DOI:
10.1039/C1AN15833C
(Paper)
Analyst, 2012,
137, 170-185
Metabolomics study on Fuzi and its processed products using ultra-performance liquid-chromatography/electrospray-ionization synapt high-definition mass spectrometry coupled with pattern recognition analysis
Received
7th September 2011
, Accepted 26th September 2011
First published on 26th October 2011
Abstract
The lateral root of Aconitum carmichaelii Debx is named “Fuzi” which is widely distributed across Asia and North America and has been used to relieve joint pain and treat rheumatic diseases for over two thousand years. However, it has very narrow therapeutic ranges and despite the toxicological risk, its usage remains very high. A traditional Chinese processing approach (Paozhi, detoxifying measure) is necessary to remove the poisonous Aconitum alkaloids mainly deriving from the diester diterpene alkaloids (DDAs) including aconitine, mesaconitine and hypaconitine. They can be decomposed into less or non-toxic derivatives through Paozhi that plays an essential role in detoxification. Processed Fuzi is mainly focused on the three main forms of Yanfuzi (YFZ), Heishunpian (HSP) and Baifupian (BFP) which are highly desirable in order to guarantee the clinical safety and their low toxicity in decoctions. The difference in metabolomic characters between Fuzi and its processed preparations is still completely unclear. Therefore, this paper was designed to investigate a comprehensive metabolome of Fuzi and its processed products by ultra-performance liquid-chromatography/electrospray-ionization synapt high-definition mass spectrometry (UPLC-Q-TOF-HDMS) combined with pattern recognition methods. The difference in metabolic profiles between Fuzi and its processed preparations was well observed by the principal component analysis (PCA) of the MS spectra. Significant changes of 19 metabolite biomarkers were detected in the Fuzi samples and three preparations. The underlying regulations of Paozhi-perturbed metabolic pathways were also discussed according to the identified metabolites. The present study proves that UPLC-Q-TOF-HDMS based metabolomic analysis greatly contributes to the investigation of Fuzi metabolism through Paozhi techniques, and provides useful information to further comprehensively understand the pharmacological activity and potential toxicity of processed Fuzi in a clinical environment.
1. Introduction
Traditional Chinese medicines (TCMs) have gained increasing acceptance worldwide in recent years and are generally considered as being natural and harmless.1–3 The Aconitum species, an essential drug in TCM, have been used for several thousand years and widely distributed across Asia and North America. As a widely used Chinese herbal medicine, the roots of Aconitum have a wide range of pharmacological effects although it has very narrow therapeutic ranges, commonly applied for various diseases, i.e., collapse, syncope, rheumatic fever, painful joints, gastroenteritis, diarrhea, oedema, bronchial asthma, various tumors, and some endocrinal disorders like irregularmenstruation.4,5 In Chinese Pharmacopoeia (CP) 2010, Aconitum carmichaeli Debx. (Fig. 1A) are recorded, extensively distributed in the Sichuan Province of China. The mother root of Aconitum carmichaelii Debx is named “Chuanwu” (CW, Fig. 1B), while the daughter or lateral root of Aconitum carmichaelii Debx is “Fuzi” (FZ, Fig. 1C). As one of the most crucial TCM drugs, FZ has been more frequently prescribed than CW to treat rheumatic diseases. It was firstly recorded in the earliest Chinese medicinal classic, Shennong's Materia Medica. There are more than 20 commonly used proprietary herbal products for muscular disorders, joint pain and arthritis, from both historical literature and modern clinical reports containing processed Fuzi as a main ingredient, such as ‘Yin Chen Si Ni Tang’ ‘Fuzi Lizhong Wan’, ‘Guifulizhong Wan’, ‘Jinguishenqi Wan’, “Shenfu Injection”.6,7
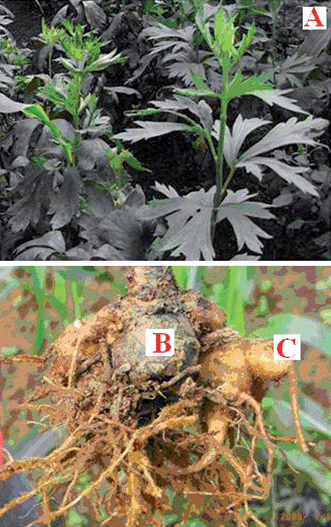 |
| | Fig. 1
Aconitum carmichaeli Debx. (A); the mother root of Aconitum carmichaelii Debx. (B); the daughter or lateral root of Aconitum carmichaelii Debx. (C). | |
However, the toxicity of Fuzi leads to a potentially high risk of severe problems that are sometimes even life-threatening.8 At present, a comparison of the metabolic profiles revealed that four major groups of alkaloids – monoester diterpene alkaloids (MDAs), amine diterpenoid alkaloids (ADAs), diester diterpene alkaloids (DDAs), and lipoalkaloids – were found in the roots of Aconitum.9 They share a common C19-norditerpenoid skeleton, in which the C8 position is occupied by a hydroxyl, an acetoxy or a fatty acid acyl group, respectively.10,11 The toxicity of Aconitum mainly derives from the DDAs including aconitine, mesaconitine, and hypaconitine.12 As we know, TCM herbal processing approaches, namely “Paozhi”, by means of the transformation of secondary plant metabolites help to reduce the toxicity of the drug, and might exert a large maximal therapeutic efficacy with minimal adverse effects.13 As a traditional processing form, Paozhi could remarkably reduce the toxicity of the Fuzi by decomposing the DDAs to the relatively lower toxic MDA. All of the processed Fuzi must be used. According to the ways of the processing form, it is converted into Fuzi preparations including Yanfuzi (YFZ), Heishunpian (HSP) and Baifupian (BFP), which are recorded in CP 2010. The Paozhi approach is a good detoxifying measure to remove the poisonous Aconitumalkaloids. However, the difference in metabolomic characters between Fuzi and its processed products is still completely unclear, restricting the further application of processed Fuzi in the clinical environment.
Metabolomics is an ‘omics’ approach that aims to analyze all metabolites in a biological sample comprehensively.14,15 The detailed metabolite profiling of thousands of the secondary metabolites has great potential for directly elucidating plant metabolic processes.16 It has brought enormous opportunities for improved detection and discovery of biomarkers, and adopts a ‘top-down’ strategy to reflect the function of organisms from terminal symptoms of the metabolic network and understand metabolic changes of a complete system caused by interventions in a holistic context.17–21 Metabolomics not only will assist in understanding the nature of plant responses to environmental change but also will provide unique insights into the fundamental nature of plant phenotypes in relation to development, physiology, resistance, biodiversity, etc. Several studies have shown that metabolomics have been used in the evaluation of differences in toxicological and pharmacological actions of aconite products when applying different TCM processing methods.22,23 A clear understanding of the differentiation mechanism of the Aconitum is essential to evaluate drug safety and for the safe use in the clinic. However, the research is still limited for content changes of several alkaloids to determine its efficacy/toxicity, while ignoring the crude and processed products as a complex group of plant metabolites. Moreover, little is known about the global change of the metabolome of processed Fuzi. With the development of new analytical techniques, metabolomics as a research platform, could provide comprehensive, detailed, reliable evidence for further study on efficacy/toxicity of processed Fuzi. The objective of ‘non-targeted’ metabolic profiling analysis can determine ‘global’ metabolites.24 Of the various profiling techniques, UPLC/MS has now become a promising tool for investigating the metabolome of plants. It is believed that metabolomics analysis will play an important role as an effective tool in terms of high-throughput elucidation of metabolic phenotypes of crude and processed preparations. Thus, this paper was designed to investigate comprehensive metabolomic characters of Fuzi and its processed products by UPLC-Q-TOF-HDMS.
2. Experimental
2.1 Chemicals and materials
Acetonitrile and methanol (HPLC grade) was purchased from Merck (Darmstadt, Germany). Distilled water was purchased from Watson's Food & Beverage Co., Ltd. (Guangzhou, China), and formic acid (HPLC grade) was purchased from the Beijing Reagent Company (Beijing, China). Leucine enkephalin was supplied by Sigma-Aldrich (MO, UK). Other chemicals, except as noted, were analytical grade. FZ were collected from the mianyang county in Sichuan province (South, China) in July 2009. They were authenticated by Prof. Xijun Wang, Department of Pharmacognosy of Heilongjiang University of Chinese Medicine (Harbin, China). The processed preparation of Fuzi was carried out according to CP 2010.5
The crude Fuzi and processed roots were pulverized into a powder, passed through a 0.45 mm sieve, and stored in a desiccator. The powder (1.0 g) was extracted with 75% methanol two times (5.0 ml and 10 min each time) by sonication at room temperature, and then vortexed for 2 min respectively. The mixture was centrifuged for 5 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C. The supernatant diluted 50 times with methanol was then filtered through a 0.22 μm-filter, the filtrate was used for UPLC analysis.
