DNAzyme-based fluorescent microarray for highly selective and sensitive detection of lead(II)†
Meiying
Liu‡
ac,
Xinhui
Lou‡
*b,
Juan
Du
ac,
Ming
Guan
b,
Jing
Wang
b,
Xiaofan
Ding
b and
Jianlong
Zhao
*a
aState Key Laboratory of Transducer Technology, Shanghai Institute of Microsystem and Information Technology, Chinese Academy of Sciences, Changning Rd. 865, Shanghai, China 200050. E-mail: jlzhao@mail.sim.ac.cn
bDepartment of Chemistry, Capital Normal University, Xisanhuan North Rd. 105, Beijing, China 100084. E-mail: xlou9999@yahoo.com; Fax: +86 10 68902320; Tel: +86 10 68902491 ext 808
cGraduate School of the Chinese Academy of Sciences, Beijing, 100049, China
First published on 28th October 2011
Abstract
A facile microarray-based fluorescent sensor for the detection of lead (II) was developed based on the catalytic cleavages of the substrates by a DNAzyme upon its binding to Pb2+. The release of the fluorophore labelled substrates resulted in the decrease of fluorescence intensity. The sensor had a quantifiable detection range from 1 nM to 1 μM and a selectivity of >20 fold for Pb2+ over other metal ions.
Because of continuing concern over lead in the environment and its deleterious effects on human health, especially on children,1 many Pb2+ detection sensors have been developed with the ultimate goal to provide approaches suitable for on-site applications. Ideal technologies for on-site applications need to meet the requirements like high sensitivity, high selectivity, easy operation, short detection time, robustness, portability and low-cost. Methods, with high-throughput and capable of multiplexed analysis, are especially in urgent request in order to significantly improve the efficiency. Traditional methods that require sophisticated instruments and rather complex laboratory techniques are not suitable for on-site applications.2 Very few sensors reported so far can meet all of the requirements for on-site detection.
Microarray technology has become a routine analytical tool for chemical and biological analysis due to its inherent advantages of high-throughput, low-manufacturing costs, simplicity and robustness. In recent years, considerable effort has been focused on the development of microarray techniques for metal ion detection.3–5 For example, Mirkin's group developed an oligonucleotide array for Hg2+ detection with a detection limit of 10 nM.6 Lee et al. demonstrated polydiacetylene liposome microarrays for selective K+ detection even in the presence of Na+.4 Wang et al. reported a sensory microarray based on conjugated polymers to detect Cu2+ ions.5
Metal-specific catalytic DNAs, DNAzymes that can catalyze the cleavage of their substrates in the presence of a particular metal ion, have emerged as a new class of metal-ion sensing molecules. DNAzymes with desired metal-ion selectivity and affinity can be obtained through a combinatorial biology method, in vitro selection.7 Using this method, lead-dependent DNAzymes such as variants of 8–17 DNAzymes, have been discovered.8,9 Upon combining with fluorophore/quencher pairs, nanomaterials (e.g.gold nanoparticles or graphene10), electro-active tags, or Raman reporters, the variants of 8–17 DNAzymes have been transformed into powerful classes of fluorescent,11–15 colorimetric,16–18 electrochemical,19,20 and SERS21 sensors for lead detection. Among them, fluorescent sensors have attracted much attention due to their high sensitivity and simplicity. Lu et al. demonstrated the first solution-based assay for highly sensitive (detection limit 10 nM) and selective detection of lead ions by labelling the DNAzyme and its substrate with a quencher and fluorophore, respectively.11 They subsequently reported the first surface-based assay,13,22 in which the fluorophore labelled substrate was immobilized on Au substrates13 and the solution containing the released substrate fragments upon incubating with Pb2+ solution was collected for fluorescence measurements. The assay improved the sensitivity to 1 nM by overcoming the inherent drawback of the solution-based assay, the high background signal from the unhybridized substrates. However, their method is not suitable for high-throughput applications since the fluorescence measurements could not be conducted directly on the surface due to the fluorescent quenching of the fluorophore by the gold surface. In addition, the DNA layers formed on the gold surface viagold–sulfur interaction are not as robust as the covalently attached ones on other surfaces, such as silylated slides. Combining the lead-specific DNAzymes with the microarray technique should enable the high-throughput and reliable determination of lead ions at minute concentrations.
