A renewable, chemoselective, and quantitative ligand density microarray for the study of biospecific interactions†‡
Abigail
Pulsipher
and
Muhammad N.
Yousaf
*
University of North Carolina at Chapel Hill, Department of Chemistry, Chapel Hill NC, USA. E-mail: mnyousaf@email.unc.edu; Fax: 91 9962 2388; Tel: 91 9843 3203
First published on 4th November 2010
Abstract
Novel renewable microarray technology has been developed to immobilize and release carbohydrates and proteins from self-assembled monolayers (SAMs) of electroactive quinone-terminated alkanethiolates on gold surfaces. This method may be applied to a variety of research fields for use in biosensor technology and the generation of renewable and tailored microarrays for biospecific cell-based assays.
Microarray-based technologies serve as high-throughput analytical tools for the evaluation of a variety of biomolecular interactions, providing a platform for rapid analysis and automation, and requiring low analyte and reagent volumes.1 Such technologies have incorporated the use of functionalized self-assembled monolayers (SAMs) of alkanethiolates on gold surfaces for the preparation of DNA, peptide, and carbohydrate microarrays to conduct studies in genomics, proteomics, and glycomics, respectively.2 SAMs can also be specifically tailored to present a number of derivatized ligands from the surface through covalent modification, affording a well-defined, biocompatible system.3 Current chemoselective conjugation strategies include Staudinger ligation,4 Click chemistry,5maleimide,6amide,7 Diels–Alder8 and oxime chemistry.9 One methodology in particular that uses electroactive, hydroquinone-terminated SAMs, has proven to be a powerful approach toward the immobilization and release of ligands.10 This strategy enables the quantification of the amount of attached ligand, as well as provides the ability to monitor the reaction progression in real-time and calculate kinetic rates.11
Carbohydrates represent the most abundant class of organic compounds found in living organisms and are known to mediate several biological processes, including neural development, signal transduction, viral invasion, and cancer metastasis.12 Microarray technologies have been designed to perform high-throughput screenings of various vital events including enzymatic glycosylation, sugar identification, and bacterial detection.13 A method based on a renewable and quantitative platform that can carry out similar analyses and also prove to be compatible with numerous biological systems, including carbohydrates, would greatly benefit a number of industries and tremendously cut down costs, materials, and time.
Herein, we present a renewable microarray technology to immobilize and release carbohydrates and proteins through control of electrochemical potential and pH. Monosaccharides were synthesized to incorporate oxyamine functionality for chemoselective conjugation to the ketone in electroactive quinone-terminated SAMs on gold. Sugar immobilization and release was monitored, and the SAM and ligand density were precisely controlled and quantified by cyclic voltammetry. In addition to the formation of mixed hydroquinone- and tetra(ethylene glycol)-terminated SAMs that provide resistance to nonspecific lectin adhesion, microarray technology allows for control over the orientation and spatial distribution of SAM molecules and ligands, criteria that are both essential for studying the biospecific interactions between molecules.3,8–11 Different microarray densities of quinone-terminated SAMs were generated, and a number of oxyamine-containing sugars were immobilized. The resultant oxime linkages were characterized by electrochemistry, X-ray photoelectron spectroscopy, and contact angle, and lectin-carbohydrate binding interactions were observed by fluorescence microscopy. This method may be used in conjunction with the tailoring of other biomolecules such as DNA, peptides, and phospholipids for studies in genomics, transcriptomics, and proteomics.
