The pattern recognition molecule deleted in malignant brain tumors 1 (DMBT1) and synthetic mimics inhibit liposomal nucleic acid delivery†
Pernille
Lund Hansen‡
ab,
Stephanie
Blaich‡
c,
Caroline
End
d,
Steffen
Schmidt
ab,
Jesper B.
Moeller
e,
Uffe
Holmskov
e and
Jan
Mollenhauer
*ab
aLundbeckfonden Center of Excellence NanoCAN, University of Southern Denmark, 5000 Odense C, Denmark. E-mail: jmollenhauer@health.sdu.dk
bMolecular Oncology Group, University of Southern Denmark, 5000 Odense C, Denmark
cUnit Cancer Genome Research, Division of Molecular Genetics, Deutsches Krebsforschungszentrum, 69120 Heidelberg, Germany
dDivision of Molecular Genome Analysis, Deutsches Krebsforschungszentrum, 69120 Heidelberg, Germany
eCardiovascular and Renal Research, University of Southern Denmark, 5000 Odense C, Denmark
First published on 7th September 2010
Abstract
Liposomal nucleic acid delivery is a preferred option for therapeutic settings. The cellular pattern recognition molecule DMBT1, secreted at high levels in various diseases, and synthetic mimics efficiently inhibit liposomal nucleic acid delivery to human cells. These findings may have relevance for therapeutic nucleic acid delivery strategies.
Liposomal nucleic acid delivery (LNAD) is a therapeutic strategy under intense investigation. It offers options for supplementation as well as for inhibition strategies viagene delivery or RNA interference, respectively. Safety concerns are substantially less than for viral nucleic acid delivery strategies. LNAD utilizes cationic lipids, which interact with the negatively charged phosphate backbone of the nucleic acids. The nucleic acid encapsulation modulates pharmacological properties in a favorable manner and enables the entry of nucleic acids into the target cells.1,2
Pattern recognition is an evolutionary ancient principle, playing a role in the defense against pathogens, regeneration, and differentiation. Deleted in Malignant Brain Tumors 1 (DMBT1, also known as gp-340) is a secreted human pattern recognition molecule with functions in bacterial and viral defense as well as in processes related to epithelial and stem cell differentiation.3,4 Dysfunction of DMBT1 has been proposed to play a role in cancer, infectious and inflammatory diseases.4–7 The various functions may at least in part rely on the recognition of poly-sulfated and -phosphated structures by a peptide motif (NH3-GRVEVLYRGSW-COOH) present up to thirteen times in the monomeric form, and probably more than about 200-times in the oligomeric form of DMBT1.8,9 Projected to the 3D-structure of its closest relatives, the human Mac2-binding protein and CD5, the motif would be predicted to provide sites for hydrogen-bonding or electrostatic interactions along a groove within each of the 13 amino-terminal scavenger receptor cysteine-rich (SRCR) domains of DMBT1 (Fig. 1).
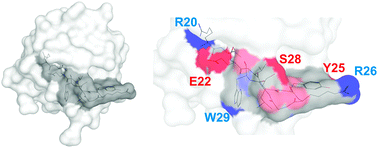 | ||
| Fig. 1 Modeling of the binding site. Left: model of one SRCR domain, i.e.SRCR3, of DMBT1 with the pattern recognition motif in dark gray. Right: exposed nitrogen and oxygen atoms of the amino acid side chains are in blue and red and of the peptide backbone are in light blue and light red. Amino acids are numbered according to their position in the domain. Modeling was performed using the automated protein structure homology modeling server SWISS-MODEL 8.05.9,10 | ||
Sulfated carbohydrates as well as DNA are among the potential ligands of DMBT1.11 Because DMBT1 is frequently upregulated in various diseases, including inflammatory bowel disease and subsets of tumors this actually raises the question as to whether the presence of the protein could affect the efficacy of LNAD, which in turn might have an impact for the corresponding therapeutic settings.
To initially test this, we first pre-incubated plasmid-DNA (pEGFP-N1, coding for a green fluorescent protein) with either human recombinant DMBT1 (hrDMBT1) or a synthetic peptide that includes the pattern recognition motif (SRCRP2 + 2N; NH3-RCQGRVEVLYRGSWGT-COOH) at final concentrations of 400 nM and 200 μM, respectively. As negative controls, we added equivalent amounts of PBS (solvent of hrDMBT1) or 10% DMSO solution (solvent of the synthetic peptides). For SRCRP2 + 2N, we further used the synthetic peptide SRCRP6 (NH3-PHNGWLSHNC-COOH) as a negative control to rule out that excess addition of peptides exerts unspecific effects. SRCRP6 comprises a distinct amino acid sequence occurring in each of the SRCR domains of DMBT1.8 It does not contain the pattern recognition motif, and does not show binding activity for SRCRP2-ligands, such as bacterial surface structures.8 Compared to the controls, both hrDMBT1 and the mimicking peptide SRCRP2 + 2N almost completely inhibited LNAD to human SW403 colon cancer cells, which was not affected by the addition of Ca2+ ions or depletion of Ca2+ ions by 10 mM EDTA (Fig. 2 left panel).
