Antibacterial action of dispersed single-walled carbon nanotubes on Escherichia coli and Bacillus subtilis investigated by atomic force microscopy
Shaobin
Liu
a,
Andrew Keong
Ng
b,
Rong
Xu
a,
Jun
Wei
c,
Cher Ming
Tan
d,
Yanhui
Yang
a and
Yuan
Chen
*a
aSchool of Chemical and Biomedical Engineering, Nanyang Technological University, Singapore 637459. E-mail: chenyuan@ntu.edu.sg; Fax: +65 67947553; Tel: +65 63168939
bDSO National Laboratories, 20 Science Park Drive, Singapore 118230
cSingapore Institute of Manufacturing Technology, Singapore 638075
dSchool of Electrical and Electronic Engineering, Nanyang Technological University, Singapore 639798
First published on 29th September 2010
Abstract
Single-walled carbon nanotubes (SWCNTs) exhibit strong antibacterial activities. Direct contact between bacterial cells and SWCNTs may likely induce cell damages. Therefore, the understanding of SWCNT–bacteria interactions is essential in order to develop novel SWCNT-based materials for their potential environmental, imaging, therapeutic, and military applications. In this preliminary study, we utilized atomic force microscopy (AFM) to monitor dynamic changes in cell morphology and mechanical properties of two typical bacterial models (gram-negative Escherichia coli and gram-positive Bacillus subtilis) upon incubation with SWCNTs. The results demonstrated that individually dispersed SWCNTs in solution develop nanotube networks on the cell surface, and then destroy the bacterial envelopes with leakage of the intracellular contents. The cell morphology changes observed on air dried samples are accompanied by an increase in cell surface roughness and a decrease in surface spring constant. To mimic the collision between SWCNTs and cells, a sharp AFM tip of 2 nm was chosen to introduce piercings on the cell surface. No clear physical damages were observed if the applied force was below 10 nN. Further analysis also indicates that a single collision between one nanotube and a bacterial cell is unlikely to introduce direct physical damage. Hence, the antibacterial activity of SWCNTs is the accumulation effect of large amount of nanotubes through interactions between SWCNT networks and bacterial cells.
Introduction
Single-walled carbon nanotubes (SWCNTs) exhibit strong antibacterial activities.1 It has been proposed that cell membrane damage, resulting from direct physical contact between bacteria and SWCNTs, is the major reason for bacterial death.1 Previous experimental results showed that the antibacterial activity of SWCNTs can be influenced by many factors: carbon nanotube (CNT) diameter,2 length,3 aggregation,3,4 concentration,4,5 and surface functional group;5 buffer solution;5 as well as contact time6 and intensity.4 In our recent study,4 we suggested that individually dispersed nanotubes could be visualized as numerous moving “nano darts” attacking bacteria in a buffer solution, degrading bacterial cell integrity and causing cell death. However, the interactions between SWCNTs and bacterial cells still remain unclear. Such information about SWCNT–bacteria interactions is essential in the development of novel SWCNT-based materials for their potential applications, such as water treatment filters7 and antibacterial coatings.8,9 Moreover, a good understanding on SWCNT–bacteria interactions may also help to elucidate the toxicology of SWCNTs, which are critical for their applications in imaging and therapeutics.10Although scanning electron microscopy (SEM) has been utilized to visualize physical damages of bacterial cells upon contact with SWCNTs,2–6 atomic force microscopy (AFM) may provide better opportunities for studying SWCNT–bacteria interactions. This is because AFM can image nanoscale ultrastructures of bacterial surfaces and measure nanomechanical properties of bacterial cells.11,12 Bacteria can be imaged in air or buffer solutions13 without complex pretreatments (such as the conductive film coating required by SEM).11 Nanomechanical properties of bacteria can also be measured.14–16 Although some air dried samples, if stored properly, can be imaged for months and even years,17 in some cases, their properties may change during air drying processes.18 To prevent such alterations to properties during drying processes, in situ measurement under physiological conditions is preferred.16,19,20 However, imaging of bacteria in buffer solutions may show reduced topographical contrast compared to images obtained on air-dried samples.17,21,22 In this preliminary study, we conducted our AFM studies on air dried samples. It should be noted that there are always artefacts associated with air drying. Imaging cells in their hydrated state is preferred, although it is technically more challenging. Numerous studies have demonstrated the effectiveness of AFM in exploring surface structure and mechanical property changes of bacterial cells upon treatment with β-lactam,23 cefodizime,24 chistosan,14,15 nitric oxide,25 and antimicrobial peptides.19,26–29
The aim of this study is to investigate dynamic changes induced by SWCNTs on cell morphology and mechanical properties of two typical bacterial models: gram-negative Escherichia coli and gram-positive Bacillus subtilis. First, a super sharp AFM tip was used to create piercings on bacterial cells, mimicking possible collisions between SWCNTs and bacteria. Subsequently, AFM was employed to image bacterial cells that were exposed to SWCNTs over different time periods. In this preliminary study, all cell imaging was conducted in air, in order to monitor morphology changes of bacteria. We also measured the bacteria mechanical properties under various pseudo-physiological conditions (0.9% NaCl solution) to avoid changes in mechanical properties during air drying process.
