Opening of DNA double helix at room temperature: Application of α-cyclodextrin self-aggregates†
Syed S.
Jaffer
a,
Prasun
Ghosh
a,
Anindita
Das
b and
Pradipta
Purkayastha
*a
aDepartment of Chemical Sciences, Indian Institute of Science Education and Research, Kolkata, Mohanpur, 741252, WB, India. E-mail: ppurkayastha@iiserkol.ac.in
bDepartment of Biological Sciences, Indian Institute of Science Education and Research, Kolkata, Mohanpur, 741252, WB, India
First published on 4th June 2010
Abstract
Self-aggregation of α-cyclodextrin (α-CD) can induce DNA opening at room temperature (25 °C) owing to the hydroxyl groups on the surface of the spherical aggregates of α-CD, which promote hydrogen bonding with the flipped-out bases in DNA duplex prohibiting them from reverting back.
The process of nucleobase flipping could represent the first step in key fundamental biological processes like replication and transcription. Thus, determination of the precise mechanism of base flipping is important in understanding processes like enzyme-catalyzed DNA methylation, repair, mismatch recognition, and initiation of transcription and replication.1 Such studies would also form the basis to elucidate the general principles that govern the protein–DNA interactions and drug development approaches targeting nucleic acids. X-Ray crystallographic studies provide the structural aspect of base flipping of cytosine 5-methyltransferase from HhaI (HhaI MTase) complexed with its DNA substrate.1a,2 The target cytosine was found to flip completely out of the helix and into the catalytic site of the binding enzyme without seriously disturbing the rest of the DNA helix. The base flipping may be induced or facilitated by enzyme binding or alternatively, the enzyme may recognize and bind a single base transiently flipped out from the helix.3a The rate constant of methyltransferase reaction is estimated to be 0.02 s−1, while the measured flipping lifetime of base pair of DNA duplex is 10 ms.3 These findings eventually indicate that an active involvement of an enzyme in accelerating base flipping appears to be unnecessary. Computational methods with full description of all chemical bonds indicate the feasibility of spontaneous base flipping in B-DNA crystal structures in the absence of an enzyme.4 In confirmation of the theoretical findings, Spies et al. mentioned two simple models for the process by which this flipping of DNA bases occurs.5 In one model, the enzyme catalyzes base flipping and in the other the enzyme relies on normal, spontaneous motions of the nucleic acid to provide it with a fully flipped-out target base.
The spontaneously flipped-out base from double helical nucleic acids, however, can be trapped by host–guest complexation with β-cyclodextrin (β-CD).5 Spies et al. suggest that α-CD does not have any effect on DNA melting due to insufficient space to encapsulate the nucleobases and γ-CD affects the DNA very little due to weak binding.5 According to them 50 mM β-CD lowers the melting temperature of DNA (Tm) by 1.4 °C (from 62.2 °C to 60.8 °C) and 50 mM γ-CD lowers the Tm by 0.5 °C. The work highlighted the encapsulation of the nucleobases inside the CD cavities and analyzed the observations.
However, during the progress of research on CDs, facts like formation of self-aggregate and molecule induced nanotubular aggregates by them have been observed.6 CDs are known to form spherical self-aggregates and the driving force for the self-assembly of cyclodextrin molecules can be considerably ascribed to hydrogen bonding.6 Moreover, the solubility of β-CD is much less in water at room temperature (∼10 mM) and has a nephrotoxic effect.7 The natural α-CD and β-CD, unlike γ-CD, cannot be hydrolyzed by human salivary and pancreatic amylases. However, both α-CD and β-CD can be fermented by the intestinal microflora. α-CD and γ-CD have moderately good solubility in water, but γ-CD is much more expensive than α-CD and β-CD.7a CDs have hydroxyl groups on both the rims and they form the aggregates through hydrogen bond formation.6a Consequently, the aggregates have a good number of hydroxyl groups on the surface. So, if the nucleobases flip and the CD aggregates are close enough then they may form hydrogen bonds with the hydroxyl groups present on the surface of the aggregates (Fig. 1).