Chromatographic analysis was performed in a Waters ACQUITY UPLC system controlled with Masslynx (V4.1, Waters Corporation, Milford, USA). An aliquot of 2 μL of sample solution was injected onto an ACQUITY UPLCTM BEH C18 column(50 mm × 2.1 mm i.d., 1.7 μm, Waters Corp, Milford, USA) held at 45 °C and the flow rate was 0.5 mL min−1. The mobile phase consisted of a linear gradient system of A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile), 0–1.5 min, B 10–35%; 1.5–2.5 min, B 35–60%; 2.5–3.0 min, B 60–67%, 3.0–4.5 min B 99%; 4.5–5.0 min, B 99–1%; 5.0–7.0 min, B 1%.
Mass spectrometry detection was performed using a Waters Micromass Q-tof micro™ Synapt High Definition Mass Spectrometer (Manchester, UK) equipped with electrospray ionization. The optimal operating conditions were as follows: the source temperature, 110 °C; desolvation gas temperature, 300 °C; cone gas flow, 50 L h−1; desolvation gas flow, 500 L h−1; capillary voltage, 2.5 kV; sampling cone voltage, 35.0 V; extraction cone voltage, 3.0 V. Data were centroided and reference mass correction on each sample was performed with a continuous infusion of a 0.2 ng mL−1 solution of leucine enkephalin at a flow rate of 100 μl min−1, generating a reference ion for positive ion mode ([M + H]+ = 556.2771) and negative ion mode ([M − H]− = 554.2614) to ensure accuracy during the MS analysis. The mass range was acquired using Masslynx™ (V4.1) software for each sample from 50 to 1000 Da with a 0.3 s scan time and a 0.1 s interscan delay over a 12 min run time in both positive modes. Analytes were identified by accurate mass detection.
000 rpm at 4 °C. The supernatant diluted 50 times with methanol was then filtered through a 0.22 μm-filter, the filtrate was used for UPLC analysis.
Chromatographic analysis was performed in a Waters ACQUITY UPLC system controlled with Masslynx (V4.1, Waters Corporation, Milford, USA). An aliquot of 2 μL of sample solution was injected onto an ACQUITY UPLCTM BEH C18 column(50 mm × 2.1 mm i.d., 1.7 μm, Waters Corp, Milford, USA) held at 45 °C and the flow rate was 0.5 mL min−1. The mobile phase consisted of a linear gradient system of A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile), 0–1.5 min, B 10–35%; 1.5–2.5 min, B 35–60%; 2.5–3.0 min, B 60–67%, 3.0–4.5 min B 99%; 4.5–5.0 min, B 99–1%; 5.0–7.0 min, B 1%.
Mass spectrometry detection was performed using a Waters Micromass Q-tof micro™ Synapt High Definition Mass Spectrometer (Manchester, UK) equipped with electrospray ionization. The optimal operating conditions were as follows: the source temperature, 110 °C; desolvation gas temperature, 300 °C; cone gas flow, 50 L h−1; desolvation gas flow, 500 L h−1; capillary voltage, 2.5 kV; sampling cone voltage, 35.0 V; extraction cone voltage, 3.0 V. Data were centroided and reference mass correction on each sample was performed with a continuous infusion of a 0.2 ng mL−1 solution of leucine enkephalin at a flow rate of 100 μl min−1, generating a reference ion for positive ion mode ([M + H]+ = 556.2771) and negative ion mode ([M − H]− = 554.2614) to ensure accuracy during the MS analysis. The mass range was acquired using Masslynx™ (V4.1) software for each sample from 50 to 1000 Da with a 0.3 s scan time and a 0.1 s interscan delay over a 12 min run time in both positive modes. Analytes were identified by accurate mass detection.
2.5 Pattern recognition analysis
The mass data acquired were imported to Markerlynx (version 4.1, Waters Corporation, MA, USA) within Masslynx software (version 4.1) for peak detection and alignment. All data were normalized to the summed total ion intensity per chromatogram, and the resultant data matrices were introduced to EZinfo 2.0 software (which is included in MarkerLynx Application Manager and can be applied directly) for principal component analysis (PCA), partial least squares-discriminant analysis (PLS-DA) and orthogonal projection to latent structures (OPLS) analysis.
3. Results and discussion
3.1 Acquisition and processing of metabolic profile data
To compare the Fuzi metabolome and its processed preparations, global profiling of its positive ion mode was analyzed by UPLC-HDMS. Using the optimal UPLC-HDMS condition described above, the representative Based Peak Intensity (BPI) chromatograms for their samples in positive ion modes are presented in Fig. 2, respectively. As can be seen from Fig. 2, the low molecular mass metabolites could be separated well in 3.5 min. In order to better visualize the subtle similarities and differences among these complex data sets, multiple pattern recognition methods were employed to phenotype their metabonome. Here, PCA, PLS and OPLS-DA were used to classify the metabolic phenotypes and identify the differentiating metabolites. In the PCA scores, each point represents an individual sample. The PCA results are displayed as score plots indicating the scatter of the samples, which indicate similar metabolomic compositions when clustered together and compositionally different metabolomes when dispersed. The PCA scores plot could divide the different samples into different blocks, suggesting that the metabolic profiles have distinguish Fuzi and its processed products (Fig. 3A). The unsupervised PCA analysis reveals a much greater metabolic signature difference from the pre- and post-treatment. It's worth noting that this change is in a diversification manner. PLS-DA was employed for classification or discrimination analyses because PLS-DA calculation yielded a significantly distinct degree of biological variation. A loadings plot predicates the list of metabolites helping in the positioning of the distance from different groups. The metabolic markers of Fuzi and its processed products were plotted by the PLS-DA, depicting the variable metabolic patterns at the phenotype (Fig. 3B). A variable importance parameters (VIP) plot was used to identify the metabolites according to the orders of their contributions to the separation of clustering. The mass spectrometry signals responsible for this differentiation are characterized by the VIP plot from PLS-DA analysis (Fig. 4, 5 and 6). The farther away from the origin, the higher value of the ions in VIP score plot. S-plot analysis represented the highest contributing signals for the pre- and post-treatment. Potential markers were extracted from S-plots constructed following the OPLS analysis, and markers were chosen based on their contribution to the variation and correlation within the data set. Based on VIP and the corresponding loadings plot, 19 metabolites were identified and were selected as the biomarkers.
