Programmable microfluidic patterning of protein gradients on hydrogels†
Simone
Allazetta
,
Steffen
Cosson
and
Matthias P.
Lutolf
*
Laboratory of Stem Cell Bioengineering, Institute of Bioengineering, School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne Station 15 Bldg. AI 1109, CH-1015 Lausanne, Switzerland. E-mail: matthias.lutolf@epfl.ch; Fax: +41 21 6939665; Tel: +41 21 6931876
First published on 8th September 2010
Abstract
Computer-controlled hydrodynamic flow focusing was utilized to generate tethered protein gradients of any user-defined shape on the surface of soft synthetic hydrogels.
Graded signalling cues govern cell behaviour in many essential biological processes.1Cells of a developing embryo, for example, interpret the concentration-dependent signalling information of proteins termed ‘morphogens’ to generate the precise patterns of specialized cell types that ultimately make up a functional tissue. The importance of biomolecule gradients in biology has spurred the intense development and application of in vitro model systems conceived to replicate the temporally and spatially controlled display of signals just like in an in vivo context. Traditionally, the focus has been on reproducing gradients of soluble signals such as chemokines by using, for example, macroscopic chambers or micropipette-based approaches.2 However, the lack of precise gradient control in these set-ups has stimulated the development of microfluidic approaches to generate gradients of tiny quantities of soluble or surface-adsorbed biomolecules with excellent spatial resolution and even temporally changing characteristics.3 Microfluidic gradients thus allow to approach the spatiotemporal complexity of graded signalling microenvironments found in vivo, but the type of substrate on which they are commonly generated is far from physiological. Since it is increasingly appreciated that cells are sensitive to the biophysical characteristics of their substrate,4 the mere use of rigid (e.g. glass or plastic) cell culture dishes could compromise the validity of an experiment. Therefore, there is an unmet need for in vitro gradient model systems that are built on materials mimicking the biophysical characteristics of the natural, soft and hydrated extracellular milieu.5–7
To address this technology gap, we have reported a microfluidic method to pattern protein gradients on biomimetic poly(ethylene glycol) (PEG)-based hydrogels.8 This system proved useful to form tethered gradients of one or more (overlapping) proteins on gel surfaces, but gradient shapes were relatively poorly controlled and limited to simple linear gradients, while in vivo gradients can often have very complex shapes.1 Consequently, here we present a scheme to form, in an automated fashion, gel-immobilized protein patterns with virtually any type of shape.
We sought to explore the versatile concept of ‘hydrodynamic flow focusing’9,10 to generate graded protein patterns on hydrogels. A microfluidic poly(dimethylsiloxane) (PDMS) chip was fabricated consisting of a channel system with three inlets, each of which was independently controllable by a pump (Fig. 1a). Software-controlled adjustment of the flow rates of the individual liquid streams thus should allow to dynamically control the position and width of the protein stream.10
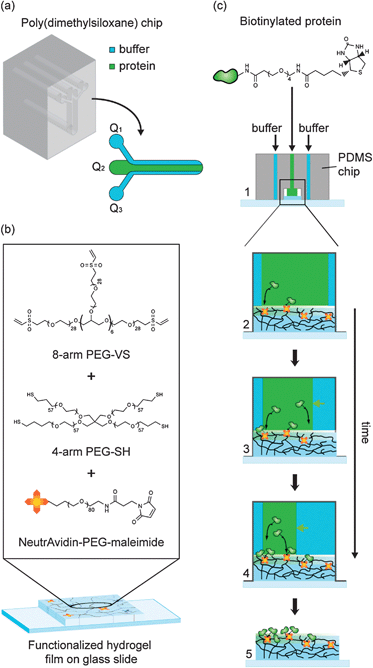 | ||
| Fig. 1 Schematic representation of the microfluidic patterning of functionalized PEG hydrogels. (a) Microfluidic chip for hydrodynamic flow focusing. (b) Formation of NeutrAvidin-conjugated hydrogels from aqueous precursor solutions containing multi-arm PEG macromers and ‘PEGylated’ NeutrAvidin. (c) Patterning of gels using hydrodynamic flow focusing. Step 1: the PDMS chip is pressed onto a glass slide bearing a thin hydrogel coating. Steps 2–4: gradient patterning by flow focusing. Step 5: the PDMS chip is removed for cell culture experiments. | ||
To capture Biotin- or Fc-labeled biomolecules from solution by affinity we generated low-swelling PEG hydrogel substrates displaying covalently bound NeutrAvidin and/or Protein A (Fig. 1b). Michael-type addition was used to form polymer networks by stepwise copolymerization of vinyl sulfone (VS)-terminated 8arm-PEG macromers and thiol (SH)-terminated 4arm-PEG macromers at equimolar amounts of functional groups.11 In the same reaction step, the gels were bioconjugated with NeutrAvidin and/or Protein A using a heterofunctional NHS-PEG-maleimide linker. Thin hydrogel films (concentration: 5% w/v) were cast on microscopic glass slides that were modified with 3-mercaptopropyl-trimethoxylsilane to expose free thiol groups. The gel films were equilibrated in PBS overnight prior to protein patterning.
Graded protein patterns were generated by dynamic flow focusing and simultaneous protein capture from the central protein stream (Fig. 1c). Starting from an almost completely filled channel (Fig. 1c, step 1), the width of the protein stream was decreased in a step-wise fashion by increasing the buffer flow rates on one or both sides (Fig. 1c, steps 3 & 4). Finally, the PDMS chip was removed from the patterned gel for further cell culture experiments (Fig. 1c, step 5).