Experimental
Bacterial cell preparation
E. coli K12 (ATCC 25404) and B. subtilis (ATCC 6051) were grown in Luria-Bertani (LB) broth at 37 °C and 30 °C overnight, respectively, and harvested in the mid-exponential growth phase. Cells were isolated by centrifugation at 6000 rpm for 10 min and washed three times with a saline solution (an aqueous mixture of 0.1 wt % Tween-20 and 0.9 wt % NaCl). Tween-20 was added to enhance the dispersion of SWCNTs.4 Next, cell pellets were suspended in the saline solution. Bacterial suspensions were further diluted to obtain cell samples containing 106 to 107 colony forming units (cfu/mL). Commonly available SWCNT samples are usually mixtures of many species including metal residues, nanosize catalyst supports, amorphous carbon, carbon nanoparticles, graphite, carbon fibers, SWCNTs and multi-walled carbon nanotubes (MWCNTs). To accurately investigate the antibacterial action of SWCNTs, we employed high purity SWCNT samples with an average tube diameter of 0.83 nm and a narrow chiral structure distribution.30–33 Nanotubes were purified by a centrifugation-based method.34 To yield individually dispersed nanotube suspensions, solid samples (5 μg/mL) were dispersed in the Tween-20–saline solution by sonication in a cup-horn sonicator (SONICS, VCX-130) at 20 W for 1 h.4 SWCNT suspensions (10 mL at 5 μg/mL) were incubated with 1 mL of bacterial suspensions for up to 2 h under 250 rpm shaking speed at 37 °C for E. coli or 30 °C for B. subtilis.Cell piercing experiments
A super sharp silicon probe (SSS-NCH from Nanoworld with typical tip radius of 2 nm) was used in cell piercing experiments to examine possible nanoscale collisions between individually dispersed SWCNTs and bacteria. Fresh bacterial suspensions were drop-casted on cleaned glass slides and dried in air. We carried out the piercing experiments in air because it is easier to observe any structural changes on bacterial surface by AFM imaging in air than in liquid. The cantilever deflection sensitivity was calibrated on a hard surface (a glass slide). AFM images were first obtained in a tapping mode at 256 pixels × 256 lines with scanning speed at 1 Hz. After imaging, the tip was aligned above bacterial cells and converted to contact mode. Four different positions on the bacterial cell were continuously punctured for 200 times each, with forces of 10, 100, 1000, and 2000 nN, respectively. The cell was eventually imaged in tapping mode to illustrate morphology changes induced by physical punctures at different force loads.Estimating mass and momentum of dispersed SWCNTs
Nanotubes with (7,5) chiral structure is the main species in our SWCNT samples.4,34 Considering the fact that the C–C bond length is 0.142 nm, one unit length of the (7,5) nanotubes (4.448 nm) contains 436 C atoms (simulated by Materials Studio, Accelrys). The average length of nanotubes is assumed to be 500 nm; accordingly, the mass of one individual nanotube is m = (500 (nm, length) × 436 (C atoms)/4.448 (nm/unit length)) × (0.012 (kg/mol) /6.02 × 1023 (atoms/mol)) = 9.8 × 10−22 kg. During incubation with bacteria, the stirring speed of 250 rmp in a flask of 5 cm in diameter could result a velocity of v = 2π × 0.025 (m) × 250 rmp/60 (sec) = 0.65 m/s. Thus, the momentum of an individual nanotube is approximately at mv = 6.37 × 10−22 kg·m/s.Cell imaging by AFM
Cover glass slides were rinsed with alcohol, acetone, and deionized water in sequential steps. A cell sample of 10 μL was extracted from bacterial cell suspensions after incubation with SWCNTs for different time periods, and drop-casted directly on cleaned glass slides. To obtain AFM images of bacterial cell wall morphology, we dried and immobilized cells on glass slides under ambient air for 10 min. All samples were characterized within 30 min after drying. AFM images were obtained via a MFP3D microscope (Asylum Research, Santa Barbara, CA). To minimize lateral forces on cells during imaging, a silicon non-contact high resonance frequency cantilever (NCH type from Nanoworld with a spring constant of 42 N/m) was used to image cells during tapping mode. The scan rate was set to 1 Hz at various scan sizes, while all data resolutions were set to 256 pixels × 256 lines. To eliminate the Z offset between scan lines, we applied the first-order flattening to AFM height profiles. Then, we analyzed the AFM height profiles using the Igor Pro MFP-3D software to obtain root-mean-squared (rms) roughness on 200 nm × 200 nm areas. Five AFM height profiles were scanned within 30 min, and five different areas on the height profiles of each cells were randomly selected to calculate their rms roughness values.Force measurement on immobilized bacteria