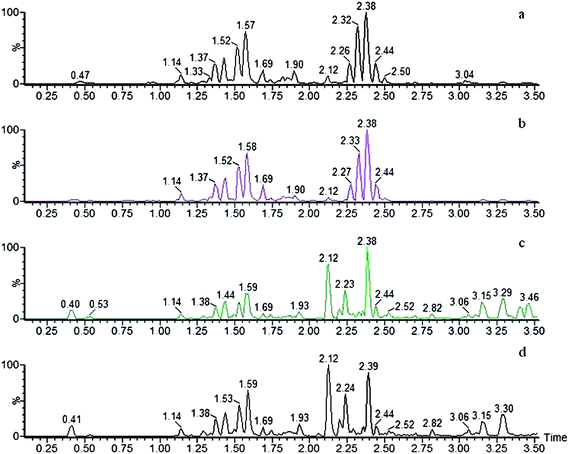 |
| | Fig. 2 Typical total ion chromatogram of Fuzi and its processed products by UPLC-HDMS in positive ESI mode. a, Fuzi; b, Yanfuzi; c, Heishunpian; d, Baifupian. | |
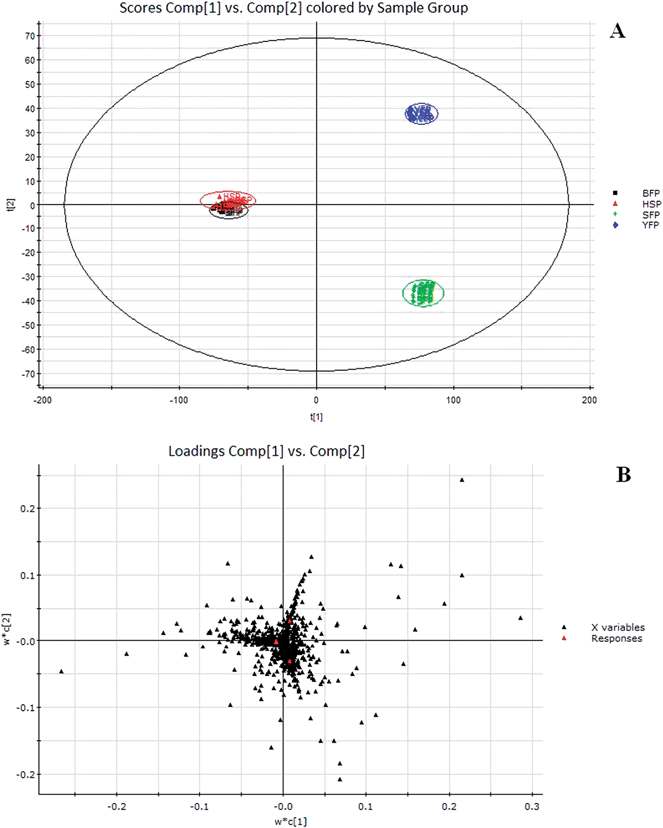 |
| | Fig. 3
PCA score plots of Fuzi and its processed products by UPLC-HDMS in positive ESI mode (A) and corresponding PLS-DA analysis (B) for biomarker recognition. SFP, Fuzi; YFP, Yanfuzi; HSP, Heishunpian; BFP, Baifupian. | |
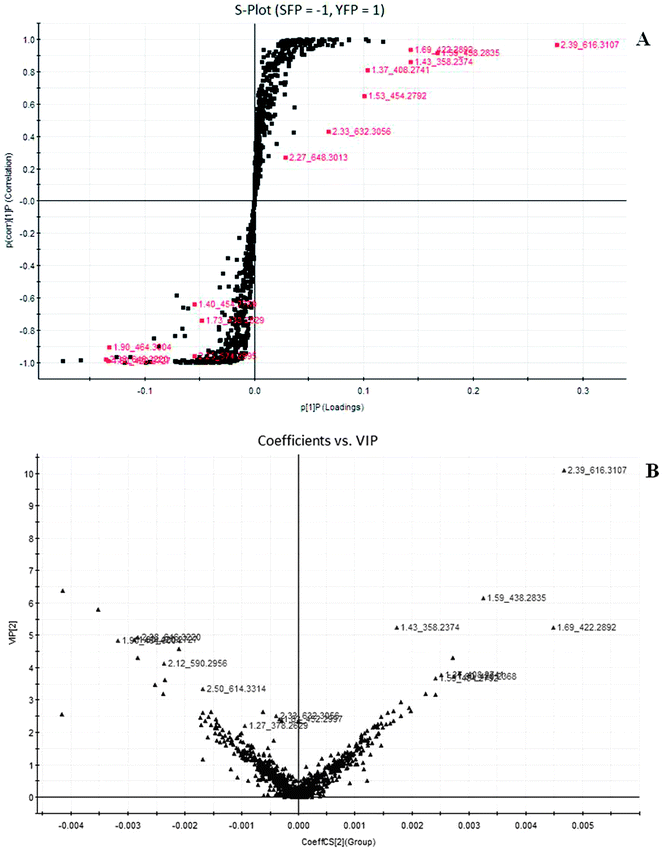 |
| | Fig. 4 Potential biomarkers in the loading S- and variable important (VIP)-plot of OPLS-DA model between Fuzi and Yanfuzi resulting from the UPLC/MS spectra in positive ESI mode. S-plot analysis representing the highest contributing signals for the pre- and post-treatment. SFP, Fuzi; YFP, Yanfuzi. | |
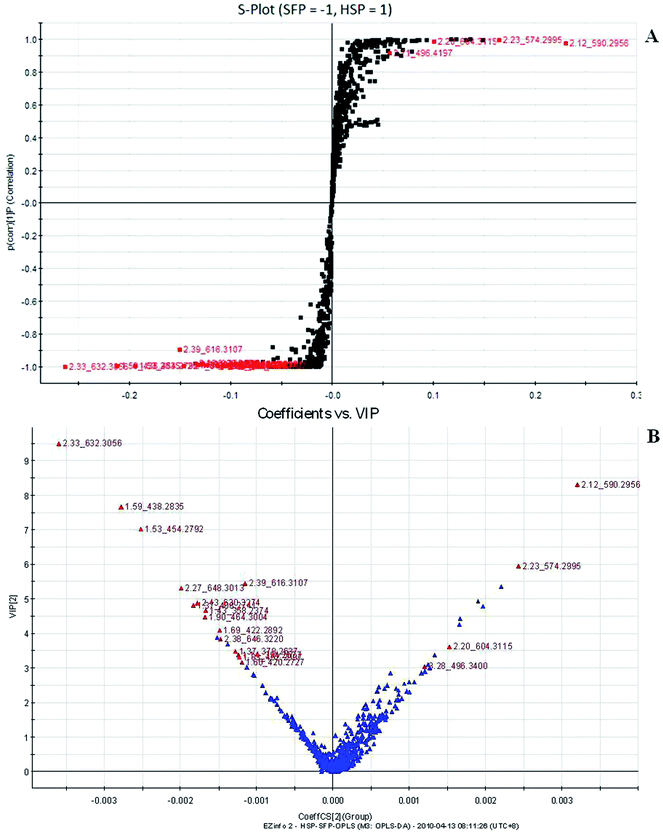 |
| | Fig. 5 Potential biomarkers in the loading S- and VIP-plot of OPLS-DA model between Fuzi and Heishunpian resulting from the UPLC/MS spectra in positive ESI mode. SFP, Fuzi; HSP, Heishunpian. | |
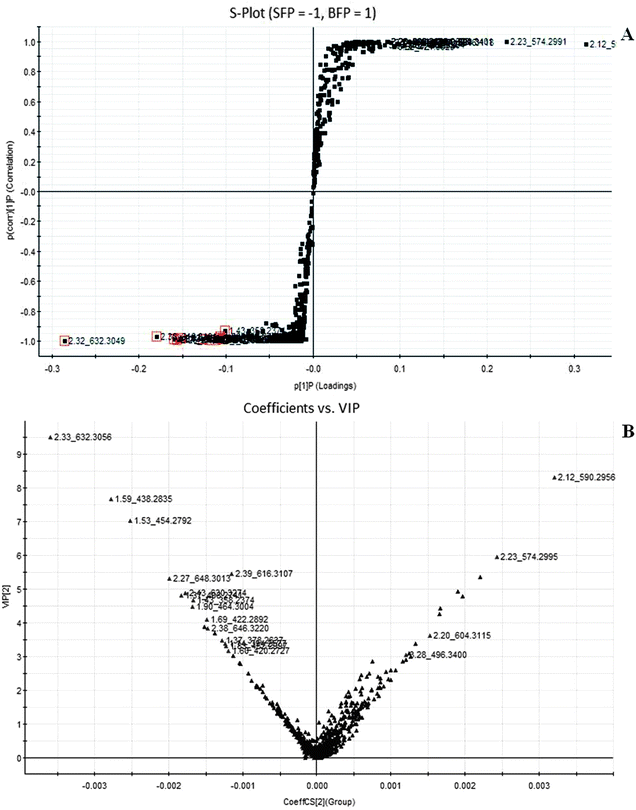 |
| | Fig. 6 Potential biomarkers in the loading S- and variable important (VIP)-plot of OPLS-DA model between Fuzi and Baifupian resulting from the UPLC/MS spectra in positive ESI mode. SFP, Fuzi; BFP, Baifupian. | |
3.2
Biomarker characterization
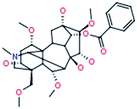
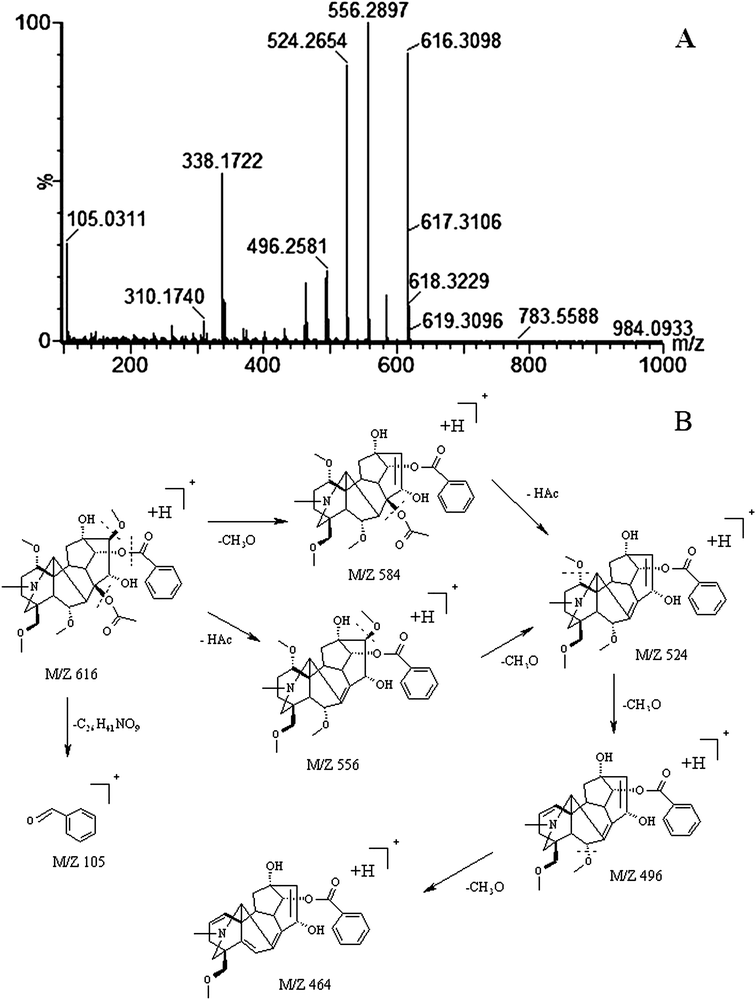
The robust UPLC-HDMS analysis that was performed in positive mode using the aforementioned protocol indicated the retention time and precise molecular mass and provided the MS/MS data that was necessary for the structural identification of the biomarkers. PCA is a data visualization method useful for overviewing relationships or ‘groupings’ within multivariatedata.25 The results of PCA displayed as score plots enabled us to compare the metabolite profiles of different groups. PLS-DA analysis showed distinct metabolites clustered according to the characteristic change of control or processed samples profiles.26 The precise molecular mass was determined within a reasonable degree of measurement error (<5 ppm) using Q-TOF, and the potential element composition, degree of unsaturation and fractional isotope abundance of the compounds were also obtained. We searched for the presumed molecular formula ChemSpider (http://www.chemspider.com), MassBank (http://www.massbank.jp/), SMPD (http://www.smpdb.ca/) and other databases to confirm possible chemical compositions, and the MS/MS data were screened to determine the potential structures of the ions. All the detected ions were arranged according to VIP value (>2), and there were 19 ions indentified in positive mode. What follows is a description of the identification procedure taking hypaconitine ion as an example. Compound (VIP 7.2912, Table 1) exhibited [M + H]+ ions at m/z 616. It gave product ions of 584 [M + H–CH4O]+, 556 [M + H–C2H4O2]+, 524 [M + H–C3H8O3]+, 494 [M + H–C4H10O4]+, 105 [M + H–C26H41NO9]+ (Table 4). The molecular formula of the aconitine compound was determined as C33H45NO10 on the basis of UPLC-MS data and the analysis of its elemental composition and fractional isotope abundance. These data indicated that it was plausibly characterized as hypaconitine based on a database search (Fig. 7). According to the protocol above, 11 potential biomarkers between Fuzi and YFZ were identified (Table 1). They are hypaconitine, neoline, songorine, talatisamine, aconitine, 14-acetylkarakoline, 14-acetyl-talatisamine, fuziline, isotalatizidine, benzoylmesaconine, mesaconitine. 19 potential biomarkers were identified between Fuzi and HSP (Table 2), including mesaconitine, benzoylmesaconine, neoline, fuziline, benzolhypacone, 10-OH-mesaconitine, deoxyaconitine, isotalatizidine, hypaconitine, 14-acetyl-talatisamine, songorine, 8-acetyl-15-hydroxyneoline, benzoylaconine, aconitine, talatisamine, chasmanine, karacoline, karacolidine, 14-acetylkarakoline. 17 potential biomarkers were identified between Fuzi and BFP (Table 3), such as benzoylmesaconine, mesaconitine, benzolhypacone, hypaconitine, neoline, 10-OH-mesaconitine, fuziline, deoxyaconitine, benzoylaconine, 14-acetyl-talatisamine, isotalatizidine, aconitine, talatisamine, songorine, 14-acetylkarakoline, chasmanine, 8-acetyl-15-hydroxyneoline.
Table 1 Potential biomarkers identified between Fuzi and Yanfuzi in positive ESI modea
|
VIP
|
Rt |
m/z [M + H]+ |
Identified compounds |
Elemental composition |
Structure type |
Content variance |
| Measured m/z |
Theoretical m/z |
|
Note: ↑, content increased; ↓, content decreased. MDAs, monoester diterpene alkaloids; ADAs, amine diterpenoid alkaloids; DDAs, diester diterpene alkaloids.
|
| 7.2912 |
2.38 |
616.3051 |
616.3043 |
Hypaconitine
|
C33H45NO10 |
DDAs |
↑ |
| 4.5883 |
1.57 |
438.2761 |
438.2777 |
Neoline
|
C24H39NO6 |
ADAs
|
↑ |
| 3.8287 |
1.43 |
358.2317 |
358.2304 |
Songorine |
C22H31NO3 |
Veatchine |
↑ |
| 3.7463 |
1.67 |
422.2839 |
422.2828 |
Talatisamine
|
C24H39NO5 |
ADAs
|
↑ |
| 3.3732 |
2.38 |
646.3151 |
646.3149 |
Aconitine
|
C34H47NO11 |
DDAs |
↓ |
| 3.3519 |
1.60 |
420.2645 |
420.2672 |
14-Acetylkarakoline
|
C24H37NO5 |
MDAs |
↓ |
| 3.2856 |
1.90 |
464.2936 |
464.2934 |
14-Acetyl-talatisamine
|
C26H41NO6 |
MDAs |
↓ |
| 2.9356 |
1.52 |
454.2723 |
454.2727 |
Fuziline |
C24H39NO7 |
ADAs
|
↑ |
| 2.8417 |
1.36 |
408.2673 |
408.2672 |
Isotalatizidine
|
C23H37NO5 |
ADAs
|
↑ |
| 2.8241 |
2.12 |
590.2895 |
590.2883 |
Benzoylmesaconine
|
C31H43NO10 |
MDAs |
↓ |
| 2.3077 |
2.32 |
632.2998 |
632.2993 |
Mesaconitine
|
C33H45NO11 |
DDAs |
↓ |
Table 2 Potential biomarkers identified between Fuzi and Heishunpian in positive ESI modea
|
VIP
|
Rt |
m/z [M + H]+ |
Identified compounds |
Elemental composition |
Structure type |
Content variance |
| Measured m/z |
Theoretical m/z |
|
Note: ↑, content increased; ↓, content decreased. MDAs, monoester diterpene alkaloids; ADAs, amine diterpenoid alkaloids; DDAs, diester diterpene alkaloids.
