A short, unsupported Cu(I)⋯Cu(I) interaction, 2.65 Å, in a dinuclear guanidine chloride complex†
Gina M.
Chiarella
a,
Doris Y.
Melgarejo
a,
Alex
Rozanski
a,
Pierre
Hempte
b,
Lisa M.
Perez
c,
Christian
Reber
b and
John P.
Fackler Jr.
*a
aDepartment of Chemistry, Texas A&M University, College Station, Texas 77843-3255, USA. E-mail: fackler@mail.chem.tamu.edu; Fax: +1-979-845-2373; Tel: +1-979-845-0648
bDépartement de chimie, Université de Montréal, Succ. Centre-ville Montréal QC H3C 3J7, Canada. E-mail: christian.reber@umontreal.ca; Fax: +1-514-343-7586; Tel: +1-514–343-7332
cLaboratory for Molecular Simulation, Texas A&M University, College Station, Texas 77843-3255, USA. E-mail: Perez@mail.chem.tamu.edu; Fax: +1-979-845-2971; Tel: +1-979-845-9384
First published on 13th November 2009
Abstract
The reaction of CuCl with the neutral guanidine Hhpp ligand (1,3,4,6,7,8-hexahydropyrimido[1,2-a]pyrimidine) yields two crystalline polymorphs of the neutral dimer, bis(1,3,4,6,7,8-hexahydropyrimido[1,2-a]pyrimidine) dichloro-di-copper(I) with the shortest distance known for a bridging unsupported copper(I)–copper(I) interaction.
Binuclear copper complexes with short Cu–Cu distances have important functional roles in metalloproteins.1 Examples include the bridged CuA site2 in cytochrome C oxidase, catechol oxidase, N2O reductase, nitrite reductase,3–11 and in the the non-bridged Cu–Cu site in the methane monooxygenase enzyme (p-MMO),12 wherein crystallographic and spectroscopic data have suggested the presence of a dinuclear center. There is controversy in this latter case, about whether this is a Cu–Cu or Fe–Fe center. It is unresolved due to the lack of model compounds to compare structural and functional behaviors.13 The significance of unsupported Cu–Cu bonding in general is still not totally clear in these metalloenzymes but may be related to the reversibility of redox activity.
Bonding interactions between heavier group 11 atoms are attributed to relativistic and correlation effects.14 Aurophilic Au(I)–Au(I) interactions are common and generally as strong as H-bonds. Argentophilic Ag(I)–Ag(I) bonds are less common and certainly weaker than the aurophilic bonds since relativistic effects increase with nuclear charge.14 Thus with dinuclear Cu(I) complexes “cuprophilicity” based upon relativistic effects should be very small or non-existent.15 Yet there are several reports of short Cu(I)–Cu(I) distances lending support to the existence of “cuprophilic” bonding.16 However, most of these compounds with short Cu(I)⋯Cu(I) distances are bridged dimers.17 The number of compounds with unsupported Cu(I)–Cu(I) interactions is quite small, with Cu–Cu distances ranging from 2.7154(5) in the ion-pair complex [Cu(hppMe)2][CuCl2]18 to 3.0248 Å in a neutral [Cu(NH3)]2 stabilized by a supramolecular framework.19
Here we report the synthesis and characterization of two polymorphs of an unsupported di-copper(I) Hhpp chloride molecule (Fig. 1) showing that short Cu–Cu interactions may occur without strong bridging ligands connecting the centers. Cu–Cu distances of 2.6725(16) and 2.6517(14) Å are observed.‡
![Crystal structure of bis(1,3,4,6,7,8-hexahydropyrimido[1,2-a]pyrimidine) dichloro-di-copper(i), thermal ellipsoids (30%). Distances, Å: Cu(1)–Cu(1)#1: 1, 2.6725(16); 2, 2,6517(14). Cu(1)–Cl(2): 1, 2.1008(12); 2, 2.1118(11). Cu(1)–N(1): 1, 1.874(3); 2,1.867(3). Cl2⋯N2A: 1, 3.326(4); 2, 3.327(3). Angles, deg.: N(1)–Cu(1)–Cu(1)#1: 1, 95.37(10); 2, 93.24(12). Cl(2)–Cu(1)–Cu(1)#1: 1, 92.79(4); 2, 94.57(3). N(1)–Cu(1)–Cl(2): 1, 171.83(10); 2, 172.14(12).](/image/article/2010/CC/b918912b/b918912b-f1.gif) | ||
| Fig. 1 Crystal structure of bis(1,3,4,6,7,8-hexahydropyrimido[1,2-a]pyrimidine) dichloro-di-copper(I), thermal ellipsoids (30%). Distances, Å: Cu(1)–Cu(1)#1: 1, 2.6725(16); 2, 2,6517(14). Cu(1)–Cl(2): 1, 2.1008(12); 2, 2.1118(11). Cu(1)–N(1): 1, 1.874(3); 2,1.867(3). Cl2⋯N2A: 1, 3.326(4); 2, 3.327(3). Angles, deg.: N(1)–Cu(1)–Cu(1)#1: 1, 95.37(10); 2, 93.24(12). Cl(2)–Cu(1)–Cu(1)#1: 1, 92.79(4); 2, 94.57(3). N(1)–Cu(1)–Cl(2): 1, 171.83(10); 2, 172.14(12). | ||
These materials were prepared by reacting stoichiometric amounts of copper(I) chloride and Hhpp at different times, resulting in two types of crystals: racemic conglomerates 1 and racemic crystals 2.§ These spectroscopic characterizations were carried out on crystals of 1.