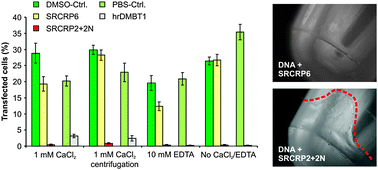 | ||
Fig. 2 Inhibition of LNAD by hrDMBT1 and mimics. Left panel: 0.5 μg plasmid DNA (pEGFP-N1) was pre-incubated for 30 min at 37 °C in a total volume of 50 μl with the synthetic peptides SRCRP2 + 2N (20 μg) or SRCRP6 (negative control; 20 μg), a corresponding volume of 10% dimethylsulfoxide in H2O (solvent of the peptides; DMSO-Ctrl.; diluted to match the final DMSO concentrations of the peptide dilutions), with hrDMBT1 (5 μg) in phosphate-buffered saline (PBS, solvent of the protein) or with a corresponding volume of PBS alone (PBS-Ctrl.). Analyses were performed in the presence or absence of 1 mM CaCl2 or 10 mM ethylenediaminetetraacetic acid (EDTA), respectively. Centrifugation depicts an experiment, in which pre-incubated plasmid DNA was centrifuged (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C for 15 min) and supernatants were used for consecutive transfection. Centrifugation resulted in precipitates, when incubation was performed with SRCRP2 + 2N (right panel). After the addition of Lipofectamine 2000, the cells (SW403) were transfected and the percentage of green fluorescent cells was determined 24 h post transfection. Data are from at least three triplicate experiments. Error bars are SEM. 000 rpm, 4 °C for 15 min) and supernatants were used for consecutive transfection. Centrifugation resulted in precipitates, when incubation was performed with SRCRP2 + 2N (right panel). After the addition of Lipofectamine 2000, the cells (SW403) were transfected and the percentage of green fluorescent cells was determined 24 h post transfection. Data are from at least three triplicate experiments. Error bars are SEM. | ||
Upon pre-incubation with plasmid DNA and subsequent centrifugation, SRCRP2 + 2N but not SRCRP6 or hrDMBT1 gave rise to precipitates (Fig. 2, right panel and not shown). Using the supernatants obtained after centrifugation likewise resulted in a nearly complete inhibition of LNAD (Fig. 2 left panel). We hypothesize that the observed pellets are bona fide SRCRP2 + 2N-DNA precipitates, which would support a physical interaction.
We next determined the active inhibitory concentrations of hrDMBT1 and of the mimicking SRCRP2-2N peptide using mouse Colo26 cells (colon epithelial origin) in addition to human SW403 cells. A concentration-dependent inhibition of LNAD was confirmed for both SRCRP2 + 2N and hrDMBT1 in human and mouse epithelial cells (Fig. 3). The inhibitory effect of 2.5 μg (∼200 nM final concentration) hrDMBT1 corresponded to that of 10 μg (∼100 μM final concentration) SRCRP2 + 2N. Considering that monomeric DMBT1 contains 13 of the peptide motifs, the protein is about 38.5-fold more effective in LNAD inhibition than the mimicking peptide under these experimental conditions.
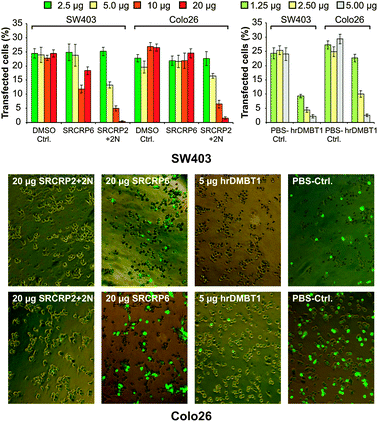 | ||
| Fig. 3 Concentration-dependent inhibition of LNAD to human and mouse cells. Human SW403 and mouse Colo26 cells were treated as described in Fig. 2 (without CaCl2 or EDTA) using dilution series of the peptide SRCRP2 + 2N and of hrDMBT1. Upper panels: transfection efficacy (fluorescent cells) in percent. Data are from three triplicate experiments. Error bars are SEM. Lower panel: exemplary images taken from the highest concentrations and respective controls (magnification: 200-fold). | ||
The pre-incubation of plasmid DNA creates favorable conditions for adsorption, which might differ from the situation encountered when DMBT1 is secreted by cells and intact DNA-cationic lipid assemblies are used. We thus constructed HEK293 cells with tetracycline-inducible expression of DMBT1 analogous to the procedure described earlier for the construction of DMBT1-expressing CHO cells.12 Inducible expression of DMBT1 and its secretion were confirmed by Western blotting of cell culture supernatants using the monoclonal antibody anti-DMBT1h12 (Fig. 4 left panel). We next seeded equal cell numbers (150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in a 6-well plate). After 24 h, DMBT1 expression was induced with 1 μg ml−1tetracycline for 48 h to allow for DMBT1 expression and accumulation in the cell culture medium (supplemented with 10% FCS). DMBT1 expressing and DMBT1-negative control cells were treated identically. Afterwards we transfected the cells with a yellow fluorescent protein (YFP) coding expression plasmid (pEYFP-N1) mixed with 1 μl Lipofectamine 2000 and scored the percentage of YFP-positive cells after 24 h. DMBT1 expressing HEK293 cells displayed an about 30% decreased efficacy of LNAD compared to DMBT1-negative cells (P = 0.0248; one-tailed Student's t-test; Fig. 4, right panel).