E. coli and B. subtilis were immobilized on poly(L-lysine) coated cover glass slides, as described in our previous publication.4 In a nutshell, 10 μL of poly (L-lysine) solution (Sigma, 0.01% solution) was dropped on a clean glass slide, and the slide was dried at room temperature. A bacterial suspension of 50 μL of was drop-casted on the poly(L-lysine) coated surface. The bacterial suspension liquid drop stood still for 10 min to anchor cells on the surface. Then, the slide was rinsed in 0.9 wt % NaCl solution to remove unattached cells. Cell attached slides were imbedded in ∼100 μL of saline solution during force measurement. The spring constant (kc) of AFM tips was measured in saline solution by a thermal method in the Igor Pro MFP 3D software. The location of bacterial cells was identified by AFM imaging in contact mode (10 × 10 μm2 with 128 × 128 pixels or 15 × 15 μm2 with 256 × 256 pixels) using Silicon probes (ContAI-G, Budget Sensors). Reference force curves were first obtained on bare spots on the poly (L-lysine) coated surface (not covered with bacteria). Next, AFM tips were aligned at different positions on bacterial cells. The positions were all set near the center of bacteria to avoid variations induced by the edge effect.35 The force trigger point was set to 3 nN, and the tip approaching and retraction rates were 500 nm/s. At least five random locations on the same cell and five different bacterial cells on each glass slide were chosen to record force–distance (f–d) curves. Subsequently, one additional AFM image was taken to confirm that the structure and location of the cells remain unchanged during the force measurement. Spring constants (kf) of bacterial cells were determined by the slope (s) of the linear portion of the approaching branch of f–d curves using the expression: kf = kcs/(1 − s).36–38Results and discussion
Multiple piercing of bacterial cells
When bacteria are incubated with SWCNT dispersions, cells may collide with numerous SWCNTs in solution. The collision between bacterial cells and SWCNTs may damage bacterial cells. Physical piercing on cell walls by a nanoscale tip can be used to simulate such collisions, and it is interesting to find out the force needed by the tip to induce damages on cell surfaces. We chose a super sharp tip with 2 nm radius to mimic the physical structure of SWCNTs, which would interact with bacteria in solution. As described in the previous section, the tip continuously pierced the cell surface under four different force loads (10, 100, 1000, and 2000 nN). Fig. 1 shows images of bacterial cells before and after pierced. When the force is higher than 100 nN, clear holes can be found on cell surfaces after punctures; the holes remain unchanged after the piercing experiment had ended for 30 min. For both E. coli and B. subtilis, the hole dimensions increase with the raise of applied force from 100 to 2000 nN. Conversely, no damages were noted when the force was at 10 nN. We further lowered the force to 3 nN, and no changes on the bacterial morphology were observed after multiple puncturings. Suo et al.39 illustrated that a live gram negative bacterium Salmonella typhimurium can survive 20 to 40 puncturings of 4 nN by a sharp AFM tip, and the authors concluded that phospholipid molecules may undergo rapid reorientation in response to AFM tip penetration, sealing holes that are caused by the tip. In the same fashion, the holes, which were caused by force at or below 10 nN, on E. coli and B. subtilis were covered back. However, the reorientation of phospholipid molecules is unable to reseal holes induced by forces above 10 nN.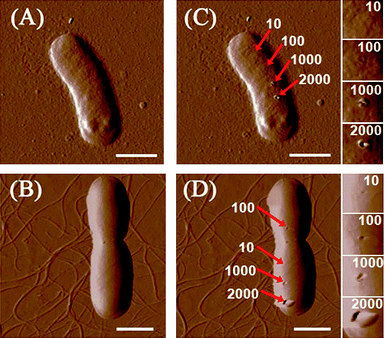 | ||
| Fig. 1 AFM amplitude images of (A) E. coli and (B) B. subtilis. (C) and (D) are, respectively, E. coli and B. subtilis after piercing by a 2 nm AFM tip for 200 times at different locations. Small images on the extreme right are enlarged images of areas punctured by the tip. The deflection set points of the AFM tip are 10, 100, 1000, and 2000 nN. All scale bars are 1 μm. | ||
An intriguing question is whether SWCNTs dispersed in solution could induce large forces (>10 nN) on bacterial cell surfaces? If we assume that the relative velocity of nanotubes to bacteria would decrease from 0.65 m/s to 0 m/s upon collision under the stirring speed of 250 rmp, the momentum of one nanotube (6.37 × 10−22 kg·m/s) could induce a force of 10 nN only when the collision impact time is as short as 6.37 × 10 −14 s (t = Δmv/F), which is too small to be realistic. Even if nanotube–surfactant assemblies in solution can be much heavier than one individual nanotube, the momentum change is still unlikely to generate a force higher than 10 nN. On the other hand, a study has shown that Brownian motion velocity of a 300 nm nanoparticle is only 4–5 μm/s.40 The momentum of SWCNTs attributed to their Brownian motion should be much smaller than that induced by stirring. Based on the above outcomes, it is unlikely that a single collision between an individual SWCNT and a bacterium would produce a force that can cause direct physical damages to the bacterial surface.