The general strategy for developing a chemoselective, renewable, and quantitative, ligand density microarray for biological analysis is described in Fig. 1. We have chosen to use different ratios of hydroquinone-(H2Q, 5, supporting information, Scheme S2) and tetra(ethylene glycol)-(EG4SH, 4, supporting information, Scheme S2) terminated alkanethiolates to construct our microarrays. Alkanethiolate solutions are transferred to a bare gold substrate using an automated microarray method, and microspots of SAMs are rapidly formed. The remaining unpatterned surface is then backfilled with EG4SH, oxidized to reveal Q, and biological ligands are then reversibly conjugated to the microarray. Depending on the particular biomolecule presented from the substrate, the corresponding bioassay can be performed and analyzed. The original microarray can then be regenerated for a subsequent round of immobilization, analysis, and release. The surface chemistry and renewable nature of this microarray technology is illustrated in and characterized by CV (Fig. 2). Mixed SAMs of H2Q/EG4SH are electrochemically oxidized (1 M HClO4, pH = 0, scan rate of 100 mV/s) to Q-terminated groups which can then chemoselectively react with oxyamine-functionalized compounds. In this particular study, our ligands of interest include galactose-, glucose-, and mannose-oxyamine (Gal-ONH2, 1; Glc-ONH2, 2; and Man-ONH2, 3, respectively, supporting information Scheme S1 and S2).
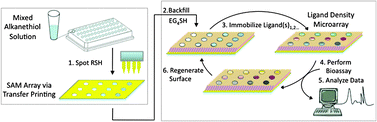 | ||
| Fig. 1 Strategy to develop a renewable and quantitative, ligand density microarray for biological analysis. (1) Different mixtures of alkanethiolates are transferred from a 96-well microplate to a bare gold substrate via automated array technology, and microspots of SAMs are formed. (2) The substrate is backfilled with EG4SH. (3) Biological ligands are then immobilized to the array. (4) The corresponding bioassay is performed, (5) analyzed, and (6) the original microarray is regenerated for a subsequent cycle of ligand immobilization and analysis. | ||
![Electrochemical characterization. (A) This CV shows distinctive redox peaks (blue) of a H2Q/Q-terminated SAM at [Q] = 630 mV and [H2Q] = −24 mV. Once an oxyamine-containing carbohydrate is immobilized, (B) the redox peaks (green) shift to [Qox] = 610 mV and [H2Qox] = 283 mV, indicating the presence of an oxime (ox) bond. (C) The bound carbohydrate (green) can be released by electrochemical reduction (20 scans), and the original H2Q/Q SAM (blue, 60 scans) can be regenerated. The two blue traces in A and C show the same peaks, which correspond to H2Q/Q before carbohydrate immobilization and after its release.](/image/article/2011/CC/c0cc01509a/c0cc01509a-f2.gif) | ||
| Fig. 2 Electrochemical characterization. (A) This CV shows distinctive redox peaks (blue) of a H2Q/Q-terminated SAM at [Q] = 630 mV and [H2Q] = −24 mV. Once an oxyamine-containing carbohydrate is immobilized, (B) the redox peaks (green) shift to [Qox] = 610 mV and [H2Qox] = 283 mV, indicating the presence of an oxime (ox) bond. (C) The bound carbohydrate (green) can be released by electrochemical reduction (20 scans), and the original H2Q/Q SAM (blue, 60 scans) can be regenerated. The two blue traces in A and C show the same peaks, which correspond to H2Q/Q before carbohydrate immobilization and after its release. | ||
After the monosaccharide is covalently bound to the surface through an oxime linkage, its corresponding lectin, or sugar-binding protein, is added. The protein then adheres to and interacts with its recognition signal and is able to be characterized by fluorescence microscopy. To regenerate the surface for another subsequent cycle of ligand immobilization, sugars and lectins are released by applying a constant potential under reducing conditions (PBS, pH = 7, 60 cyclic scans, 15 min). The result consists of a reduced alcohol and renewed H2Q-terminated SAM; this cycle may be performed again. As previously mentioned, CV was employed to characterize ligand immobilization and release from Q-terminated SAMs on gold (Fig. 2). Distinctive H2Q/Q redox peaks were observed at −24 and 630 mV, respectively (blue trace, Fig. 2A). Following four hours of sugar–oxyamine conjugation (90 mM in MeOH), peaks were completely shifted to 283 and 610 mV (green trace, Fig. 2B), indicating full oxime (H2Qox/Qox) bond formation. It is also important to note that the sugar-Q reaction was also monitored at various time points to determine the percentage of ligand bound (supporting information, Fig. S2), as well as the approximate rate of product conversion (data not shown). To release the ligand and regenerate the original H2Q/Q-terminated SAM (blue, Fig. 2C), potential was applied for 60 cyclic scans, or 15 min, under reducing conditions (PBS, pH = 7). Approximately one-third of the carbohydrate ligand was released following 20 scans, as shown in the green trace in Fig. 2C, with the presence of H2Q/Q production.