000 cells per well in a 6-well plate). After 24 h, DMBT1 expression was induced with 1 μg ml−1tetracycline for 48 h to allow for DMBT1 expression and accumulation in the cell culture medium (supplemented with 10% FCS). DMBT1 expressing and DMBT1-negative control cells were treated identically. Afterwards we transfected the cells with a yellow fluorescent protein (YFP) coding expression plasmid (pEYFP-N1) mixed with 1 μl Lipofectamine 2000 and scored the percentage of YFP-positive cells after 24 h. DMBT1 expressing HEK293 cells displayed an about 30% decreased efficacy of LNAD compared to DMBT1-negative cells (P = 0.0248; one-tailed Student's t-test; Fig. 4, right panel).
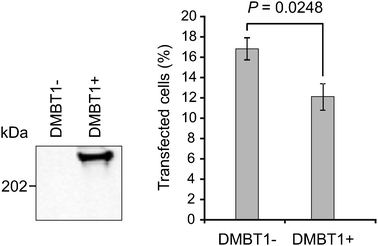 | ||
| Fig. 4 Efficacy of LNAD is decreased in DMBT1-positive cells. Left panel: Western blotting of cell culture supernatants from HEK293 cells without and with stable insertion of a DMBT1 expression plasmid after 48 h induction with 1 μg ml−1tetracycline. Right panel: percentage of fluorescent cells (mean of three experiments). Error bars are SEM. Statistical test: one-tailed Student's t-test. | ||
The data suggest that the cellular pattern recognition molecule DMBT1 and the synthetic peptide SRCRP2 + 2N, previously shown to mimic its binding activity,8 decrease the efficacy of LNAD to human and mouse cellsin vitro. Opposed to interactions with bacteria, where binding and aggregation viaDMBT1 and peptide mimics is enhanced by divalent Ca2+ ions,8 inhibition of LNAD seems not to be substantially influenced by Ca2+ ions.
Pre-incubation of DNA with synthetic mimics represents a highly reductionistic approach. However, due to the observation that SRCRP2 + 2N forms precipitates after centrifugation and that the supernatant consecutively lacks efficient LNAD, we speculate that a physical interaction between the peptide and the DNA takes place, resulting in co-precipitation and depletion of the DNA. Such precipitates were not observed for hrDMBT1, which could be due to the orientation of the binding sites in the protein not allowing for a high degree of cross-linking. On the other hand, it seemingly allows for a more effective binding of the protein, which is 38.5-fold increased compared to SRCRP2 + 2N under these experimental conditions.
The studies performed with HEK293 cells might more closely resemble the situation within a diseased tissue, because no favorable pre-adsorption was performed, pre-assembled DNA-cationic lipid complexes were used, and serum proteins were present. These data suggested that still a substantial reduction of LNAD efficacy of about 30% takes place in the presence of DMBT1. Still, the conditions in an intact tissue might differ with the potential to either decrease or increase the observed effects. The local concentration of DMBT1 fixed in the mucus or in the extracellular matrix could be substantially higher than in the cell culture supernatants, which would be expected to increase its inhibitory effects. On the other hand, DMBT1 might be bound to natural ligands, which could render it inactive in inhibiting LNAD. Having said this, sulfated glycosaminoglycans (GAGs) could represent natural ligands for DMBT1 in the intact tissue. DMBT1 shows binding activity for heparan sulfate and chondroitin sulfate.11 It has, however, also been proposed that net positively charged cationic lipid–DNA complexes might utilize sulfated GAGs for cellular entry.1,2 Thus, alternatively or in addition to LNAD inhibition via DNA-interaction, DMBT1 could interfere with docking of the cationic lipid–DNA complexes to cellular binding sites. This hypothetical mechanism warrants further studies. It could also be of importance for the functions of DMBT1 in viral defense, because viruses may utilize sulfated GAGs for host cell infection.