Bacterial cell morphology changes
Our previous study indicated that 58.1 ± 5.0% of E. coli and 87.5 ± 6.5% of B. subtilis were killed after 2 h of incubation with SWCNTs, and SEM images also showed that bacterial cells are seriously damaged after exposure to SWCNTs.4 Since a direct single collision has little chance in damaging cells, it is essential to investigate how SWCNTs cause dynamic changes to cell wall architecture and cell mechanical properties, as well as to examine the dependence of such changes on interaction time and intensity. Fig. 2 renders AFM amplitude images of E. coli and B. subtilis observed after incubation with SWCNTs for 0, 10, 60, and 120 min. As illustrated in Fig. 2A, the surface of E. coli is relatively smooth before interacting with SWCNTs. No obvious nano-pores or physical damages were found, implying that both drying in air and tapping mode do not alter cell wall architecture. In this study, no peritrichous flagella structures were observed on E. coli. On the contrary, gram-positive bacterium B. subtilis (see Fig. 2B) possesses distinctive peritrichous flagella structures with smooth surfaces. After incubation with SWCNTs for 10 min, E. coli is winded with nanoscale tube-like structures (see Fig. 2C). A close examination of the cell surface in Fig. 3 suggests that these tube-like structures partially cover cell surface. We further measured the height of the structures on the flat glass surface. As clearly depicted in Fig. 3B, heights of these small bundles range from 4 to 10 nm, which are similar to those of SWCNT bundles. Since no peritrichous flagella structures were found on E. coli and no other tube-like materials exist in our cell cultures, we conclude that these tube-like structures are SWCNT bundles.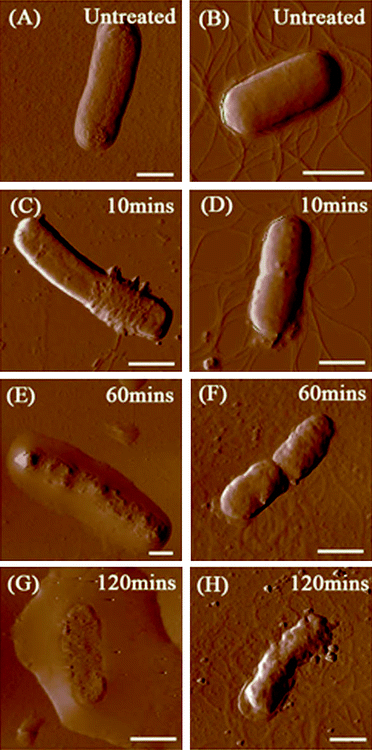 | ||
| Fig. 2 AFM amplitude images of E. coli and B. subtilis (1 mL of bacterial suspensions, 106–107 cfu/mL) after incubation with SWCNT dispersions (10 mL, 5 μg/mL) over different time periods. (A) E. coli before contact with SWCNTs, (B) B. subtilis before contact with SWCNTs, (C, E and G) E. coli after incubated with SWCNTs for 10, 60 and 120 min, respectively, and (D, F and H) B. subtilis after incubated with SWCNTs for 10, 60 and 120 min, respectively. All scale bars are 1 μm. | ||
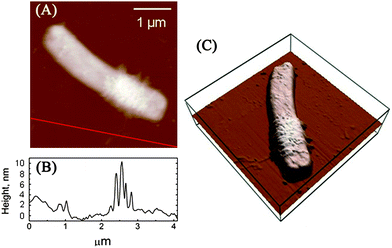 | ||
| Fig. 3 (A) AFM height image of the E. coli cell (the same cell shown on Fig. 1C) after incubation with SWCNT dispersions for 10 min. (B) The height profile along the line in part A. (C) 3D height image of the E. coli cell. | ||
The surface of bacterial cells became uneven after incubation for 60 min. Fig. 2E shows a damaged E. coli. Grooves and holes were randomly distributed on the cell surface. The cell wall collapsed and the cell height decreased, releasing its cytoplasm. The cell damages were more apparent and severe after the cell was incubated for 120 min (see Fig. 2G); the cell was almost flattened, and cytoplasm was found all around the cell. AFM image in Fig. 2G confirmed that SWCNTs can fragment bacterial membranes, leading to a severe volume reduction. The surface roughness of E. coli outer membranes was also monitored. The average rms roughness of E. coli before contact with SWCNTs was 3.6 ± 0.5 nm. After incubation with SWCNTs for 10 min, the rms of cell surface with and without SWCNT networks were correspondingly 9.9 ± 1.6 nm and 4.0 ± 0.9 nm. The surface roughness further increased to an average of 16.8 ± 1.8 nm after 60 min of incubation with SWCNTs.
The same cell morphology changes were observed on B. subtilis. The cell surface becomes rougher after 10 min of incubation with SWCNTs (see Fig. 2D). It is difficult to identify tube-like structures on B. subtilis due to the existence of peritrichous flagella structures, which look similar to nanotube bundles. Some solid residues can be identified near the edge of the cell. Serious bacterial membrane destruction and leakage of cytoplasm was observed after exposure to SWCNTs for 60 and 120 min (see Fig. 2F and 2H). The morphology observation for B. subtilis was consistent with that for E. coli. The rms roughness of the untreated B. subtilis was 4.9 ± 1.1 nm, and the roughness increased to 7.3 ± 2.0 nm, 15.8 ± 4.6 nm, and 22.3 ± 2.7 nm after 10, 60, and 120 min of exposure, correspondingly.
Mechanical properties of bacterial cells
Apart from morphology changes, mechanical properties of bacteria should also be changed after exposure to SWCNTs. To prevent the mechanical properties from changing during our AFM characterization, we conducted all the following measurements in saline solution. A silicon probe (ContAI-G, Budget) was selected to assess the mechanical properties of bacterial cells.41 Several studies have demonstrated that spring constants of AFM cantilevers would not have significant influences on the measurement of mechanical properties of bacterial cells.19,37 In this study, the AFM probe has a spring constant of 0.35 ± 0.03 N/m, which was determined by the thermonoise method.42 F–d curves were used to reveal cell hardness changes, as illustrated in Fig. 4 for E. coli, and B. subtilis before and after treatment with SWCNTs. For glass surfaces, the slope of f–d curves equals to one, because no indentation was induced by the AFM tip. Conversely, any deformations in cell membranes can cause f–d curve slopes to be less than one. In line with our previous study,4Fig. 4 shows that f–d curves of B. subtilis exhibit smaller slopes than those of E. coli, suggesting that B. subtilis are softer than E. coli. Although gram-positive cells are generally stiffer than gram-negative cells43 due to their thick peptidoglycan layers (20–80 nm vs. 7–8 nm), our