Development, 2006, 133(13), 2487–2495 CrossRef CAS.
- C. W. Frevert, et al., Measurement of cell migration in response to an evolving radial chemokine gradient triggered by a microvalve, Lab Chip, 2006, 6(7), 849–856 RSC.
- B. G. Chung, F. Lin and N. L. Jeon, A microfluidic multi-injector for gradient generation, Lab Chip, 2006, 6(6), 764–768 RSC.
- T. Thorsen, S. J. Maerkl and S. R. Quake, Microfluidic large-scale integration, Science, 2002, 298(5593), 580–584 CrossRef CAS.
- G. M. Walker, et al., Effects of flow and diffusion on chemotaxis studies in a microfabricated gradient generator, Lab Chip, 2005, 5(6), 611–618 RSC.
- C. R. Cabrera and P. Yager, Continuous concentration of bacteria in a microfluidic flow cell using electrokinetic techniques, Electrophoresis, 2001, 22(2), 355–362 CrossRef CAS.
- A. Hatch, et al., A rapid diffusion immunoassay in a T-sensor, Nat. Biotechnol., 2001, 19(5), 461–465 CrossRef CAS.
- M. A. Holden, et al., Generating fixed concentration arrays in a microfluidic device, Sens. Actuators B: Chem., 2003, 92(1–2), 199–207 CrossRef.
- A. E. Kamholz, et al., Quantitative analysis of molecular interaction in a microfluidic channel: the T-sensor, Anal. Chem., 1999, 71(23), 5340–5347 CrossRef CAS.
- K. Macounova, C. R. Cabrera and P. Yager, Concentration and separation of proteins in microfluidic channels on the basis of transverse IEF, Anal. Chem., 2001, 73(7), 1627–1633 CrossRef CAS.
- E. A. Schilling, A. E. Kamholz and P. Yager, Cell lysis and protein extraction in a microfluidic device with detection by a fluorogenic enzyme assay, Anal. Chem., 2002, 74(8), 1798–1804 CrossRef CAS.
- N. L. Jeon, et al., Generation of solution and surface gradients using microfluidic systems, Langmuir, 2000, 16(22), 8311–8316 CrossRef CAS.
- D. Irimia, D. A. Geba and M. Toner, Universal microfluidic gradient generator, Anal. Chem., 2006, 78(10), 3472–3477 CrossRef CAS.
- H. B. Mao, P. S. Cremer and M. D. Manson, A sensitive, versatile microfluidic assay for bacterial chemotaxis, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(9), 5449–5454 CrossRef CAS.
- H. Wu, B. Huang and R. N. Zare, Generation of complex, static solution gradients in microfluidic channels, J. Am. Chem. Soc., 2006, 128(13), 4194–4195 CrossRef CAS.
- W. Saadi, et al., Generation of stable concentration gradients in 2D and 3D environments using a microfluidic ladder chamber, Biomed. Microdev., 2007, 9(5), 627–635 Search PubMed.
- S. Paliwal, et al., MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast, Nature, 2007, 446(7131), 46–51 CrossRef CAS.
- C. W. Li, R. Chen and M. Yang, Generation of linear and non-linear concentration gradients along microfluidic channel by microtunnel controlled stepwise addition of sample solution, Lab Chip, 2007, 7(10), 1371–1373 RSC.
- B. Mosadegh, et al., Generation of Stable Complex Gradients Across Two-Dimensional Surfaces and Three-Dimensional Gels, Langmuir, 2007, 23(22), 10910–10912 CrossRef.
- S. Y. Cheng, et al., A hydrogel-based microfluidic device for the studies of directed cell migration, Lab Chip, 2007, 7(6), 763–769 RSC.
| This journal is © The Royal Society of Chemistry 2008 |