Our data indicate that high DMBT1 levels, as observed in various disease conditions, including chronic inflammatory disease (Crohn's disease, ulcerative colitis) and some cancers, could have an impact for LNAD efficacy. Should the inhibitory mechanism be based on an interaction of DMBT1 with sulfated GAGs, this could also be relevant for the liposomal delivery of drugs other than nucleic acids. Providing that studies of drug response in relation to DMBT1 levels confirm clinical relevance, the determination of DMBT1 levels in a diseased tissue in conjunction with genetic analyses of the number of SRCR exons within the gene could represent a possible means to correspondingly adjust the dosage for liposomal drug delivery.
This work was supported by Lundbeckfonden (Grant for Center of Excellence in Nanomedicine NanoCAN), the Ingeborg og Leo Dannins Fondens Legat 2009, the Novo Nordisk Fonden, and the Kong Christian Den Tiendes Fond.
Notes and references
- S. Simões, A. Filipe, H. Faneca, M. Mano, N. Penacho, N. Düzgünes and M. P. de Lima, Expert Opin. Drug Delivery, 2005, 2, 237 Search PubMed.
- E. Ashihara, E. Kawata and T. Maekawa, Curr. Drug Targets, 2010, 11, 345 Search PubMed.
- J. Mollenhauer, S. Wiemann, W. Scheurlen, B. Korn, Y. Hayashi, K. K. Wilgenbus, A. von Deimling and A. Poustka, Nat. Genet., 1997, 17, 32 CrossRef CAS.
- J. Mollenhauer, C. End, M. Renner, S. Lyer and A. Poustka, Inmunologia, 2007, 26, 193 Search PubMed.
- E. Stoddard, H. Ni, G. Cannon, C. Zhou, N. Kallenbach, D. Malamud and D. Weissman, J. Virol., 2009, 83, 8596 CrossRef CAS.
- M. Renner, G. Bergmann, I. Krebs, C. End, S. Lyer, F. Hilberg, B. Helmke, N. Gassler, F. Autschbach, F. Bikker, O. Strobel-Freidekind, S. Gronert-Sum, A. Benner, S. Blaich, R. Wittig, M. Hudler, A. J. Ligtenberg, J. Madsen, U. Holmskov, V. Annese, A. Latiano, P. Schirmacher, A. V. Nieuw Amerongen, M. D'Amato, P. Kioschis, M. Hafner, A. Poustka and J. Mollenhauer, Gastroenterology, 2007, 133, 1499 CrossRef CAS.
- S. Tchatchou, A. Riedel, S. Lyer, J. Schmutzhard, O. Strobel-Freidekind, S. Gronert-Sum, C. Mietag, M. D'Amato, B. Schlehe, K. Hemminki, C. Sutter, N. Ditsch, A. Blackburn, L. Zhai Hill, D. J. Jerry, P. Bugert, B. H. F. Weber, D. Niederacher, N. Arnold, R. Varon-Mateeva, B. Wappenschmidt, R. K. Schmutzler, C. Engel, A. Meindl, C. R. Bartram, J. Mollenhauer and B. Burwinkel, Hum. Mutat., 2010, 31, 60 CrossRef CAS.
- F. J. Bikker, A. J. M. Ligtenberg, K. Nazmi, E. C. I. Veerman, W. van't Hof, J. G. M. Bolscher, A. Poustka, A. V. Nieuw Amerongen and J. Mollenhauer, J. Biol. Chem., 2002, 277, 32109 CrossRef CAS.
- K. Arnold, L. Bordoli, J. Kopp and T. Schwede, Bioinformatics, 2006, 22, 195 CAS.
- F. Kiefer, K. Arnold, M. Künzli, L. Bordoli and T. Schwede, Nucleic Acids Res., 2009, 37, D387 CrossRef CAS.
- C. End, F. J. Bikker, M. Renner, G. Bergmann, S. Lyer, S. Blaich, M. Hudler, B. Helmke, N. Gassler, F. Autschbach, A. J. Ligtenberg, A. Benner, U. Holmskov, P. Schirmacher, A. V. Nieuw Amerongen, P. Rosenstiel, C. Sina, A. Franke, M. Hafner, P. Kioschis, S. Schreiber, A. Poustka and J. Mollenhauer, Eur. J. Immunol., 2009, 39, 833 CrossRef CAS.
- C. End, S. Lyer, M. Renner, C. Stahl, J. Ditzer, A. Holloschi, H. M. Kuhn, H. T. Flammann, A. Poustka, M. Hafner, J. Mollenhauer and P. Kioschis, Protein Expression Purif., 2005, 41, 275 CrossRef CAS.
Footnotes |
| † This article is part of the ‘Emerging Investigators’ themed issue for ChemComm. |
| ‡ The authors contributed equally and should both be considered as first authors. |
| This journal is © The Royal Society of Chemistry 2011 |