|
| 6.6783 |
2.32 |
632.2998 |
632.2993 |
Mesaconitine
|
C33H45NO11 |
DDAs |
↓ |
| 6.4895 |
2.12 |
590.2885 |
590.2883 |
Benzoylmesaconine
|
C31H43NO10 |
MDAs |
↑ |
| 5.0967 |
1.57 |
438.2761 |
438.2777 |
Neoline
|
C24H39NO6 |
ADAs
|
↓ |
| 4.7443 |
1.52 |
454.2723 |
454.2727 |
Fuziline |
C24H39NO7 |
ADAs
|
↓ |
| 4.6270 |
2.23 |
574.2953 |
574.2938 |
Benzolhypacone
|
C31H43NO9 |
MDAs |
↑ |
| 3.6771 |
2.26 |
648.2942 |
648.2942 |
10-OH-mesaconitine
|
C33H45NO12 |
DDAs |
↓ |
| 3.2174 |
2.44 |
630.3209 |
630.3200 |
Deoxyaconitine
|
C34H47NO10 |
DDAs |
↓ |
| 3.1976 |
1.36 |
408.2673 |
408.2672 |
Isotalatizidine
|
C23H37NO5 |
ADAs
|
↓ |
| 3.1282 |
1.9 |
464.2936 |
464.2934 |
14-Acetyl-talatisamine
|
C26H41NO6 |
MDAs |
↓ |
| 2.9466 |
1.43 |
358.2314 |
358.2304 |
Songorine |
C22H31NO3 |
Veatchine |
↓ |
| 2.8202 |
2.21 |
604.3033 |
604.3043 |
Benzoylaconine
|
C32H45NO10 |
MDAs |
↑ |
| 2.7664 |
1.67 |
422.2819 |
422.2828 |
Talatisamine
|
C24H39NO5 |
ADAs
|
↓ |
| 2.6718 |
2.38 |
646.3151 |
646.3149 |
Aconitine
|
C34H47NO11 |
DDAs |
↓ |
| 2.3515 |
3.28 |
496.2889 |
496.2832 |
8-acetyl-15-hydroxyneoline
|
C26H41NO8 |
MDAs |
↑ |
| 2.3400 |
2.38 |
616.3051 |
616.3043 |
Hypaconitine
|
C33H45NO10 |
DDAs |
↓ |
| 2.2912 |
1.82 |
452.2936 |
452.2934 |
Chasmanine |
C25H41NO6 |
ADAs
|
↓ |
| 2.2846 |
1.36 |
378.2569 |
378.2566 |
Karacoline
|
C22H35NO4 |
ADAs
|
↓ |
| 2.2828 |
1.13 |
394.2514 |
394.2515 |
Karacolidine
|
C22H35NO5 |
ADAs
|
↓ |
| 2.2245 |
1.6 |
420.2645 |
420.2672 |
14-acetylkarakoline
|
C24H37NO5 |
MDAs |
↓ |
Table 3 Potential biomarkers identified between Fuzi and Baifupian in positive ESI modea
|
VIP
|
Rt |
m/z [M + H]+ |
Identified compounds |
Elemental composition |
Structure type |
Content variance |
| Measured m/z |
Theoretical m/z |
|
Note: ↑, content increased; ↓, content decreased. MDAs, monoester diterpene alkaloids; ADAs, amine diterpenoid alkaloids; DDAs, diester diterpene alkaloids.
|
| 7.6957 |
2.12 |
590.2899 |
590.2883 |
Benzoylmesaconine
|
C31H43NO10 |
MDAs |
↑ |
| 7.0069 |
2.32 |
632.2998 |
632.2993 |
Mesaconitine
|
C33H45NO11 |
DDAs |
↓ |
| 5.4621 |
2.23 |
574.2943 |
574.2938 |
Benzolhypacone
|
C31H43NO9 |
MDAs |
↑ |
| 4.4166 |
2.38 |
616.3051 |
616.3043 |
Hypaconitine
|
C33H45NO10 |
DDAs |
↓ |
| 3.9260 |
1.57 |
438.2761 |
438.2777 |
Neoline
|
C24H39NO6 |
ADAs
|
↓ |
| 3.8299 |
2.26 |
648.2942 |
648.2942 |
10-OH-mesaconitine
|
C33H45NO12 |
DDAs |
↓ |
| 3.7518 |
1.52 |
454.2723 |
454.2727 |
Fuziline |
C24H39NO7 |
ADAs
|
↓ |
| 3.7263 |
2.44 |
630.3209 |
630.3200 |
Deoxyaconitine
|
C34H47NO10 |
DDAs |
↓ |
| 3.4060 |
2.21 |
604.3066 |
604.3043 |
Benzoylaconine
|
C32H45NO10 |
MDAs |
↑ |
| 3.1107 |
1.9 |
464.2936 |
464.2934 |
14-Acetyl-talatisamine
|
C26H41NO6 |
MDAs |
↓ |
| 2.9309 |
2.38 |
646.3151 |
646.3149 |
Aconitine
|
C34H47NO11 |
DDAs |
↓ |
| 2.6283 |
1.67 |
422.2873 |
422.2828 |
Talatisamine
|
C24H39NO5 |
ADAs
|
↓ |
| 2.6138 |
1.36 |
408.2373 |
408.2672 |
Isotalatizidine
|
C23H37NO5 |
ADAs
|
↓ |
| 2.4717 |
1.43 |
358.2314 |
358.2304 |
Songorine |
C22H31NO3 |
Veatchine |
↓ |
| 2.1994 |
1.6 |
420.2645 |
420.2672 |
14-acetylkarakoline
|
C24H37NO5 |
MDAs |
↓ |
| 2.1964 |
1.82 |
452.2936 |
452.2934 |
Chasmanine |
C25H41NO6 |
ADAs
|
↓ |
| 2.1082 |
3.28 |
496.2899 |
496.2832 |
8-acetyl-15-hydroxyneoline
|
C26H41NO8 |
MDAs |
↑ |
3.3 Changes of relative intensity of metabolic biomarkers
PCA based on the metabolite accumulation patterns clearly showed differences among Fuzi and its processed products by UPLC-HDMS analysis. In examining the multivariate data in more detail, clear clustering into four parts from PCA was observed, indicating that Paozhi makes its components change. A comparison of metabolite profiles was conducted to determine the variation extent between Fuzi and its processed products. In order to more clearly characterize the action of Paozhi, the expression level of markers identified in the different processed groups was analyzed to illustrate how Paozhi exhibits detoxification (Fig. 8, 9 and 10). One advantage of the metabolomics approach is that it can give marker metabolites that contribute to the differentiation. At present, a comparison of the key metabolites in Fuzi revealed that relative concentrations of DDAs, i.e., aconitine, mesaconitine, hypaconitine, deoxyaconitine, 10-OH-mesaconitine, in HSP and BFP, were significantly decreased, while that of YFZ was increased (Fig. 8). It is already known that the detoxification of its roots is bound to a hydrolysis procedure. HSP and BFP is thoroughly boiled with salt solution or steamed over a few days. In general, the DDAs content of unprocessed roots is higher than that of processed roots. YFZ had lower concentrations of MDAs, i.e., benzolhypacone, benzoylmesaconine, benzolhypacone, and HSP and BFP had higher concentrations than did the Fuzi group. However, YFZ had higher concentrations of MDAs, i.e., 14-acetyl-talatisamine, 8-acetyl-15-hydroxyneolin, chasmanine, 14-acetylkarakolin, and HSP and BFP had lower concentrations than did the Fuzi group (Fig. 9). Compared to Fuzi and its processed products, the relative level of ADAs (except fuziline) in the processed HSP and BFP was increased (Fig. 10). These results showed that Paozhi could play a key role in detoxification. Metabolomic analysis represents the highest contributing signals for Paozhi that could decompose the poisonous DDAs into less or non-toxic derivatives.