The 1H-NMR at room temperature in deuterated benzene shows a slight shift downfield with higher concentration, from 1.26 to 6.30 mM of 0.153–0.104 ppm, but the low solubility precludes further interpretation at this time. A faintly green colored solution is obtained in THF, at 4 mM concentration, with an absorption band at 396 nm having moderate absorptivity (ε) of 917.5 cm−1 M−1. This band may arise from M–L charge transfer after partial oxidation. Further dilution causes additional change reflecting some oxidation or unresolved solvent interaction.
The IR spectrum shows a shift to lower wave number for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretch with respect to the free ligand, from 1644 to 1634 cm−1.20
N stretch with respect to the free ligand, from 1644 to 1634 cm−1.20
The Raman spectrum, Fig. 2, was recorded in the solid state at 80 K, in order to obtain the Cu⋯Cu stretching frequency. Using Woodruff’s rules,21 a frequency of 209 cm−1 is predicted for this interaction at the distance observed. Applying uncertainties to the data, four reasonable candidates for the Cu–Cu stretch were found.
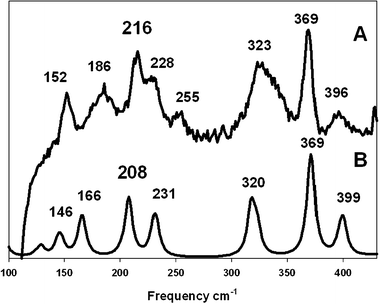 | ||
| Fig. 2 Comparison between the experimental Raman spectrum (A, λexc = 785 nm) and the computational simulation (B). | ||
The Raman spectrum for the optimized molecular structure of 1 was calculated† using DFT (MPW1PW91). These calculations generated a Cu–Cu distance of 2.87 Å; the closest stretching vibrational mode for the Cu–Cu stretch was sited at 208 cm−1 (very close to the value predicted by Woodruff equations). This DFT simulation is compared with the actual Raman spectrum (Fig. 2), matching the observed peak at 216 cm−1, thus attributed to the Cu(I)–Cu(I) stretching frequency.
In both structures, 1 and 2, the two Cu(Hhpp)Cl units are arranged almost perpendicular to one another with dihedral angles of 90.32 (0.10) and 92.28 (0.09)°. The Hhpp guanidine is attached through a N atom to each copper(I); the N–C distances in the guanidine reveal considerable localization22 of the pi electron density on the C1–N1 bond with N1 attached to the copper atom, Table 1. Structures 1 and 2 show a short N1–C1 bond distance relative to the N(2,3)–C1 distance as expected for the more localized pi double bond of the guanidine ligand. This distance is significantly shorter than in the hpp− anion (1.339(5) Å), and in the [Cu(hpp)]223 dimer where there is almost no difference between N1–C1 and N2–C2 distances. Using the quantitative criteria for delocalization of the pi electron-pair suggested by Coles et al.,24 the ΔCN/Å values for structure 1, 0.042, and for 2, 0.062, are very large relative to the value of 0.007 in the delocalized [Cu(hpp)]2.
| 1 | 2 | [Cu(hpp)]2 | |
|---|---|---|---|
| Bond | Distance | Distance | Distance |
| N1–C1 | 1.313(4) | 1.304(4) | 1.339(5) |
| N2–C1 | 1.355(4) | 1.366(4) | 1.346(5) |
| N3–C1 | 1.338(4) | 1.359(4) | 1.371(5) |
| ΔCN/Å | 0.042 | 0.062 | 0.007 |
There is no driving force from bridging ligands or ion pair formation5 in forming the short copper(I)–copper(I) interaction found in 1 and 2, although there may be some weak H-bonding between the N–H on the guanidine attached to one Cu(I) atom and the Cl attached to the second Cu(I) atom (vide infra). The existence of similar structures has been reported6,25 where the Cu(I)–Cu(I) interactions have been explained on the basis of stabilization through supramolecular frameworks or pi stacking interactions, but none of these compounds have a Cu–Cu distance as short as those reported here.