unexpected outcome may likely be rationalised as follows: (1) B. subtilis possesses a lesser resistance to the AFM tip than E. coli because the former bacterial cell does not have an extra layer, the outer membrane, and/or (2) the particular strain of B. subtilis used in this study is softer than the strain of E. coli. Similar results have been observed in previous AFM studies.15,41 Nevertheless, f–d curve slopes of both B. subtilis and E. coli decrease with increasing incubation time. The effective spring constants of bacterial cell surfaces can be calculated by the linear part of f–d curves, and the results are listed in Table 1. The effective spring constants of E. coli and of B. subtilis decrease from 0.172 ± 0.044 to 0.088 ± 0.036 N/m and from 0.102 ± 0.036 to 0.034 ± 0.025 N/m, respectively, after incubation with SWCNTs for 120 min. Spring constants of bacterial cells correspond to their turgor pressure.36 Hence, the spring constant decrement observed in this study can be attributable to the leakage of cytoplasm;44 the turgor pressure of bacterial cells decreases when the dense cytoplasm is released. As evidenced in Fig. 2, the cell membrane is damaged and a large amount of cytoplasm is visible around the cell body. The significant changes found in spring constants of E. coli and of B. subtilis are consistent with their respective morphology changes.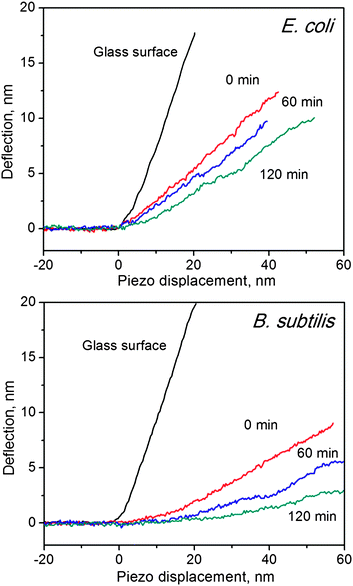 | ||
| Fig. 4 Force–distance curves of glass surface, E. coli (top) and B. subtilis (bottom), before and after incubation with SWCNTs at different time durations. Slopes indicate the hardness of the measured surface. | ||
| Incubation time (min) | E. coli | B. subtilis | ||
|---|---|---|---|---|
| slope | k b (N/m) | slope | k b (N/m) | |
| 0 | 0.32 ± 0.06 | 0.172 ± 0.044 | 0.23 ± 0.06 | 0.102 ± 0.036 |
| 60 | 0.24 ± 0.07 | 0.114 ± 0.034 | 0.14 ± 0.05 | 0.059 ± 0.026 |
| 120 | 0.19 ± 0.06 | 0.088 ± 0.036 | 0.09 ± 0.06 | 0.034 ± 0.025 |
In our previous study, we demonstrated that the antibacterial activity of SWCNTs can be improved by dispersing nanotubes individually, increasing nanotube concentration and elevating the shaking speed during incubation. Well-dispersed nanotubes are more mobile in solution and have a higher chance of colliding with bacterial cells. Higher nanotube concentration and shaking speed can also enhance the interaction between SWCNTs and cells. Other studies have observed that CNT diameter,2 length,3 aggregation,3,4 concentration,4,5 and surface functional groups;5 buffer solutions;5 as well as contact time6 and intensity4 can affect CNT antibacterial activity. All these factors apparently influence interactions between bacteria and nanotubes in solution. According to the earlier experimental results for AFM tip piercing, one individual nanotube (at 0.65 m/s) under a stirring speed of 250 rmp is not likely to directly damage the cell. The relative speed between suspended cells and SWCNTs could be even smaller. Furthermore, considering the possibility of “head on” vs. “side on” collisions, the chances of a SWCNT “piercing” bacteria are negligible. AFM images in Fig. 2 show that nanotube networks partially cover cell surfaces, signifying that bacterial death is associated with the accumulation effect of large amount of nanotubes. We propose the following scenario to explain our results. The SWCNT concentration is 5 μg/mL, and the mass of individual nanotubes is 9.8 × 10−22 kg; accordingly, there are about 5.1 × 1013 nanotubes in the 10 mL SWCNT suspension interacting with 106 to 107 bacterial cells. Cell surfaces may collide with SWCNTs millions of times during incubation. It has been reported that glycoproteins can disperse SWCNTs in aqueous solutions,45 and SWCNTs also interact strongly with polysaccharides, such as chitosan.46 Thus, individual nanotubes may adsorb on cell surfaces, and more nanotubes on the cell surface would bundle together owing to the strong van der Waals attractions between nanotubes.47,48 On top of the strong van der Waals attractions, a recent theoretical study also highlighted that collisions with bacterial surface and rotational Brownian motion can lead to the accumulation of microswimmers near cell surfaces.49 SWCNTs dispersed in solution, therefore, may have high tendency to accumulate near cell surfaces and to be trapped into the SWCNT networks formed on cell surfaces. For the above mentioned scenario, the interaction between SWCNT networks and cells is expected to be more intense than that of individual nanotubes, giving rise to the destruction of cell envelope. Once the cell envelope is damaged, it will result in cell death. Experimental parameters, such as dispersion, concentration, contact time and intensity, etc., which enhance the adsorption and accumulation of SWCNTs on cell surfaces, are able to enhance the antibacterial activities of SWCNTs.
In this study, a super sharp silicon probe was used to investigate the interactions of SWCNTs with bacteria, however, because the silicon probe has different chemistry/reactivity than SWCNTs, the “chemical” effects of SWCNT–bacteria interactions cannot be probed. As recommended by previous studies,1,2 SWCNTs may generate bacterial oxidative stress responses. The close contacts between SWCNT networks and cells can also possibly enhance the oxidative stress. However, some recent studies suggested that carbon nanotubes can also behave as free-radical scavengers.50–52 The exact chemical or biological roles of SWCNTs require further investigation. It should be noted that the results in this study have clarified that the physical piercing by individual SWCNTs is not the feasible antibacterial mechanism under incubation conditions.