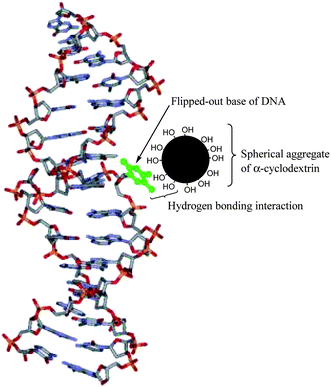 | ||
| Fig. 1 Nucleobase flipping out of the DNA helix (shown in green) interacts with the spherical aggregate of α-CD through hydrogen bonding. The α-CD aggregate has large number of hydroxyl groups on its surface. | ||
To this end, we explored DNA opening due to the influence of the spontaneously developing α-CD nanoaggregates in aqueous buffer (pH 7.6). Since the formation of the α-CD aggregates is a natural phenomenon,6a we intended to apply the concept toward DNA opening at room temperature (25 °C). Melting of calf thymus DNA at 25 °C due to complexation with Cu2+ ions was proposed long before.8 However, this method may not be particularly applicable physiologically because of the toxic effects of the metal ion and other side effects.9 Search for melting DNA at a lower temperature (preferably room temperature) is always an important task toward application in DNA healing.
Works on the natural tendency of the CDs to form aggregates has been the topic of research for many groups. We ventured to apply the concept in opening the double stranded DNA (ds-DNA). The plot of the absorbance of 20 μg ml−1 of ds-DNA in 10 mM Tris-HCl buffer (pH 7.6) at 260 nm against increasing concentration of α-CD shows a typical melting profile of the DNA (Fig. 2). A closer look at the plot reflects that the perturbation of the ds-DNA starts right from the time of initial addition of α-CD, probably because the CD molecules start self-aggregating from the beginning. The phenomenon points towards the increase in the ds-DNA opening process with the building up of the aggregates. The typical concentration of α-CD that induces the opening of the DNA helix seems to be over 12 mM, a concentration where β-CD saturates its solvent (water) at room temperature. Under this circumstance one may have to sonicate the saturated solution rigorously, which may break the β-CD aggregates. At a higher concentration of α-CD (above 33 mM) the absorbance increases linearly reflecting the appearance of single strand DNA (see the ESI†).
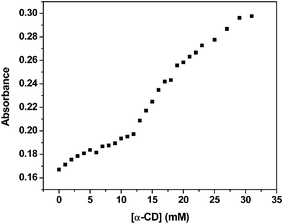 | ||
| Fig. 2 ds-DNA opens up at an α-CD concentration of ∼15 mM at 25 °C. The absorbance values were taken at 260 nm. | ||
In a recent report Spies et al. mentioned that the melting temperature of ds-DNA could be reduced using the guest–host chemistry, i.e., by the encapsulation of the nucleobases by β-CD.5 However, they went to a high temperature of more than 50 °C to observe the melting process and also took 50 mM CD solution. At such a high temperature the solubility of β-CD may increase in water and the possibility of the existence of the self-aggregates may become smaller. Moreover, they mentioned that because of the smaller size of the inner cavity of α-CD molecules, they cannot encapsulate the flipped out nucleobases and thus cannot influence the melting temperature at all. The bigger cavity size of γ-CD also fails to influence the phenomenon too much because of weak binding of the guest.5
Our present concept is different from that of Spies et al. in the sense that we focused on the self-aggregation of the CD molecules in aqueous buffer and their consequent influence on the opening of the ds-DNA at room temperature (25 °C). The purity of DNA was verified by monitoring the ratio of absorbance at 260 nm to that at 280 nm, which was in the range 1.8–1.9. This indicates that the structure of the ds-DNA is intact. The spherical aggregates exist and grow naturally in aqueous medium at ordinary temperature. Hence, with reference to Fig. 1, we propose the mechanism of opening of the ds-DNA with the aid of α-CD aggregates ornamented with a large number of hydroxyl groups on the surface. Due to “breathing” of the DNA, the bases flip out and during that process they find the hydroxyl groups on the α-CD aggregates in the vicinity to re-establish hydrogen bonding. This prevents the bases from reverting back to reconstruct the DNA helix. The opening of the ds-DNA, thus, is found to occur at a much lower concentration (∼15 mM) of the CD and also at room temperature (25 °C).