Additional surface characterization was conducted to confirm that consistent SAMs of H2Q/Q were being formed and that oxyamine-functionalized carbohydrates were being immobilized after reaction. The static contact angle of water (10 μL) was measured on bare gold substrates, as well as on surfaces with SAMs of Q-, H2Q-, and glucose-presenting groups (supporting information, Fig. S1B). There was a significant decrease in the contact angle, from 64.19° to 33.32°, for unreacted Q-terminated SAMs and Q-containing SAMs reacted with glc-ONH2 (90 mM in MeOH, 4 h), respectively. The large difference in contact angle is accounted for in the increase of wettability and cohesive forces between the water and hydrophilic hydroxyl groups, indicating there was successful attachment of carbohydrate–oxyamine ligands. X-ray photoelectron spectroscopy (XPS) was also performed to confirm oxyamine conjugation to substrates bearing Q-terminated SAMs. The 1s nitrogen peak observed at 398 eV, corresponds to the presence of glc-ONH2 immobilized to the surface (green trace, supporting information, Fig. S1A), while there is no nitrogen present on substrates with Q-terminated SAMs, as shown by the black trace. SAM and ligand density were also quantified by CV (supporting information, Fig. S2).
Although our renewable and quantitative ligand microarray strategy is designed to be extended toward a number of biological systems, we chose to represent the biospecific interactions between carbohydrate ligands and lectins. Fig. 3 displays three cycles of ligand immobilization and release. Automated microarray technology was utilized to generate a 24 x 24 microarray of 100 μm in diameter spot-arrays of H2Q/Eg4SH-containing alkanethiolates (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1 mM in EtOH) on a bare gold substrate. Glc- and gal-ONH2 (10 μL, 90 mM in MeOH) were transferred to the microarray of SAMs at random using the same program. The surface was then backfilled with EG4SH (1 mM in EtOH, 16 h), and a mixture containing fluorescently-conjugated Concanavalin A (ConA, 1 mg/mL in H2O, 2 h) and Peanut Agglutinin (PNA, 1 mg/mL in H2O, 2 h) was allowed to bind. ConA and PNA are lectins, known to specifically recognize and bind to glucose and galactose residues, respectively. Carbohydrate ligands and lectins were then released from the substrate after 60 cyclic scans of electric potential (PBS, pH = 7, 15 min), renewing the original mixed H2Q/EG4SH-terminated SAM microarray, which was also determined by electrochemistry. Glc- and gal-ONH2 were again randomly delivered to the SAM microarray, followed by reaction with ConA and PNA for two more cycles. Carbohydrate–protein binding was then analyzed by fluorescence microscopy. As a control, lectins were added to H2Q-presenting microarrays with no immobilized sugars, and minimal fluorescence was detected. To assure selectivity of ConA for mannose and glucose and PNA for galactose residues, carbohydrates were randomly tethered to the surface and a mixture containing both proteins was allowed to react. Images displayed color separation, with little to no overlap, indicating that the lectins were able to recognize and successfully interact with their respective adhesive signal.
10, 1 mM in EtOH) on a bare gold substrate. Glc- and gal-ONH2 (10 μL, 90 mM in MeOH) were transferred to the microarray of SAMs at random using the same program. The surface was then backfilled with EG4SH (1 mM in EtOH, 16 h), and a mixture containing fluorescently-conjugated Concanavalin A (ConA, 1 mg/mL in H2O, 2 h) and Peanut Agglutinin (PNA, 1 mg/mL in H2O, 2 h) was allowed to bind. ConA and PNA are lectins, known to specifically recognize and bind to glucose and galactose residues, respectively. Carbohydrate ligands and lectins were then released from the substrate after 60 cyclic scans of electric potential (PBS, pH = 7, 15 min), renewing the original mixed H2Q/EG4SH-terminated SAM microarray, which was also determined by electrochemistry. Glc- and gal-ONH2 were again randomly delivered to the SAM microarray, followed by reaction with ConA and PNA for two more cycles. Carbohydrate–protein binding was then analyzed by fluorescence microscopy. As a control, lectins were added to H2Q-presenting microarrays with no immobilized sugars, and minimal fluorescence was detected. To assure selectivity of ConA for mannose and glucose and PNA for galactose residues, carbohydrates were randomly tethered to the surface and a mixture containing both proteins was allowed to react. Images displayed color separation, with little to no overlap, indicating that the lectins were able to recognize and successfully interact with their respective adhesive signal.