We report here a facile microarray-based fluorescent sensor intended for on-site Pb(II) detection using the 17E variant of the 8–17 DNAzyme.8 The typical trans-cleaving 17E consists of a catalytic DNA strand and a DNA/RNA chimera substrate containing a single, scissile ribo-adenine (rA). The design of the fluorescent lead sensor is shown in Scheme 1. The sensor consists of a 5′-amine-modified catalytic DNA strand (1) (5′-NH2-T12-TCTCTTCTCCGAG CCGGTCGAAATAGTGAGT-3′) and a 5′-Cy5-modified substrate (2) (5′-Cy5-ACTCACTATrAGGAAGAGATG-3′). A 12-T linker was inserted between the amine group and the enzyme sequence for improving the hybridization efficiency. The catalytic DNA strand (1) is immobilized on the surface of the slide via an amine–aldehyde interaction. Then the Cy5 tagged substrate (2) is assembled onto the surface through a hybridization reaction to prepare the sensor with a strong fluorescence (64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 3000). When the sensor is incubated with solutions containing Pb2+, the catalytic strand carries out catalytic reactions for hydrolytic cleavage of the substrate strand at the scissile rA (indicated by a red arrow in Scheme 1). Then the substrate is broken into two pieces and dissociates from the catalytic strand, resulting in the decrease of the surface fluorescence intensities. The experimental details are provided in the ESI†.
000 ± 3000). When the sensor is incubated with solutions containing Pb2+, the catalytic strand carries out catalytic reactions for hydrolytic cleavage of the substrate strand at the scissile rA (indicated by a red arrow in Scheme 1). Then the substrate is broken into two pieces and dissociates from the catalytic strand, resulting in the decrease of the surface fluorescence intensities. The experimental details are provided in the ESI†.
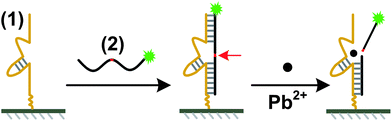 | ||
| Scheme 1 Schematic of the microarray-based fluorescent lead(II) sensor immobilization and Pb(II) reaction process. The 5′-amine-modified catalytic DNA strand (1) is immobilized on the aldehyde-coated glass slides through covalent reaction. Via hybridization, Cy5-labeled DNA/RNA chimera substrate (2) is attached to the microarray. The substrate is cleaved in the presence of Pb(II). The scissile ribo-adenine (rA) is indicated by a red arrow. | ||
To evaluate the sensitivity of the fluorescent sensor, the microarray was incubated with different concentrations of Pb2+ (from 0 to 10 μM) at 4 °C for 1 h. Fig. 1A shows the fluorescence scanometric images of the microarray after incubation with Pb2+ solution. A plot of fluorescence decrease (%) vs.Pb2+ concentration is shown in Fig. 1B. As the concentration of Pb2+ increased, the fluorescence decrease (%) increased. When the Pb2+ concentration was increased to over 1 μM, the fluorescence decrease (%) began to level off (average fluorescence intensity 2870), suggesting that more than 95% of the substrates confined on the slide surface had been hydrolyzed and released. The detection limit (3 times of the standard deviation of the buffer control) was determined to be 1 nM, the same as that of the gold surface-based detection methods13 and much better than those of the colorimetric17 and other electrical biosensors for lead (II).19 A linear calibration was obtained over nearly three decades: 1 μM ≥ [Pb(II)] ≥ 1 nM. Please note that the choice of 1 h incubation time was based on the reported kinetic experiment on Au surface.13 A shorter incubation time could be allowed under optimized conditions.