Transmission electron microscopy (TEM) confirms our findings and demonstrates that, indeed, the approach and existence of the α-CD aggregates near the vicinity of the DNA is very important for the process of opening of DNA (Fig. 3). The spherical self-aggregates are stable at 25 °C and can serve as an agent to pull the DNA strands apart when they come in an optimum distance of the ds-DNA to establish hydrogen bonds with the flipped out nucleobases. The nucleobases seem to cling to the surface of the CD aggregate due to the formation of hydrogen bonds and never go back to re-establish the inter-nucleobase hydrogen bonds situated in the ds-DNA. Because the surface of the α-CD nanoaggregate contains a sufficiently high number of hydroxyl groups, the interaction between the nucleobases and the said surface becomes strong enough to pull the strands of the ds-DNA apart. Panel B of Fig. 3 shows that in the presence of a cluster of such aggregates along the vicinal side of the ds-DNA, the strands can be pulled apart in an extensive region. The observations indicate that with an increase in the population of such spherical α-CD aggregates, the ds-DNA can possibly be converted to single strand.
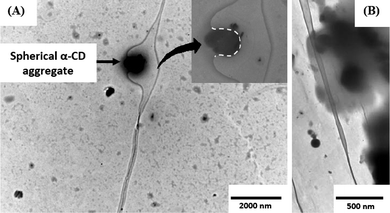 | ||
| Fig. 3 (A) Transmission electron micrograph demonstrating the phenomenon of opening of the ds-DNA. The spherical α-CD aggregate allows the flipped-out nucleobases to cling to its surface, which in turn, does not allow them to flip back and restore the double helix. The inset shows a zoomed image of the bound part of the ds-DNA (The dotted line is to clarify the motif of binding). The α-CD aggregate has a diameter of ∼650 nm. (B) Approach of a cluster of such spherical α-CD aggregates and separating the DNA strands (upper part of the image), whereas when the aggregates are away from the ds-DNA no opening is observed (the lower part of the image). α-CD concentration was taken to be 14 mM. | ||
In summary, it can be stated that opening of ds-DNA at room temperature may have huge implications in biological and pharmaceutical research where DNA repairing becomes important.10 Processes adopted in DNA damage control may require the partial opening of the DNA strands so as to restore the damaged portion using DNA polymerase. Phenomena like base excision and nucleotide excision can be encouraged by the proposed mechanism of opening of ds-DNA at room temperature without inducing any toxicity to the physiological environment. Although we did not study any biological or pharmaceutical applications of this phenomenon, the principle of hydrogen bonding is the responsible factor here and we believe that the topic urges further study by biologists and pharmacists. In this communication we have emphasized on the “breathing” of the DNA nucleobases and have applied the concept of hydrogen bond formation with hydroxyl groups to induce local opening of the ds-DNA. Any microsphere of similar diameter without any hydrogen bond forming group on its surface will not be able to open the ds-DNA based on this concept.