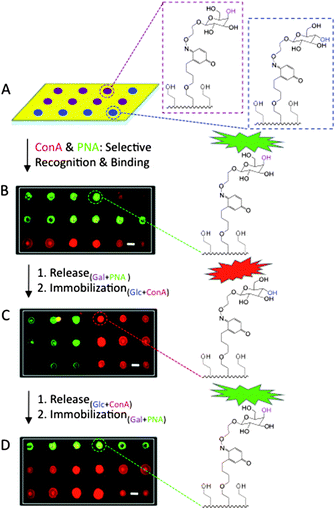 | ||
| Fig. 3 Fluorescent micrographs displaying a renewable microarray for carbohydrate immobilization and subsequent lectin recognition and adhesion. (A) A microarray of H2Q/EG4SH SAMs were generated using automated technology, and gal- (purple) and glc- (blue) ONH2 were immobilized, followed by subsequent adhesion of PNA (green) and ConA (red). Gal monomers were spotted and bound to where PNA (green) appears, and Glc, where ConA (red) appears. (B) After electrochemical release of all ligands and regeneration of the original surface, gal and glc were again immobilized. (C) PNA and ConA were then added for specific carbohydrate recognition. The ligands were again released to renew the substrate, and (D) the immobilization process was again performed. The scale bars represent 100 μm. | ||
In summary, we have developed a renewable, chemoselective, and quantitative ligand density microarray for the rapid analysis of biological interactions. Automated technology was used to transfer different mixtures of H2Q- and EG4SH-terminated alkanethiols to a bare gold substrate, resulting in the tailoring of SAM microspots (100 μm diameter). Following electrochemical oxidation of H2Q to the corresponding Q, oxyamine compounds were reacted and immobilized to the microarray. A number of monosaccharides (mannose, glucose, and galactose) were synthesized to incorporate oxyamine functionality and tethered to the surface. The oxime linkages were confirmed and characterized by CV, XPS, and contact angle. By applying constant potential (15 min) under reducing conditions (PBS, pH = 7), sugar ligands were released, renewing the original H2Q-terminated array for another round of immobilization and release. The extent of the molecule reaction, as well as its release and amount bound was monitored and quantified by CV. Fluorescently-conjugated, carbohydrate-binding proteins ConA and PNA were added to the microarray following man-, glc-, and gal-ONH2 reaction, and substrates were visualized by fluorescence microscopy. The images displayed distinctly colored spots, corresponding to specific signal (sugar) recognition, indicating successful observation of ligand–receptor interactions on our platform. This method may be applied to a variety of scientific fields for use in biosensor technology and the generation of renewable and tailored microarrays for biospecific ligand–receptor interaction assays.
Notes and references
- N. Laurent, R. Haddoub and S. L. Flitsch, Trends Biotechnol., 2008, 26, 328 CrossRef CAS; G. Ramsey, Nat. Biotechnol., 1998, 16, 40 CrossRef CAS; C. M. Perou, Nature, 2000, 406, 747 CrossRef CAS; N. Laurent, R. Haddoub, J. Voglmeir, S. C. C. Wong, S. J. Gaskell and S. L. Flitsch, ChemBioChem, 2008, 9, 2592 CrossRef CAS.
- Z.-L. Zhi, N. Laurent and J. E. Turnbull, ChemBioChem, 2008, 9, 1568 CrossRef CAS; S. Park, M.-R. Lee and I. Shin, J. Am. Chem. Soc., 2004, 126, 4812 CrossRef CAS; B. T. Houseman, E. S. Gawalt and M. Mrksich, Langmuir, 2003, 19, 1522 CrossRef CAS; D.-H. Min, J. Su and M. Mrksich, Angew. Chem., Int. Ed., 2004, 43, 5973 CrossRef CAS.