To generate predictable gradients of various shapes by automated flow focusing, we first determined the capture kinetics of Biotin-tagged proteins on NeutrAvidin-modified PEG gels. Substrates were exposed for variable periods of time to a constant, focused stream of fluorescein- and Biotin-labelled bovine serum albumin (FITC-BSA-Biotin) model protein. Fluorescence intensities were plotted as a function of the exposure time to obtain the capture kinetics at a particular flow rate, BSA and NeutrAvidin concentration (data not shown here). These curves served as a basis to generate a desired gradient type by step-wise flow focusing (Fig. 2, left columns): we first defined a gradient of interest by a mathematical equation of the intensity as a function of discrete steps n (typically nmax = 10) performed by the syringe pumps during gradient patterning. Utilizing the experimentally determined immobilization curves, the intensity value of each step was converted into a speficic exposure time of the protein to the gel surface. Finally, the flow rates were calculated imposing a constant velocity of the protein stream from one step to the following one, allowing to program the syringe-pumps for all the steps.
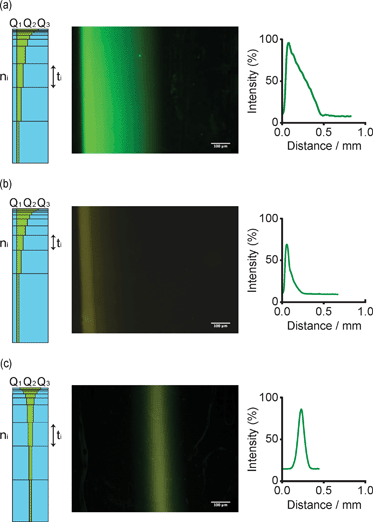 | ||
| Fig. 2 Generation of three types of model gradients by hydrodynamic flow focusing of FITC-BSA-Biotin on NeutrAdvidin-conjugated PEG gels. Fluorescent micrographs and graphical representations obtained by image analysis of a linear (a), exponential (b) and Gaussian (c) gradients (scale bar = 100 μm). Schemes on the left of each gradient depict the step-wise flow focusing process with the two buffer streams (Q1 and Q3) flanking the protein stream Q2. Flow rates were maintained constant for a time ti for each step ni. | ||
As instructive examples, we programmed the syringe pumps to obtain linear, exponential and Gaussian gradients (Fig. 2a–c). Indeed, fluorescent microscopy and subsequent image analyses showed reproducible patterning for all three gradients along the entire length of the hydrogel (9 mm). Notably, we also overlapped two different protein gradients by sequential flow focusing on PEG gels functionalized with both NeutrAvidin and Protein A. An example of dual, overlapping gradient patterning is shown in Fig. 3.
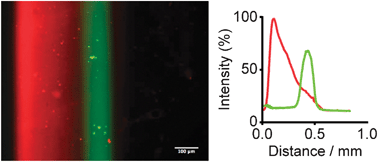 | ||
| Fig. 3 Generation of gel-immobilized overlapping gradients by sequential hydrodynamic flow focusing. Fluorescence micrograph and measured fluorescence intensities showing a linear gradient of tethered DsRed-BSA-Biotin combined with a Gaussian gradient of FITC-labelled IgG (scale bar =100 μm). | ||
In summary, here we demonstrate the generation of user-defined graded protein patterns on biofunctionalized hydrogels. Quantification of the capture kinetics of tagged biomolecules on these gels allows us to generate by automated hydrodynamic flow focusing nearly any type of protein profile with a simple microfluidic channel system. Indeed, the types of gradients and gradient combinations that can be generated are only limited by the imagination of the user. Since the microfluidic chip can be removed after patterning, the presented technique should be of interest for multiple cell-based assays.
This work was supported by the SNSF grant FN 205321-112323/1 and by a EURYI award to M.P.L.
Notes and references
- H. L. Ashe and J. Briscoe, Development (Cambridge, UK), 2006, 133, 385–394 CrossRef CAS.
- T. M. Keenan and A. Folch, Lab Chip, 2008, 8, 34–57 RSC.
- G. Velve-Casquillas, M. Le Berre, M. Piel and P. T. Tran, Nano Today, 2010, 5, 28–47 CrossRef CAS.
- D. E. Discher, P. Janmey and Y. L. Wang, Science, 2005, 310, 1139–1143 CrossRef CAS.
- J. Genzer and R. R. Bhat, Langmuir, 2008, 24, 2294–2317 CrossRef CAS.
- J. A. Burdick, A. Khademhosseini and R. Langer, Langmuir, 2004, 20, 5153–5156 CrossRef.
- S. A. DeLong, J. J. Moon and J. L. West, Biomaterials, 2005, 26, 3227–3234 CrossRef.
- S. Cosson, S. A. Kobel and M. P. Lutolf, Adv. Funct. Mater., 2009, 19, 3411–3419 CrossRef CAS.
- J. B. Knight, A. Vishwanath, J. P. Brody and R. H. Austin, Phys. Rev. Lett., 1998, 80, 3863–3866 CrossRef CAS.
- W. Georgescu, J. Jourquin, L. Estrada, A. R. A. Anderson, V. Quaranta and J. P. Wikswo, Lab Chip, 2008, 8, 238–244 RSC.
- M. P. Lutolf and J. A. Hubbell, Biomacromolecules, 2003, 4, 713–722 CrossRef CAS.
Footnote |
| † This article is part of the ‘Emerging Investigators’ themed issue for ChemComm. |
| This journal is © The Royal Society of Chemistry 2011 |