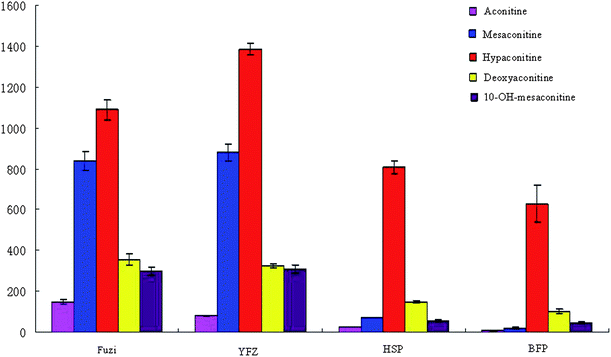 |
| | Fig. 8 Graphical representation of relative intensity of biomarkers diester diterpene alkaloids (DDAs) between Fuzi and its processed products. The graph displays significant changes in the metabolite level (relative changes; data are standardized). | |
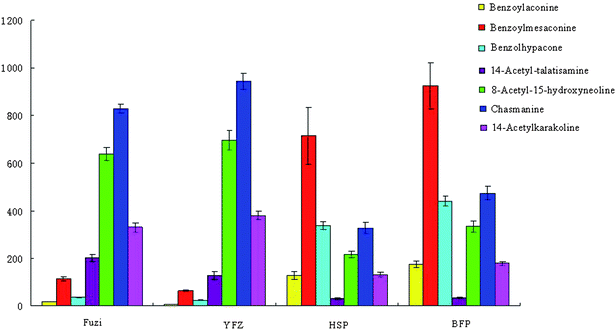 |
| | Fig. 9 The relative mean height intensity of biomarkers MDAs between Fuzi and its processed products. The graph displays significant changes in the metabolite level (relative changes; data are standardized). | |
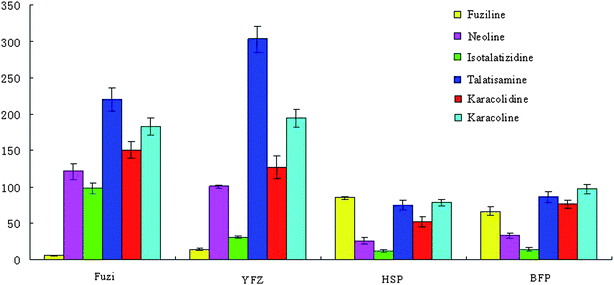 |
| | Fig. 10 Graphical representation of relative changes of biomarkers ADAs between Fuzi and its processed products. The graph displays significant changes in the metabolite level (relative changes; data are standardized). | |
In the post genomics era, metabolomics is fast emerging as a vital source of information to aid in solving systems biology puzzles with an emphasis on the metabolome of a biological sample.27–29 Coverage of the entire plant metabolome is a daunting task as it is estimated that there are over 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different metabolites within the plant kingdom.30 Metabolomics is a whole metabolite profiling approach for plant samples. It is often combined with multivariate statistical analysis to efficiently assess principal factors contributing to the phenotypic changes and to monitor the concentration changes of metabolite and profiles in response to environmental challenges.31,32 At present, metabolomics has provide information about the basic toxicity mechanism of Fuzi and its processed products, as well as potential metabolic biomarkers which can be used for safety evaluation processes.
000 different metabolites within the plant kingdom.30 Metabolomics is a whole metabolite profiling approach for plant samples. It is often combined with multivariate statistical analysis to efficiently assess principal factors contributing to the phenotypic changes and to monitor the concentration changes of metabolite and profiles in response to environmental challenges.31,32 At present, metabolomics has provide information about the basic toxicity mechanism of Fuzi and its processed products, as well as potential metabolic biomarkers which can be used for safety evaluation processes.
Conclusion
In this study, the comprehensive metabolomic characters of Fuzi and its processed products by UPLC-Q-TOF-HDMS combined with pattern recognition methods was investigated and highly desirable in order to guarantee the clinical safety and its low toxicity in decoctions. The present study proves that metabolomics based UPLC-Q-TOF-HDMS coupled with multivariate analyses successfully identify specific changes of metabolites between Fuzi and its processed preparations, and 19 key biomarkers which are responsible for the detoxifying measure of Paozhi were discovered. The underlying regulations of Paozhi-perturbed metabolic pathways were discussed according to the identified metabolites. The UPLC-Q-TOF-HDMS analysis revealed that the levels of these metabolites as biomarkers were significantly changed after Paozhi. Traditional Chinese processing approach, Paozhi can decompose the poisonous DDAs into less or non-toxic derivatives. Metabolomic analysis represents the highest contributing signals for the pre- and post-treatment. This method is fast and efficient, with more accurate characterization in the detoxification by traditional Paozhi. It will be helpful to investigate Fuzi in toxicology and pharmacology. Further, it can be expected that the Paozhi approach provides an effective and essential tool for detoxification of Chinese toxicity medicines.
Table 4 Main MS/MS fragmentations of potential biomarkers of Fuzi and its processed products in positive modea
| No |
Compound |
Proposed structure |
MS/MS
|
Type |
|
Note: MDAs, monoester diterpene alkaloids; ADAs, amine diterpenoid alkaloids; DDAs, diester diterpene alkaloids.
|
| 1 |
Benzoylmesaconine
|
|
590[M+H]+ |
MDAs |
| |
|
572[M+H–H2O]+ |
|
| |
|
558[M+H–CH4O]+ |
|
| |
|
540[M+H–CH6O2]+ |
|
| |
|
508[M+H–C2H10O3]+ |
|
| |
|
386[M+H–C9H16O5]+ |
|
| |
|
105[M+H–C24H39NO9]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 2 |
Mesaconitine
|
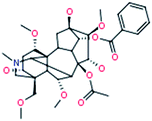
|
632[M+H]+ |
DDAs |
| |
|
602[M+H–CH2O]+ |
|
| |
|
572[M+H–C2H4O2]+ |
|
| |
|
540[M+H–C3H8O3]+ |
|
| |
|
512[M+H–C4H8O4]+ |
|
| |
|
496[M+H–C5H12O4]+ |
|
| |
|
390[M+H–C11H14O6]+ |
|
| |
|
105[M+H–C26H41NO10]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 3 |
Benzolhypacone
|
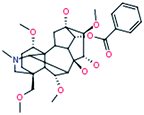
|
574[M+H]+ |
MDAs |
| |
|
542[M+H–CH4O]+ |
|
| |
|
510[M+H–C2H8O2]+ |
|
| |
|
386[M+H–C8H12O5]+ |
|
| |
|
105[M+H–C24H39NO8]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 4 |
Hypaconitine
|
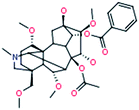
|
616[M+H]+ |
DDAs |
| |
|
584[M+H–CH4O]+ |
|
| |
|
556[M+H–C2H4O2]+ |
|
| |
|
524[M+H–C3H8O3]+ |
|
| |
|
494[M+H–C4H10O4]+ |
|
| |
|
105[M+H–C26H41NO9]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 5 |
Neoline
|
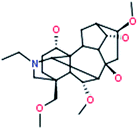
|
438[M+H]+ |
ADAs
|
| |
|
420[M+H–H2O]+ |
|
| |
|
402[M+H–H4O2]+ |
|
| |
|
388[M+H–CH6O2]+ |
|
| |
|
370[M+H–CH8O3]+ |
|
| |
|
342[M+H–C3H12O3]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 6 |
10-OH-Mesaconitine
|
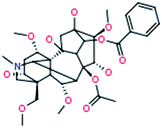
|
648[M+H]+ |
DDAs |
| |
|
616[M+H–CH4O]+ |
|
| |
|
598[M+H–CH6O2]+ |
|
| |
|
590[M+H–C2H2O2]+ |
|
| |
|
556[M+H–C3H8O3]+ |
|
| |
|
538[M+H–C3H10O4]+ |
|
| |
|
528[M+H–C4H8O4]+ |
|
| |
|
506[M+H–C4H14O5]+ |
|
| |
|
105[M+H–C26H41NO11]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 7 |
Fuziline |
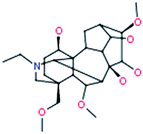
|
454[M+H]+ |
ADAs
|
| |
|
436[M+H–H2O]+ |
|
| |
|
422[M+H–CH4O]+ |
|
| |
|
408[M+H–C2H6O]+ |
|
| |
|
404[M+ H–CH6O2]+ |
|
| |
|
374[M+H–C2H8O3]+ |
|
| |
|
344[M+H–C4H14O3]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 8 |
Deoxyaconitine
|
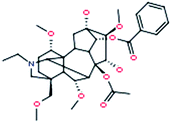
|
630[M+H]+ |
DDAs |
| |
|
570[M+H–C2H4O2]+ |
|
| |
|
538[M+H–C3H8O3]+ |
|
| |
|
510[M+H–C4H8O4]+ |
|
| |
|
478[M+H–C5H12O5]+ |
|
| |
|
416[M+H–C10H14O5]+ |
|
| |
|
388[M+H–C11H14O6]+ |
|
| |
|
105[M+H–C27H43NO9]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 9 |
Benzoylaconine
|
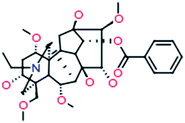
|
604[M+H]+ |
MDAs |
| |
|