Recent literature has shown that when bacteria were filtered through nanotube based membranes, many cells were killed.7 We reason that the filtration process is different from incubation. In a filtration process, cells are dragged to the nanotube network. The force, which a bacterial cell experiences during filtration, could be much higher than 10 nN. Such heavy forces are capable of directly damaging bacterial cells. The direct physical piecing can be the dominant factor in the antibacterial mechanism under filtration conditions. On the other hand, the latest report showed that bacteria in contact with a deposit layer of SWCNTs under static conditions produce much less biofilm.53 This suggests that multiple contact between SWCNTs and bacteria is necessary for bacterial death, which is consistent with our observations under incubation conditions.
Conclusions
The present study is the first AFM visual demonstration of SWCNT antibacterial effects on bacterial cells. For both E. coli and B. subtilis, AFM images obtained on air dried samples showed that bacterial envelopes were severely damaged with leakage of intracellular contents after incubation with SWCNTs. The volume and height of bacterial cells also decreased. The rms roughness of bacterial surfaces increased with the incubation time, which was in agreement with the observed cell destruction. The spring constants of bacterial surfaces after exposure to SWCNTs over different time periods were measured in saline solution. The effective spring constants of E. coli and B. subtilis declined from 0.172 ± 0.044 to 0.088 ± 0.036 N/m and 0.102 ± 0.036 to 0.034 ± 0.025 N/m, respectively, after incubation with SWCNTs for 120 min. The decrement of spring constants can be associated with the destruction of cell envelope and the release of cytoplasm. Physical piercings on bacterial cells by the 2 nm AFM tip can result in significant damages when the applied force was higher than 10 nN. The collision force between an individual nanotube and a bacterial cell during incubation is much weaker as compared to the tested force under piercing conditions. Moreover, SWCNT network structures were found on E. coli surface, further implying that bacterial death is related to the accumulation effect of large amount of nanotubes.Acknowledgements
This work was supported by National Research Foundation, Singapore (NRF-CRP2-2007-02), and Environment & Water Industry Development Council, Singapore (0802-IRIS-12).References
- S. Kang, M. Pinault, L. D. Pfefferle and M. Elimelech, Single-walled carbon nanotubes exhibit strong antimicrobial activity, Langmuir, 2007, 23(17), 8670–8673 CrossRef CAS.
- S. Kang, M. Herzberg, D. F. Rodrigues and M. Elimelech, Antibacterial Effects of Carbon Nanotubes: Size Does Matter, Langmuir, 2008, 24(13), 6409–6413 CrossRef CAS.
- S. Kang, M. S. Mauter and M. Elimelech, Physicochemical Determinants of Multiwalled Carbon Nanotube Bacterial Cytotoxicity, Environ. Sci. Technol., 2008, 42(19), 7528–7534 CrossRef CAS.
- S. B. Liu, L. Wei, L. Hao, N. Fang, M. W. Chang, R. Xu, Y. H. Yang and Y. Chen, Sharper and Faster “Nano Darts” Kill More Bacteria: A Study of Antibacterial Activity of Individually Dispersed Pristine Single-Walled Carbon Nanotube, ACS Nano, 2009, 3(12), 3891–3902 CrossRef CAS.
- L. R. Arias and L. J. Yang, Inactivation of Bacterial Pathogens by Carbon Nanotubes in Suspensions, Langmuir, 2009, 25(5), 3003–3012 CrossRef CAS.
- S. Kang, M. S. Mauter and M. Elimelech, Microbial Cytotoxicity of Carbon-Based Nanomaterials: Implications for River Water and Wastewater Effluent, Environ. Sci. Technol., 2009, 43(7), 2648–2653 CrossRef CAS.
- A. S. Brady-Estevez, S. Kang and M. Elimelech, A Single-Walled-Carbon-Nanotube Filter for Removal of Viral and Bacterial Pathogens, Small, 2008, 4(4), 481–484 CrossRef CAS.
- M. S. Mauter and M. Elimelech, Environmental Applications of Carbon-Based Nanomaterials, Environ. Sci. Technol., 2008, 42(16), 5843–5859 CrossRef CAS.
- S. J. Klaine, P. J. J. Alvarez, G. E. Batley, T. F. Fernandes, R. D. Handy, D. Y. Lyon, S. Mahendra, M. J. McLaughlin and J. R. Lead, Nanomaterials in the Environment: Behavior, Fate, Bioavailability, and Effects, Environ. Toxicol. Chem., 2008, 27(9), 1825–1851 CrossRef CAS.
- K. Kostarelos, A. Bianco and M. Prato, Promises, facts and challenges for carbon nanotubes in imaging and therapeutics, Nat. Nanotechnol., 2009, 4(10), 627–633 CrossRef CAS.
- Y. F. Dufrene, Towards nanomicrobiology using atomic force microscopy, Nat. Rev. Microbiol., 2008, 6(9), 674–680 Search PubMed.
- P. Hinterdorfer and Y. F. Dufrene, Detection and localization of single molecular recognition events using atomic force microscopy, Nat. Methods, 2006, 3(5), 347–355 CrossRef CAS.
- B. Drake, C. B. Prater, A. L. Weisenhorn, S. A. C. Gould, T. R. Albrecht, C. F. Quate, D. S. Cannell, H. G. Hansma and P. K. Hansma, Imaging Crystals, Polymers, and Processes in Water with the Atomic Force Microscope, Science, 1989, 243(4898), 1586–1589 CrossRef CAS.