Cyclic guanidines such as Hhpp are strong bases (pK > 10) and Y-shaped molecules.20 They resemble the arginine’s side chain functional group structure that plays an important role in many enzyme processes such as production of NO and transfer of hydrogen in cytochrome P450..26 Guanidines are also excellent electron donor species with the ability to act as Lewis bases.20 The anionic guanidinates function as bridging and chelating ligands to many metals, providing very strong N–metal bonds. The double bond in the free neutral ligand27 is localized on the imino nitrogen which binds terminally to a metal ion, transferring significant electron density and shortening the N–C bond of the N coordinated to the metal.
An interesting feature in those guanidine structures is the bending toward the chlorine atom of the hydrogen atom attached to N2 (the position of that hydrogen is found from the differential Fourier synthesis map), suggesting a weak N–H⋯Cl hydrogen bond. Theory at the MP2 level calculated the Cu–Cu distance in 1 to be longer, 3.01 Å, than found in the DFT, further suggesting that the N–H⋯Cl interaction (N2⋯Cl2, 3.326 Å) is important. Structural disorder caused by puckering of the rings makes further crystallographic analysis questionable although the Cu–Cu distance in 2 is slightly shorter, 0.02 Å, than in 1 while the N2⋯Cl2 distance is slightly longer, 3.327 Å. No significant interactions are found along the Cu–Cu axis in the packing diagrams.
The compounds reported here model very well the distances of the dinuclear copper center of pMMO. This protein has three metal centers: one mononuclear Cu(II), one dinuclear Cu(I)–Cu(I), or a Cu(I)–Cu(II). The third site is known to be copper in M. trichosporium OB3b pMMO, but is unknown in other bacteria. In the dinuclear copper site one Cu atom is coordinated to an N-terminal nitrogen of a histidine and the side chain δ nitrogen residue of the same histidine. The other copper atom is ligated by the δ nitrogen and a nitrogen atom of two different histidines.
An intriguing feature of the pMMO dinuclear site is that it shows no metal–metal bridging group, in spite of the short Cu–Cu distance. Other copper enzymes, e.g. at the purple CuA site of cytochrome C oxidase, have bridging groups. For the closed shell 3d10 Cu(I) ion this metal–metal interaction was unexpected. However the bonding characteristics of the pMMO histidine and the guanidines in the compounds reported here are similar enough to suggest that the Cu(I)⋯Cu(I) contact is reasonable, and that there is a stabilization by formation of a weak but real metal–metal interaction.
In conclusion the synthesis and characterization of the [Cu(Hhpp)Cl]2 reported here with its short unsupported Cu–Cu interaction emphasizes the role of guanidines, guanidinates and other strongly basic N pi-delocalized ligands in having an ability to mimic biologically relevant nitrogen bonding in metalloenzymes. The work highlights the ability of the closed shell M(I) ions in group 11 of the periodic table to produce unsupported metal–metal interactions.
The Welch Foundation of Houston, TX is acknowledged for support of this work through grant No. A-0960. A Dreyfus Senior Mentor Award to JPF supported the studies of AR. Discussions with Amy Rosenzweig and David Phillips were helpful.
Notes and references
- (a) T. D. P. Stack, Dalton Trans., 2003, 1881 RSC; (b) R. L. Richards and M. C. Durrant, J. Chem. Res., Synop., 2002, 95 Search PubMed.
- K. R. Williams, D. R. Gamelin, L. B. LaCroix, R. P. Houser, W. B. Tolman, T. C. Mulder, S. de Vires, B. Hedman, K. O. Hodgson and E. I. Solomon, J. Am. Chem. Soc., 1997, 119, 613 CrossRef.
- C. R. Andrew, P. Lappalainen, M. Saraste, M. T. Hay, Y. Lu, C. Dennison, G. W. Canters, J. A. Fee, C. E. Slutter, N. Nakamura and J. Sanders-Loehr, J. Am. Chem. Soc., 1995, 117, 10759 CrossRef CAS.