586[M+H–H2O]+ |
|
| |
|
572[M+H–CH4O]+ |
|
| |
|
496[M+H–C4H12O3]+ |
|
| |
|
105[M+H–C25H41NO9]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 10 |
14-Acetyl-talatisamine
|
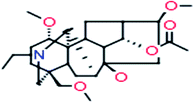
|
464[M+H]+ |
MDAs |
| |
|
432[M+H–CH4O]+ |
|
| |
|
372[M+H–C4H12O2]+ |
|
| |
|
314[M+H–C6H14O4]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 11 |
Aconitine
|
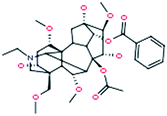
|
646[M+H]+ |
DDAs |
| |
|
586[M+H–C2H4O2]+ |
|
| |
|
556[M+H–C3H6O3]+ |
|
| |
|
522[M+H–C4H12O4]+ |
|
| |
|
404[M+H–C11H14O6]+ |
|
| |
|
105[M+H–C27H43NO10]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 12 |
Talatisamine
|
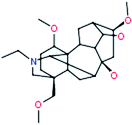
|
422[M+H]+ |
ADAs
|
| |
|
390[M+H–CH4O]+ |
|
| |
|
358[M+H–C2H8O2]+ |
|
| |
|
344[M+H–C3H10O2]+ |
|
| |
|
326[M+H–C3H12O3]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 13 |
Isotalatizidine
|
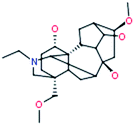
|
408[M+H]+ |
ADAs
|
| |
|
390[M+H–H2O]+ |
|
| |
|
358[M+H–CH6O2]+ |
|
| |
|
340[M+H–CH8O3]+ |
|
| |
|
328[M+H–C2H8O3]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 14 |
Songorine |
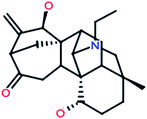
|
358[M+H]+ |
Veatchine |
| |
|
340[M+H–H2O]+ |
|
| |
|
330[M+H–C2H4]+ |
|
| |
|
322[M+H–H4O2]+ |
|
| |
|
298[M+H–C3H8O]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 15 |
14-Acetylkarakoline
|
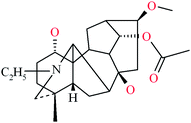
|
420[M+H]+ |
MDAs |
| |
|
402[M+H–H2O]+ |
|
| |
|
388[M+H–CH4O]+ |
|
| |
|
370[M+H–CH6O2]+ |
|
| |
|
356[M+H–C2H8O2]+ |
|
| |
|
340[M+H–C2H8O3]+ |
|
| |
|
328[M+H–C4H12O2]+ |
|
| |
|
398[M+H–C4H10O4]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 16 |
Chasmanine |
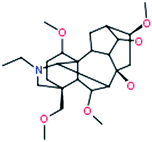
|
452[M+H]+ |
ADAs
|
| |
|
420[M+H–CH4O]+ |
|
| |
|
388[M+H–C2H8O2]+ |
|
| |
|
370[M+H–C2H10O3]+ |
|
| |
|
328[M+H–C5H16O3]+ |
|
| |
|
312[M+H–C5H16O4]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 17 |
8-Acetyl-15-hydroxyneoline
|
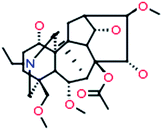
|
496[M+H]+ |
MDAs |
| |
|
436[M+H–C2H4O2]+ |
|
| |
|
404[M+H–C3H8O3]+ |
|
| |
|
376[M+H–C4H8O4]+ |
|
| |
|
360[M+H–C4H8O5]+ |
|
| |
|
358[M+H–C4H10O5]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 18 |
Karacoline
|
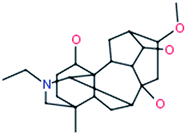
|
378[M+H]+ |
ADAs
|
| |
|
360[M+H–H2O]+ |
|
| |
|
342[M+H–H4O2]+ |
|
| |
|
328[M+H–C2H10O]+ |
|
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| 19 |
Karacolidine
|
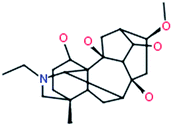
|
376[M+H]+ |
ADAs
|
| |
|
358[M+H–H4O2]+ |
|
| |
|
328[M+H–CH6O3]+ |
|
| |
|
322[M+H–C4H8O]+ |
|
Abbreviations
| UPLC-Q-TOF-HDMS | ultra-performance liquid-chromatography/electrospray-ionization synapt high-definition mass spectrometry |
|
TCM
| traditional Chinese medicine |
| Fuzi | lateral root of Aconitum carmichaelii Debx |
|
CW
| Chuanwu |
| DDAs |
diester
diterpene alkaloids
|
| MDAs |
monoester
diterpene alkaloids
|
|
ADAs
|
Amine
diterpenoid
alkaloids
|
|
YFZ
| Yanfuzi |
|
HSP
| Heishunpian |
|
BFP
| Baifupian |
| CP | Chinese Pharmacopoeia |
|
PCA
| the principal component analysis |
|
PLS-DA
| partial least squares-discriminant analysis |
| OPLS | orthogonal projection to latent structures analysis |
|
BPI
| based peak intensity |
|
VIP
| variable importance plot. |
Acknowledgements
This work was supported by grants from the Key Program of the Natural Science Foundation of the State (Grant No. 90709019), the National Key Program on the Subject of Drug Innovation (Grant No. 2009ZX09502-005), the National Specific Program on the Subject of Public Welfare (Grant No. 200807014), and the National Program for Key Basic Research Projects in China (Grant No. 2005CB523406).
References
- D. Normile, Asian medicine, the new face of traditional Chinese medicine, Science, 2003, 299, 188–190 CrossRef CAS.
- R. Stone, Biochemistry. Lifting the veil on traditional Chinese medicine, Science, 2008, 319, 709–710 CrossRef CAS.
- Y. Yuan, Y. Wang, R. Xu, M. Huang and H. Zeng, Application of ionic liquids in the microwave-assisted extraction of podophyllotoxin from Chinese herbal medicine, Analyst, 2011, 136, 2294–2305 RSC.
- R. Kawasaki, W. Motoya, T. Atsumi, C. Mouri, N. Kakiuchi and M. Mikage, The relationship between growth of the aerial part and alkaloid content variation in cultivated Aconitum carmichaeli Debeaux, J. Nat. Med., 2011, 65, 111–115 CrossRef CAS.
- J. Singhuber, M. Zhu, S. Prinz and B. Kopp, Aconitum in traditional Chinese medicine: a valuable drug or an unpredictable risk?, J. Ethnopharmacol., 2009, 126, 18–30 CrossRef.
- Y. Wu, Z. Y. Xia, Q. T. Meng, J. Zhu, S. Lei, J. Xu and J. Dou, Shen-Fu injection preconditioning inhibits myocardial ischemia-reperfusion injury in diabetic rats: activation of eNOS via the PI3K/Akt pathway, J. Biomed. Biotechnol., 2011, 384627 Search PubMed.
- G. Yan, H. Sun, W. Sun, L. Zhao, X. Meng and X. Wang, Rapid and global detection and characterization of aconitum alkaloids in Yin Chen Si Ni Tang, a traditional Chinese medical formula, by ultra performance liquid chromatography-high resolution mass spectrometry and automated data analysis, J. Pharm. Biomed. Anal., 2010, 53, 421–431 CrossRef CAS.
- D. Shaw, Toxicological risks of Chinese herbs, Planta Med., 2010, 76, 2012–2018 CrossRef CAS.
- B. Sun, L. Li, S. Wu, Q. Zhang, H. Li, H. Chen, F. Li, F. Dong and X. Yan, Metabolomic analysis of biofluids from rats treated with Aconitum alkaloids using nuclear magnetic resonance and gas chromatography/time-of-flight mass spectrometry, Anal. Biochem., 2009, 395, 125–133 CrossRef CAS.
- M. Liu, S. Zhang, C. Yang, Y. Xia, J. Liu and J. Liang, Isolation and structure identification of C19 diterpenoid alkaloids from Aconitum carmichaeli by high-speed counter-current chromatography, SePu, 2011, 29, 430–434 CAS.
- R. Hu, J. Zhao, L. W. Qi, P. Li, S. L. Jing and H. J. Li, Structural characterization and identification of C(19)- and C(20)-diterpenoid alkaloids in roots of Aconitum carmichaeli by rapid-resolution liquid chromatography coupled with time-of-flight mass spectrometry, Rapid Commun. Mass Spectrom., 2009, 23, 1619–1635 CrossRef CAS.
- G. Lu, Z. Dong, Q. Wang, G. Qian, W. Huang, Z. Jiang, K. S. Leung and Z. Zhao, Toxicity assessment of nine types of decoction pieces from the daughter root of Aconitum carmichaeli (Fuzi) based on the chemical analysis of their diester diterpenoid alkaloids, Planta Med., 2010, 76, 825–830 CrossRef CAS.