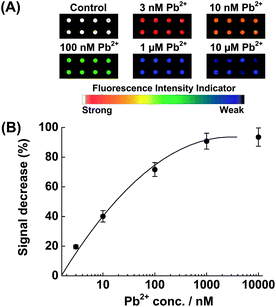 | ||
| Fig. 1 (A) Scanometric images of the microarray in the presence of various concentrations of Pb2+. (B) Relative fluorescence change (%) as a function of the Pb2+ concentration. The illustrated error bars represent the standard deviation obtained from 8 data points. | ||
A control experiment was carried out to ensure that the observed fluorescence intensity decrease was actually due to the selective cleavage and subsequent release of the substrate (2) in the presence of Pb2+, rather than the direct quenching of the fluorescence of Cy5 by Pb2+. For this experiment, the fluorescence intensities of substrate (2) in buffer solution were measured before and after adding Pb2+ at various concentrations (from 0 to 10 μM). No statistical differences in fluorescence intensity were observed at all Pb2+ concentrations (Fig. S1†). In addition, as reported and observed by us, mutant versions of the lead-dependent DNAzyme 17E, with a single base change at the wobble position (e.g. the G–T wobble pair was changed into the G–C pair) can cause the complete inhibition of the catalysis activity of the DNAzyme.8 The data clearly demonstrate that the fluorescence decrease was indeed due to the cleavage reaction catalyzed by the DNAzyme.
For the Pb(II)-specific DNAzyme sensor on the microarray surface, it is necessary to verify that selectivity is maintained in the immobilized state. An experiment was conducted to compare the activity of the DNAzyme with Pb(II) to that of its previously identified most strongly interfering divalent metal ion Zn(II)13 and other environmentally common ions, Hg(II), Mg(II), Cu(II) and Zn(II). Detections were performed as previously described, independently reacting 10 μM of each of the above listed metal ions with the sensor. Fig. 2A shows the fluorescence scanometric images of the microarray after incubation with various metal ions at 4 °C for 1 h. Fig. 2B shows that the fluorescence intensity decreases of the interfering ions are within the error of the blank (<5%). The Pb(II)-reacted samples, however, show 95 ± 5% signal decreases of the blank control. This demonstrates that the covalent immobilization has not lowered the specificity of the Pb(II) DNAzyme.14,19
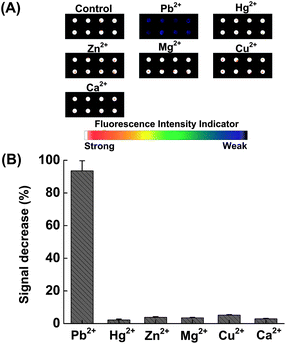 | ||
| Fig. 2 (A) Scanometric images of the microarray in the presence of various metal ions. (B) Relative fluorescence change (%) as a function of various metal ions. 10 μM of each metal ion was incubated with the sensor at 4 °C for 1 h. | ||
A major advantage of utilizing a microarray-immobilized DNAzyme for analyte recognition is the good stability of the microarray, which allows robust detection in a ready-to-use and reagentless fashion. It is well known that DNA microarrays possess sufficient shelf life and have been commercialized as routine analytical tools. In contrast, DNA immobilized gold surfaces have quite limited storage time and are not suitable for practical applications. Different from a regular DNA microarray, our sensor is duplex DNA-based. The longer length of the duplex is attributed to the better stability, which also allows that the formation of the duplex and the cleavage reaction can be performed at room temperature, instead of the much lower temperature (e.g. 4 °C).15,18 To further improve the stability of the sensor and to test the generality of the platform as well, we fabricated another sensor using a different version of the lead DNAzyme which can work at room temperature. The sensor comprises two longer probes: a catalytic DNA strand (3) (5′-CATCTCTTCTCCGAGCCGGTCGAAATAGTGAGTCAGACATA-T12-NH2-3′) and a DNA/RNA chimera substrate (4) (5′-TATGTCTGACTCACTATrAGGAAGAGATG-Cy5-3′). The increases of the probe lengths and the modification changes of the probes (amino group and Cy5 were all moved to another end of the probe) did not affect the performance of the sensor (Fig. S2†), but allows the sensor to be prepared and function at room temperature, rather than at 4 °C.