- J. C. Fernandes, P. Eaton, A. M. Gomes, M. E. Pintado and F. X. Malcata, Study of the antibacterial effects of chitosans on Bacillus cereus (and its spores) by atomic force microscopy imaging and nanoindentation, Ultramicroscopy, 2009, 109(8), 854–860 CrossRef CAS.
- P. Eaton, J. C. Fernandes, E. Pereira, M. E. Pintado and F. X. Malcata, Atomic force microscopy study of the antibacterial effects of chitosans on Escherichia coli and Staphylococcus aureus, Ultramicroscopy, 2008, 108(10), 1128–1134 CrossRef CAS.
- Y. Z. Wu and A. H. Zhou, In situ, real-time tracking of cell wall topography and nanomechanics of antimycobacterial drugs treated Mycobacterium JLS using atomic force microscopy, Chem. Commun., 2009,(45), 7021–7023 RSC.
- A. V. Bolshakova, O. I. Kiselyova and I. V. Yaminsky, Microbial surfaces investigated using atomic force microscopy, Biotechnol. Prog., 2004, 20(6), 1615–1622 CrossRef CAS.
- J. J. Thwaites and U. C. Surana, Mechanical Properties of Bacillus subtilis Cell Walls - Effects of Removing Residual Culture Medium, J. Bacteriol., 1991, 173(1), 197–203 CAS.
- N. P. Mortensen, J. D. Fowlkes, C. J. Sullivan, D. P. Allison, N. B. Larsen, S. Molin and M. J. Doktycz, Effects of Colistin on Surface Ultrastructure and Nanomechanics of Pseudomonas aeruginosa Cells, Langmuir, 2009, 25(6), 3728–3733 CrossRef CAS.
- Y. Y. Chen, C. C. Wu, J. L. Hsu, H. L. Peng, H. Y. Chang and T. R. Yew, Surface Rigidity Change of Escherichia coli after Filamentous Bacteriophage Infection, Langmuir, 2009, 25(8), 4607–4614 CrossRef CAS.
- A. V. Bolshakova, O. I. Kiselyova, A. S. Filonov, O. Y. Frolova, Y. L. Lyubchenko and I. V. Yaminsky, Comparative studies of bacteria with an atomic force microscopy operating in different modes, Ultramicroscopy, 2001, 86(1–2), 121–128 CrossRef CAS.
- D. Robichon, J. C. Girard, Y. Cenatiempo and J. F. Cavellier, Atomic force microscopy imaging of dried or living bacteria, Comptes Rendus de l Académie des Sciences - Series III - Sciences de la Vie, 1999, 322(8), 687–693 Search PubMed.
- L. Yang, K. M. Wang, W. H. Tan, X. X. He, R. Jin, J. Li and H. M. Li, Atomic force microscopy study of different effects of natural and semisynthetic beta-lactam on the cell envelope of Escherichia coli, Anal. Chem., 2006, 78(20), 7341–7345 CrossRef CAS.
- P. C. Braga and D. Ricci, Atomic force microscopy: Application to investigation of Escherichia coli morphology before and after exposure to cefodizime, Antimicrob. Agents Chemother., 1998, 42(1), 18–22 CAS.
- S. M. Deupree and M. H. Schoenfisch, Morphological analysis of the antimicrobial action of nitric oxide on Gram-negative pathogens using atomic force microscopy, Acta Biomater., 2009, 5(5), 1405–1415 CrossRef CAS.
- A. Meincken, D. L. Holroyd and M. Rautenbach, Atomic force microscopy study of the effect of antimicrobial peptides on the cell envelope of Escherichia coli, Antimicrob. Agents Chemother., 2005, 49(10), 4085–4092 CrossRef CAS.
- A. da Silva and O. Teschke, Dynamics of the antimicrobial peptide PGLa action on Escherichia coli monitored by atomic force microscopy, World J. Microbiol. Biotechnol., 2005, 21(6–7), 1103–1110 CrossRef.
- J. E. Shaw, J. R. Alattia, J. E. Verity, G. G. Prive and C. M. Yip, Mechanisms of antimicrobial peptide action: Studies of indolicidin assembly at model membrane interfaces by in situ atomic force microscopy, J. Struct. Biol., 2006, 154(1), 42–58 CrossRef CAS.
- G. Rossetto, P. Bergese, P. Colombi, L. E. Depero, A. Giuliani, S. F. Nicoletto and G. Pirri, Atomic force microscopy evaluation of the effects of a novel antimicrobial multimeric peptide on Pseudomonas aeruginosa, Nanomed.: Nanotechnol., Biol. Med., 2007, 3(3), 198–207 CrossRef CAS.
- Y. Chen, D. Ciuparu, S. Lim, G. L. Haller and L. D. Pfefferle, The effect of the cobalt loading on the growth of single wall carbon nanotubes by CO disproportionation on Co-MCM-41 catalysts, Carbon, 2006, 44(1), 67–78 CrossRef CAS.
- Y. Chen, D. Ciuparu, S. Lim, Y. Yang, G. L. Haller and L. Pfefferle, Synthesis of Uniform Diameter Single-Wall Carbon Nanotubes in Co-MCM-41: Effects of the Catalyst Prereduction and Nanotube Growth Temperatures, J. Catal., 2004, 225(2), 453–465 CrossRef CAS.
- Y. Chen, D. Ciuparu, S. Lim, Y. Yang, G. L. Haller and L. Pfefferle, Synthesis of Uniform Diameter Single Wall Carbon Nanotubes in Co-MCM-41: Effects of CO Pressure and Reaction Time, J. Catal., 2004, 226(2), 351–362 CrossRef CAS.