Acknowledgements
This work was supported by CSIR (01(2261)/08-EMR-II), New Delhi and IISER, Kolkata. S. S. J. and P. G. gratefully acknowledge the fellowships from CSIR. The authors thank the Director of Indian Institute of Chemical Biology, Kolkata, India for granting permissions to perform the TEM experiments. They also are grateful to Dr Paulraj Balaji who provided them with purified hsDNA.References
- (a) R. J. Roberts, Cell, 1995, 82, 9 CrossRef CAS; (b) G. Slupphaug, C. D. Mol, B. Kavli, A. S. Arvai, H. E. Krokan and J. A. Tainer, Nature, 1996, 384, 87 CrossRef CAS; (c) D. G. Vassylyev and K. Morikawa, Curr. Opin. Struct. Biol., 1997, 7, 103 CrossRef CAS; (d) B. Allan, N. Reich and J. Beechem, Biochemistry, 1999, 38, 5308 CrossRef CAS; (e) J. P. Jost and H. P. Saluz, in DNA Methylation: Molecular Biology and Biological Significance, Birkhauser, Basel, Switzerland, 1993 Search PubMed.
- (a) S. Klimasauskas, S. Kumar, R. J. Roberts and X. Cheng, Cell, 1994, 76, 357 CrossRef CAS; (b) M. O'Gara, S. Klimasauskas, R. J. Roberts and X. Cheng, J. Mol. Biol., 1996, 261, 634 CrossRef CAS; (c) S. Kumar, X. Cheng, S. Klimasauskas, S. Mi, J. Posfai, R. J. Roberts and G. G. Wilson, Nucleic Acids Res., 1994, 22, 1 CAS.
- (a) F. K. Winkler, Structure, 1994, 2, 79 CrossRef CAS; (b) J. C. Wu and D. V. Santi, J. Biol. Chem., 1987, 262, 4778 CAS; (c) M. Guéron, M. Kochoyan and J. L. Leroy, Nature, 1987, 328, 89 CrossRef CAS.
- (a) Y. Z. Chen, V. Mohan and R. H. Griffey, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 2000, 62, 1133 CrossRef CAS; (b) P. Várnai and R. Lavery, J. Am. Chem. Soc., 2002, 124, 7272 CrossRef CAS.
- M. A. Spies and R. L. Schowen, J. Am. Chem. Soc., 2002, 124, 14049 CrossRef.
- (a) Y. He, P. Fu, X. Shen and H. Gao, Micron, 2008, 39, 495 CrossRef CAS; (b) A. Wu, X. Shen and Y. He, J. Colloid Interface Sci., 2006, 297, 525 CrossRef CAS; (c) S. S. Jaffer, S. K. Saha, G. Eranna, A. K. Sharma and P. Purkayastha, J. Phys. Chem. C, 2008, 112, 11199 CrossRef CAS; (d) S. S. Jaffer, S. K. Saha and P. Purkayastha, J. Colloid Interface Sci., 2009, 337, 294 CrossRef CAS; (e) G. G. Gaitano, W. Brown and G. Tardajos, J. Phys. Chem. B, 1997, 101, 710 CrossRef CAS.
- (a) S. Polarz, B. Smarsly, L. Bronstein and M. Antonietti, Angew. Chem., Int. Ed., 2001, 40, 4417 CrossRef CAS; (b) T. Loftsson, What are cyclodextrins (http://eurocdsoc.com) Search PubMed; (c) H. W. Frijlink, A. C. Eissens, N. R. Hefting, K. Poelstra, C. F. Lerk and D. K. F. Meijer, Pharm. Res., 1991, 8, 9–16 CrossRef CAS.
- K. K. Deb, Spectrosc. Lett., 1981, 14, 385 CrossRef CAS.
- (a) V. V. Shutova, V. V. Revin and Y. A. Myakushina, Appl. Biochem. Microbiol., 2008, 44, 619 Search PubMed; (b) T. C. Cheng and J. T. Sullivan, Comparative Gen. Pharm., 1973, 4, 37 Search PubMed.
- Z. Livneh, J. Biol. Chem., 2001, 276, 25639 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details; absorbance data of ds-DNA at higher concentration of α-CD. See DOI: 10.1039/c0nr00184h |
| This journal is © The Royal Society of Chemistry 2010 |