In summary, we have demonstrated a facile microarray-based fluorescent sensor for Pb2+ detection that meets all of the requirements for on-site detection: our sensor exhibited a quantifiable detection range from 1 nM to 1 μM and a selectivity of >20 fold for Pb2+ over other metal ions. Our ready-to-use sensor is operated in a very simple fashion: incubation, wash and direct imaging of the microarray, without the need to collect solutions for individual fluorescence measurements, precise temperature control and a further signal amplification step. Our sensor is quite robust due to the stable surface-immobilization of the probesvia covalent bonds. Our sensor is capable of high-throughput and multiplexed analysis because of the direct imaging of the whole microarray, which is not possible for other detection platforms. Our sensor is technically compatible with miniaturization technologies, which is currently an important trend in environmental monitoring, food safety and clinical toxicology. We believe that our work represents a new progress in the field of metal ion detection. The sensor could be practically used for real and more complex samples such as food samples or industrial wastewater. The detection platform should be able to extend to other metal ions and components to develop a facile detection platform for parallel analysis of multi-targets, which is highly desired for significantly improving the detection efficiency. Finally, even though the sensor demonstrated in this work is a signal-off sensor, the sensor theoretically could be changed into a signal-on sensor by simply labelling the catalytic DNA strand with the fluorophore and the substrate with the quencher, respectively, or by subsequently hybridizing with nanoparticle labelled probes to the positions previously occupied by the cleaved substrates.
This work was supported by National Natural Science Foundation (20975108), Shanghai Pujiang Program (09PJ1411800), Science and Technology Commission of Shanghai Municipality (09JC1416500, 10391901600, 09391911500), and Funding Project for Academic Human Resources Development in Institutions of Higher Learning under the Jurisdiction of the Beijing Municipality (PHR20100718).
Notes and references
- N. Rifai, G. Cohen, M. Wolf, L. Cohen, C. Faser, J. Savory and L. DePalma, Ther. Drug Monit., 1993, 15, 71–74 CrossRef CAS; H. A. Godwin, Curr. Opin. Chem. Biol., 2001, 5, 223–227 CrossRef.
- D. T. Miller, D. C. Paschal, E. W. Gunter, P. E. Stroud and J. D'Angelo, Analyst, 1987, 112, 1701–1704 RSC; H. Longerich, B. Fryer and D. Strong, Spectrochim. Acta, Part B, 1987, 42, 39–48 CrossRef CAS; H. W. Liu, S. J. Jiang and S. H. Liu, Spectrochim. Acta, Part B, 1999, 54, 1367–1375 CrossRef; M. S. Chan and S. D. Huang, Talanta, 2000, 51, 373–380 CrossRef.
- C. M. Wu and L. Y. Lin, Biosens. Bioelectron., 2004, 20, 864–871 CrossRef CAS; Z. Wang, M. A. Palacios and P. Anzenbacher, Jr, Anal. Chem., 2008, 80, 7451–7459 CrossRef; J. Lee, H. Jun and J. Kim, Adv. Mater., 2009, 21, 3674–3677 CrossRef; P. Zuo, B. C. Yin and B. C. Ye, Biosens. Bioelectron., 2009, 25, 935–939 CrossRef.
- J. Lee, H. J. Kim and J. Kim, J. Am. Chem. Soc., 2008, 130, 5010–5011 CrossRef CAS.
- F. Lv, X. Feng, H. Tang, L. Liu, Q. Yang and S. Wang, Adv. Funct. Mater., 2011, 21, 845–850 CrossRef CAS.