- D. Ciuparu, Y. Chen, S. Lim, G. L. Haller and L. Pfefferle, Uniform-Diameter Single-Walled Carbon Nanotubes Catalytically Grown in Cobalt-Incorporated MCM-41, J. Phys. Chem. B, 2004, 108(2), 503–507 CrossRef CAS.
- L. Wei, B. Wang, Q. Wang, L. J. Li, Y. H. Yang and Y. Chen, Effect of Centrifugation on the Purity of Single-Walled Carbon Nanotubes from MCM-41 Containing Cobalt, J. Phys. Chem. C, 2008, 112(45), 17567–17575 CrossRef CAS.
- S. B. Velegol, S. Pardi, X. Li, D. Velegol and B. E. Logan, AFM imaging artifacts due to bacterial cell height and AFM tip geometry, Langmuir, 2003, 19(3), 851–857 CrossRef CAS.
- M. Arnoldi, M. Fritz, E. Bauerlein, M. Radmacher, E. Sackmann and A. Boulbitch, Bacterial turgor pressure can be measured by atomic force microscopy, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 2000, 62(1), 1034–1044 CrossRef.
- S. B. Velegol and B. E. Logan, Contributions of bacterial surface polymers, electrostatics, and cell elasticity to the shape of AFM force curves, Langmuir, 2002, 18(13), 5256–5262 CrossRef CAS.
- S. K. Das, A. R. Das and A. K. Guha, Adsorption Behavior of Mercury on Functionalized Aspergillus versicolor Mycelia: Atomic Force Microscopic Study, Langmuir, 2009, 25(1), 360–366 CrossRef CAS.
- Z. Y. Suo, R. Avci, M. Deliorman, X. H. Yang and D. W. Pascual, Bacteria Survive Multiple Puncturings of Their Cell Walls, Langmuir, 2009, 25(8), 4588–4594 CrossRef CAS.
- Y. J. Fan, H. J. Sheen, C. J. Hsu, C. P. Liu, S. M. Lin and K. C. Wu, A quantitative immunosensing technique based on the measurement of nanobeads' Brownian motion, Biosens. Bioelectron., 2009, 25(4), 688–694 CrossRef CAS.
- C. C. Perry, M. Weatherly, T. Beale and A. Randriamahefa, Atomic force microscopy study of the antimicrobial activity of aqueous garlic versus ampicillin against Escherichia coli and Staphylococcus aureus, J. Sci. Food Agric., 2009, 89(6), 958–964 CrossRef CAS.
- J. L. Hutter and J. Bechhoefer, Calibration of Atomic-Force Microscope Tips, Rev. Sci. Instrum., 1993, 64(7), 1868–1873 CrossRef CAS.
- C. Shiu; Z. Zhang; C. R. Thomas., A Comparison of the Mechanical Properties of Different Bacterial Species. In Applied Microbiology, Durieux, A.; Simon, J.-P., ed. Kluwer Academic Publishers: Netherlands, 2001; pp 155–162 Search PubMed.
- D. E. Bradley and C. Ann Dewar, Intracellular Changes in Cells of Escherichia coli Infected with a Filamentous Bacteriophage, J. Gen. Virol., 1967, 1(2), 179–188 Search PubMed.
- B. Wegenhart, L. Tan, M. Held, M. Kieliszewski and L. Chen, Aggregate structure of hydroxyproline-rich glycoprotein (HRGP) and HRGP assisted dispersion of carbon nanotubes, Nanoscale Res. Lett., 2006, 1(2), 154–159 Search PubMed.
- L. Y. Yan, Y. F. Poon, M. B. Chan-Park, Y. Chen and Q. Zhang, Individually dispersing single-walled carbon nanotubes with novel neutral pH water-soluble chitosan derivatives, J. Phys. Chem. C, 2008, 112(20), 7579–7587 CrossRef CAS.
- L. A. Girifalco, M. Hodak and R. S. Lee, Carbon nanotubes, buckyballs, ropes, and a universal graphitic potential, Phys. Rev. B: Condens. Matter Mater. Phys., 2000, 62(19), 13104–13110 CrossRef CAS.
- F. Wang, G. Dukovic, L. E. Brus and T. F. Heinz, The optical resonances in carbon nanotubes arise from excitons, Science, 2005, 308(5723), 838–841 CrossRef CAS.
- G. Li and J. X. Tang, Accumulation of Microswimmers near a Surface Mediated by Collision and Rotational Brownian Motion, Phys. Rev. Lett., 2009, 103(7), 078101-1–078101-4.
- A. Galano, Carbon nanotubes: promising agents against free radicals, Nanoscale, 2010, 2(3), 373–380 RSC.
- A. Martinez and A. Galano, Free Radical Scavenging Activity of Ultrashort Single-Walled Carbon Nanotubes with Different Structures through Electron Transfer Reactions, J. Phys. Chem. C, 2010, 114(18), 8184–8191 CrossRef CAS.
- I. Fenoglio, M. Tomatis, D. Lison, J. Muller, A. Fonseca, J. B. Nagy and B. Fubini, Reactivity of carbon nanotubes: Free radical generation or scavenging activity?, Free Radical Biol. Med., 2006, 40(7), 1227–1233 CrossRef CAS.
- D. F. Rodrigues and M. Elimelech, Toxic Effects of Single-Walled Carbon Nanotubes in the Development of E. coli Biofilm, Environ. Sci. Technol., 2010, 44(12), 4583–4589 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2010 |