- C. R. Andrew, R. Fracziewicz, R. S. Czernuszewicz, P. Lappalainen, M. Saraste and J. Sanders-Loehr, J. Am. Chem. Soc., 1996, 118, 10436 CrossRef CAS.
- J. L. Cole, P. A. Clark and E. I. Solomon, J. Am. Chem. Soc., 1990, 112, 9534 CrossRef CAS.
- T. E. Meyer, A. Marchesini, M. A. Cusanovich and G. Tollin, Biochemistry, 1991, 30, 4619 CrossRef CAS.
- S. E. Wallace-Williams, C. A. James, S. DeVries, M. Saraste, P. Lappalainen, J. van der Oost, M. Fabian, G. Palmer and W. H. Woodruff, J. Am. Chem. Soc., 1996, 118, 3986 CrossRef CAS.
- D. W. Randall, D. R. Gamelin, L. B. LaCroix and E. I. Solomon, J. Inorg. Biochem., 2000, 5, 16 CAS.
- J. A. Farrar, F. Neese, P. Lappalainen, P. M. H. Kroneck, M. Saraste, W. G. Zumft and A. J. Thomson, J. Am. Chem. Soc., 1996, 118, 11501 CrossRef CAS.
- N. J. Blackburn, S. de Vries, M. E. Barr, R. P. Houser, W. B. Tolman, D. Sanders and J. A. Fee, J. Am. Chem. Soc., 1997, 119, 6135 CrossRef CAS.
- P. M. H. Kroneck, W. E. Antholine, D. H. W. Kastrau, G. Buse, G. C. M. Steffens and W. G. Zumft, FEBS Lett., 1990, 268, 274 CrossRef CAS.
- R. L. Lieberman and A. C. Rosenzweig, Nature, 2005, 434, 177 CrossRef CAS.
- A. S. Hakemian, K. C. Kondapalli, J. Telser, B. M. Hoffman, T. L. Stemmler and A. C. Rosenzweig, Biochemistry, 2008, 47, 6793 CrossRef CAS.
- P. Pyykkö, Angew. Chem., Int. Ed., 2004, 43, 4412 CrossRef.
- C. L. Collins, K. G. Dyall and H. F. Schaefer III, J. Chem. Phys., 1995, 102, 2024 CrossRef CAS.
- P. Pyykkö, Chem. Rev., 1988, 88, 563 CrossRef CAS.
- M. A. Carvajal, S. Alvarez and J. J. Novoa, Chem.–Eur. J., 2004, 10, 2117 CrossRef CAS.
- S. H. Oakley, M. P. Coles and P. B. Hitchcock, Inorg. Chem., 2004, 43, 5168 CrossRef CAS.
- S.-L. Zheng, M. Messerschmidt and P. Coppens, Angew. Chem., Int. Ed., 2005, 44, 4614 CrossRef CAS.
- P. J. Bailey and S. Pace, Coord. Chem. Rev., 2001, 214, 91 CrossRef CAS.
- R. B. Dyer and W. H. Woodruff, Applications of Physical Methods to Inorganic and Bioinorganic Chemistry, John Wiley & Sons Ltd., Chichester, UK, 2007, vol. 489, p. 489 Search PubMed.
- M. S. Khalaf, M. P. Coles and P. B. Hitchcock, Dalton Trans., 2008, 4288 RSC.
- F. A. Cotton, X. Feng and D. Timmons, Inorg. Chem., 1998, 37, 4066 CrossRef CAS.
- M. P. Coles and P. B. Hitchcock, Organometallics, 2003, 22, 5201 CrossRef CAS.
- K. Singh, J. R. Long and P. Stavropoulos, J. Am. Chem. Soc., 1997, 119, 2942 CrossRef CAS.
- B. Meunier, S. P. de Visser and S. Shaik, Chem. Rev., 2004, 104, 3947 CrossRef CAS.
- S. H. Oakley, D. B. Soria, M. P. Coles and P. B. Hitchcock, Dalton Trans., 2004, 537 RSC.