- G. Cao, H. Cai, Y. Zhang, X. Cong, C. Zhang and B. Cai, Identification of metabolites of crude and processed Fructus Corni in rats by microdialysis sampling coupled with electrospray ionization linear quadrupole ion trap mass spectrometry, J. Pharm. Biomed. Anal., 2011, 56, 118–125 CrossRef CAS.
- A. Sreekumar, L. M. Poisson, T. M. Rajendiran, A. P. Khan, Q. Cao, J. Yu, B. Laxman, R. Mehra, R. J. Lonigro, Y. Li, M. K. Nyati, A. Ahsan, S. Kalyana-Sundaram, B. Han, X. Cao, J. Byun, G. S. Omenn, D. Ghosh, S. Pennathur, D. C. Alexander, A. Berger, J. R. Shuster, J. T. Wei, S. Varambally, C. Beecher and A. M. Chinnaiyan, Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression, Nature, 2009, 457, 910–914 CrossRef CAS.
- A. Zhang, H. Sun, Z. Wang, W. Sun, P. Wang and X. Wang, Metabolomics: towards understanding traditional Chinese medicine, Planta Med., 2010, 76, 2026–2035 CrossRef CAS.
- A. E. Allen, C. L. Dupont, M. Oborník, A. Horák, A. Nunes-Nesi, J. P. McCrow, H. Zheng, D. A. Johnson, H. Hu, A. R. Fernie and C. Bowler, Evolution and metabolic significance of the urea cycle in photosynthetic diatoms, Nature, 2011, 473, 203–207 CrossRef CAS.
- X. Wang, H. Sun, A. Zhang, W. Sun, P. Wang and Z. Wang, Potential role of metabolomics apporoaches in the area of traditional Chinese medicine: as pillars of the bridge between Chinese and Western medicine, J. Pharm. Biomed. Anal., 2011, 55, 859–868 CrossRef CAS.
- A. Sreekumar, L. M. Poisson, T. M. Rajendiran, A. P. Khan, Q. Cao, J. Yu, B. Laxman, R. Mehra, R. J. Lonigro, Y. Li, M. K. Nyati, A. Ahsan, S. Kalyana-Sundaram, B. Han, X. Cao, J. Byun, G. S. Omenn, D. Ghosh, S. Pennathur, D. C. Alexander, A. Berger, J. R. Shuster, J. T. Wei, S. Varambally, C. Beecher and A. M. Chinnaiyan, Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression, Nature, 2009, 457, 910–914 CrossRef CAS.
- L. Lin, Q. Yu, X. Yan, W. Hang, J. Zheng, J. Xing and B. Huang, Direct infusion mass spectrometry or liquid chromatography mass spectrometry for human metabonomics? A serum metabonomic study of kidney cancer, Analyst, 2010, 135, 2970–2978 RSC.
- F. Courant, G. Pinel, E. Bichon, F. Monteau, J. P. Antignac and B. Le Bizec, Development of a metabolomic approach based on liquid chromatography-high resolution mass spectrometry to screen for clenbuterol abuse in calves, Analyst, 2009, 134, 1637–1646 RSC.
- C. Junot, G. Madalinski, J. C. Tabet and E. Ezan, Fourier transform mass spectrometry for metabolome analysis, Analyst, 2010, 135, 2203–2219 RSC.
- L. Li, B. Sun, Q. Zhang, J. Fang, K. Ma, Y. Li, H. Chen, F. Dong, Y. Gao, F. Li and X. Yan, Metabonomic study on the toxicity of Hei-Shun-Pian, the processed lateral root of Aconitum carmichaelii Debx. (Ranunculaceae), J. Ethnopharmacol., 2008, 116, 561–568 CrossRef.
- B. Sun, S. Wu, L. Li, H. Li, Q. Zhang, H. Chen, F. Li, F. Dong and X. Yan, A metabolomic analysis of the toxicity of Aconitum sp. alkaloids in rats using gas chromatography/mass spectrometry, Rapid Commun. Mass Spectrom., 2009, 23, 1221–1228 CrossRef CAS.
- D. H. Kim, R. M. Jarvis, Y. Xu, A. W. Oliver, J. W. Allwood, L. Hampson, I. N. Hampson and R. Goodacre, Combining metabolic fingerprinting and footprinting to understand the phenotypic response of HPV16 E6 expressing cervical carcinoma cells exposed to the HIV anti-viral drug lopinavir, Analyst, 2010, 135, 1235–1244 RSC.
- S. Zhang, G. A. Nagana Gowda, T. Ye and D. Raftery, Advances in NMR-based biofluid analysis and metabolite profiling, Analyst, 2010, 135, 1490–1498 RSC.
- M. Basanta, R. M. Jarvis, Y. Xu, G. Blackburn, R. Tal-Singer, A. Woodcock, D. Singh, R. Goodacre, C. L. Thomas and S. J. Fowler, Non-invasive metabolomic analysis of breath using differential mobility spectrometry in patients with chronic obstructive pulmonary disease and healthy smokers, Analyst, 2010, 135, 315–320 RSC.
- A. K. Smilde, M. J. van der Werf, J. P. Schaller and C. Kistemaker, Characterizing the precision of mass- spectrometry-based metabolic profiling platforms, Analyst, 2009, 134, 2281–2285 RSC.
- M. Brown, W. B. Dunn, P. Dobson, Y. Patel, C. L. Winder, S. Francis-McIntyre, P. Begley, K. Carroll, D. Broadhurst, A. Tseng, N. Swainston, I. Spasic, R. Goodacre and D. B. Kell, Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics, Analyst, 2009, 134, 1322–1332 RSC.
- P. Gao, C. Lu, F. Zhang, P. Sang, D. Yang, X. Li, H. Kong, P. Yin, J. Tian, X. Lu, A. Lu and G. Xu, Integrated GC-MS and LC-MS plasma metabonomics analysis of ankylosing spondylitis, Analyst, 2008, 133, 1214–1220 RSC.
- A. Koglin, F. Löhr, F. Bernhard, V. V. Rogov, D. P. Frueh, E. R. Strieter, M. R. Mofid, P. Güntert, G. Wagner, C. T. Walsh, M. A. Marahiel and V. Dötsch, Structural basis for the selectivity of the external thioesterase of the surfactin synthetase, Nature, 2008, 454, 907–911 CrossRef CAS.
- H. Winning, E. Roldán-Marín, L. O. Dragsted, N. Viereck, M. Poulsen, C. Sánchez-Moreno, M. P. Cano and S. B. Engelsen, An exploratory NMR nutri-metabonomic investigation reveals dimethyl sulfone as a dietary biomarker for onion intake, Analyst, 2009, 134, 2344–2351 RSC.
- Y. Chen, R. Zhang, Y. Song, J. He, J. Sun, J. Bai, Z. An, L. Dong, Q. Zhan and Z. Abliz, RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: finding potential biomarkers for breast cancer, Analyst, 2009, 134, 2003–2011 RSC.
|
| This journal is © The Royal Society of Chemistry 2012 |
Click here to see how this site uses Cookies. View our privacy policy here. 
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C. The supernatant diluted 50 times with methanol was then filtered through a 0.22 μm-filter, the filtrate was used for UPLC analysis.
000 rpm at 4 °C. The supernatant diluted 50 times with methanol was then filtered through a 0.22 μm-filter, the filtrate was used for UPLC analysis.








![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different metabolites within the plant kingdom.30 Metabolomics is a whole metabolite profiling approach for plant samples. It is often combined with multivariate statistical analysis to efficiently assess principal factors contributing to the phenotypic changes and to monitor the concentration changes of metabolite and profiles in response to environmental challenges.31,32 At present, metabolomics has provide information about the basic toxicity mechanism of Fuzi and its processed products, as well as potential metabolic biomarkers which can be used for safety evaluation processes.
000 different metabolites within the plant kingdom.30 Metabolomics is a whole metabolite profiling approach for plant samples. It is often combined with multivariate statistical analysis to efficiently assess principal factors contributing to the phenotypic changes and to monitor the concentration changes of metabolite and profiles in response to environmental challenges.31,32 At present, metabolomics has provide information about the basic toxicity mechanism of Fuzi and its processed products, as well as potential metabolic biomarkers which can be used for safety evaluation processes.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