- P. Harder, M. Grunze, G. M. Whitesides and P. E. Laibinis, J. Phys. Chem. B, 1998, 102, 426 CrossRef; H. J. Kreuzer, R. L. C. Wang and M. Grunze, J. Am. Chem. Soc., 2003, 125, 8384 CrossRef CAS; E. Ostuni, R. C. Chapman, M. N. Liang, G. Meluleni, G. Pier, D. E. Ingber and G. M. Whitesides, Langmuir, 2001, 17, 6336 CrossRef CAS.
- A. Watzke, M. Kohn, R. Wacker, S. L. Schroder and H. Waldmann, Angew. Chem., Int. Ed., 2006, 45, 1408 CrossRef CAS; M. Kohn, R. Wacker, C. Peters, H. Schroder, L. Soulere, R. Breinbauer, C. M. Niemeyer and H. Waldmann, Angew. Chem., Int. Ed., 2003, 42, 5830 CrossRef.
- Y. Zhang and P. G. Wang, Anal. Chem., 2006, 78, 2001 CrossRef CAS.
- B. T. Houseman and M. Mrksich, Langmuir, 2003, 19, 1522 CrossRef CAS.
- J. Lahiri, L. Isaacs and G. M. Whitesides, Anal. Chem., 1999, 71, 777 CrossRef CAS.
- M. N. Yousaf, B. T. Houseman and M. Mrksich, Angew. Chem., 2001, 113, 1127 CrossRef; M. N. Yousaf, B. T. Houseman and M. Mrksich, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 5992 CrossRef CAS; M. N. Yousaf and M. Mrksich, J. Am. Chem. Soc., 1999, 121, 4286 CrossRef CAS.
- E. W. L. Chan and M. N. Yousaf, ChemPhysChem, 2007, 8, 1469 CrossRef CAS; S. Park and M. N. Yousaf, Langmuir, 2008, 24, 6201 CrossRef CAS; E. W. L. Chan and M. N. Yousaf, J. Am. Chem. Soc., 2006, 128, 15542 CrossRef CAS; N. P. Westcott and M. N. Yousaf, Langmuir, 2008, 24, 2261 CrossRef CAS.
- E. W. L. Chan, S. Park and M. N. Yousaf, Angew. Chem., Int. Ed., 2008, 47, 6267 CrossRef CAS.
- E.-J. Lee and M. N. Yousaf, ChemBioChem, 2009, 10, 1648 CrossRef CAS.
- N. E. Zachara and G. W. Hart, Chem. Rev., 2002, 102, 431 CrossRef CAS; J. Roth, Chem. Rev., 2002, 102, 285 CrossRef CAS; K. Sugahara, T. Mikami, T. Uyama, S. Mizuguchi, K. Nomura and H. Kitagawa, Curr. Opin. Struct. Biol., 2003, 13, 612 CrossRef CAS; C. I. Gama and L. C. Hsieh-Wilson, Curr. Opin. Struct. Biol., 2005, 9, 609 CAS; S. A. Whelan and G. W. Hart, Circ. Res., 2003, 93, 1047 CrossRef CAS; C. R. Bertozzi and L. L. Kiessling, Science, 2001, 291, 2357 CrossRef CAS.
- B. T. Houseman and M. Mrksich, Chem. Biol., 2002, 9, 443 CrossRef CAS; D. M. Hatch, A. A. Weiss, R. R. Kale and S. S. Iyer, ChemBioChem, 2008, 9, 2433 CrossRef CAS; K. G. I. Nilsson and C.-F. Mandenius, Nature, 1994, 12, 1376 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, carbohydrate synthesis, XPS, and contact angle data. See DOI: 10.1039/c0cc01509a |
| ‡ This article is part of the ‘Emerging Investigators’ themed issue for ChemComm. |
| This journal is © The Royal Society of Chemistry 2011 |