- G. M. Sheldrick, SHELXTL, Bruker AXS Inc., Madison, Wisconsin, USA, 2008 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Computational details. CCDC 736655 and 736656. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/b918912b |
| ‡ Single crystal data for 1 C14H26Cl2Cu2N6, M = 476.39, orthorhombic, space group Fdd2, a = 16.569(11), b = 31.152(18), c = 7.371(4) Å, V = 3805(4) Å3, Z = 8, Dc = 1.663 g cm−3, μ = 2.525 mm−1, Mo-Kα, λ = 0.71073, T = 110 K, R(F2 > 2σ) = 0.0298, Rw (F2, all data) = 0.0591, goodness-of-fit = 0.828 for all 2145 unique data (8898 measured, no. refined (1447), Rint = 0.0574, 2θ < 54.6). CCDC: 736655. Single crystal data for 2 C14H26Cl2Cu2N6, M = 476.39, monoclinic, space group C2/c, a = 15.994(7), b = 7.167(3), c = 17.257(7) Å, β = 109.497(5)°, V = 1864.7(14) Å3, Z = 4, Dc = 1.697 g cm−3, μ = 2.577 mm−1, Mo-Kα, λ = 0.71073, T = 110 K, R(F2 > 2σ) = 0.0412, Rw (F2, all data) = 0.1248, goodness-of-fit = 1.071 for all 2132 unique data (6509 measured, no. refined (1105), Rint = 0.1200, 2θ < 54.98). CCDC: 736656. Refinement. All H atoms except the ones attached to the nitrogen atoms were placed in calculated positions and were refined using a riding model (C–H distances were fixed at 0.97 Å, Uiso(H) parameters were fixed at 1.2Ueq(C)). The hydrogens attached to the nitrogen atoms were located in a difference Fourier map and refined freely (N–H distances 0.78(3) Å in structure 1 and 0.89(4) Å in structure 2). Structure 1 is disordered at C2, C3, C6 and C7; the disorder was successfully treated with 4 EADP constraints and 14 SAME/SADI restraints; the SOF of the major components are 0.64846 and 0.55835 and for the minor components 0.35154 and 0.44165, respectively. Structure 2 also is disordered at C2, C3, C6 and C7. The disorder was successfully treated with 4 EADP constraints and 10 SAME/SADI restraints; the SOF of the major components are 0.50762 and 0.52148 and for the minor components 0.49238 and 0.47852, respectively. BRUKER-NONIUS Bruker APEX-II CCD diffractometer. Programs: standard Bruker AXS control and integration software and SHELXTL.28 |
| § This compound was prepared by the reaction of 0.5 mmol (0.05 g) of CuCl previously dried with 0.5 mmol (0.0703 g) of Hhpp using 20 mL of a very well dried and degasified THF in a side arm flask with a condenser in the top. The reaction mixture was refluxed 10 days under nitrogen which produced polymorph 1, from commercial Hhpp. Polymorph 2 was obtained after 3 days reflux from sublimed Hhpp. After cooling the solution, it was filtered under nitrogen through a frit with Celite. The final solution was layered with hexane in a side arm tube. After one week the system produced colorless rod shaped enantiomeric crystals, 1, which crystallized in the orthorhombic space group Fdd2; this preparation yielded 0.45 mmol (0.107 g) 90%. Polymorph 2 was obtained after three weeks as colorless needle shaped crystals suitable for diffraction. The yield was 0.25 mmol (0.059 g) 50%. Those crystals are a racemic mixture of both enantiomers corresponding to the monoclinic space group C2/c. 1H-NMR (C6D6, 298 K): δ = 6.71 (br, s, 2H, 2NH), 2.67 (t, 3JH–H, 2.67, 8, 4CH2), (t, 3JH–H, 2.08, 8, 4CH2), (p, 5JH–H, 1.00, 8, 4CH2). The MALDI and ESI MS showed a very weak peak at m/z = 473 in the negative mode, corresponding to [Cu2Cl2(Hhpp)(hpp)]−1 and a medium peak at m/z = 413 related with [Cu2Cl2(Hhpp)(CH3NCH2Cl)]−1. Elemental analysis: Calculated: C = 35.44%, H = 5.48%, N = 17.71%; found: C = 35.57%, H = 5.41%, N = 17.63%. FTIR in KBr shows the following peaks: 3271, 3211, 2953, 2818, 1634, 1539, 1441, 1365, 1321, 1196, 1113, 1067, 1020, 710, 553, 517 cm−1. The UV-visible spectrum shows two main peaks at 393 and 292 nm. The Raman spectrum in the near infra-red excitation (λexc = 785 nm) gave the following peaks: 152, 186, 216, 228, 255, 323, 369, 396, 428, 446, 461, 469, 536, 550, 595, 668, 698, 718, 743, 752. |
| This journal is © The Royal Society of Chemistry 2010 |