- J. S. Lee and C. A. Mirkin, Anal. Chem., 2008, 80, 6805–6808 CrossRef CAS.
- R. R. Breaker and G. F. Joyce, Chem. Biol., 1994, 1, 223–229 CrossRef CAS; Y. Li and R. R. Breaker, Curr. Opin. Struct. Biol., 1999, 9, 315–323 CrossRef.
- A. K. Brown, J. Li, M. B. P. Caroline and Y. Lu, Biochemistry, 2003, 42, 7152–7161 CrossRef CAS.
- Y. Lu, T. Lan and K. Furuya, Chem. Commun., 2010, 46, 3896–3898 RSC.
- X. B. Zhang, X. H. Zhao, R. M. Kong, H. M. Meng, W. N. Liu, W. H. Tan, G. L. Shen and R. Q. Yu, Anal. Chem., 2011, 83, 5062–5066 CrossRef; D. Li, Y. Q. Wen, C. Peng, L. Zhuo, S. J. He, L. H. Wang, Q. Huang, Q. H. Xu and C. H. Fan, Chem. Commun., 2011, 47, 6278–6280 RSC.
- J. Li and Y. Lu, J. Am. Chem. Soc., 2000, 122, 10466–10467 CrossRef CAS.
- I. H. Chang, J. J. Tulock, J. Liu, W. S. Kim, D. M. Cannon, Jr, Y. Lu, P. W. Bohn, J. V. Sweedler and D. M. Cropek, Environ. Sci. Technol., 2005, 39, 3756–3761 CrossRef CAS; W. H. Tan, H. Wang, Y. M. Kim, H. P. Liu, Z. Zhu and S. Bamrungsap, J. Am. Chem. Soc., 2009, 131, 8221–8226 CrossRef.
- C. B. Swearingen, D. P. Wernette, D. M. Cropek, Y. Lu, J. V. Sweedler and P. W. Bohn, Anal. Chem., 2005, 77, 442–448 CrossRef CAS.
- T. S. Dalavoy, D. P. Wernette, M. Gong, J. V. Sweedler, Y. Lu, B. R. Flachsbart, M. A. Shannon, P. W. Bohn and D. M. Cropek, Lab Chip, 2008, 8, 786–793 RSC.
- N. Nagraj, J. Liu, S. Sterling, J. Wu and Y. Lu, Chem. Commun., 2009, 4103–4105 RSC.
- J. Liu and Y. Lu, J. Am. Chem. Soc., 2003, 125, 6642–6643 CrossRef CAS; J. Liu and Y. Lu, Chem. Mater., 2004, 16, 3231–3238 CrossRef; J. Liu and Y. Lu, J. Am. Chem. Soc., 2005, 127, 12677–12683 CrossRef; D. Mazumdar, J. Liu, G. Lu, J. Zhou and Y. Lu, Chem. Commun., 2010, 46, 1416–1418 RSC.
- J. Liu and Y. Lu, J. Am. Chem. Soc., 2004, 126, 12298–12305 CrossRef CAS.
- Z. Wang, J. H. Lee and Y. Lu, Adv. Mater., 2008, 20, 3263–3267 CrossRef CAS.
- Y. Xiao, A. A. Rowe and K. W. Plaxco, J. Am. Chem. Soc., 2007, 129, 262–263 CrossRef CAS.
- L. Shen, Z. Chen, Y. Li, S. He, S. Xie, X. Xu, Z. Liang, X. Meng, Q. Li and Z. Zhu, Anal. Chem., 2008, 80, 6323–6328 CrossRef CAS.
- J. Irudayaraj and Y. L. Wang, Chem. Commun., 2011, 47, 4394–4396 RSC.
- T. J. Yim, J. Liu, Y. Lu, R. S. Kane and J. S. Dordick, J. Am. Chem. Soc., 2005, 127, 12200–12201 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section, scanometric images of the chips, solution fluorescence emission measurements. See DOI: 10.1039/c1an15633k |
| ‡ These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2012 |
