 Open Access Article
Open Access ArticleA modified guanosine phosphoramidite for click functionalization of RNA on the sugar edge†‡§
Salifu
Seidu-Larry
,
Bettina
Krieg
,
Markus
Hirsch
,
Mark
Helm
and
Olwen
Domingo
*
Institute of Pharmacy and Biochemistry, Johannes Gutenberg University, 5 Staudinger Weg, Mainz, D-55128, Germany. E-mail: dominol@uni-mainz.de; Fax: +49 6131 39 20373; Tel: +49 6131 39 24352
First published on 18th September 2012
Abstract
A propargyl containing guanosine phosphoramidite was synthesized and incorporated into siRNA, enabling click-ligation with an azido fluorophore onto the nucleobase sugar edge. Duplex stability was not affected by labeling at this new site, which allowed deconvolution of the effects of label, structure and attachment site on RNAi activity.
The labeling of biopolymers such as RNA is of outstanding importance for investigations into structure–function relationships.1 The attachment of fluorescent labels may, however, influence both structure and function of the RNA, depending on whether it involves the Watson–Crick, the Hoogsteen or the sugar edge of a given nucleoside. In the case of simple duplexes the secondary structure is essentially restricted to the Watson–Crick edge. Hence, dye labeling directly onto the Watson–Crick face of a given nucleobase is expected to be detrimental to duplex formation. On the other hand, popular labeling strategies on the Hoogsteen edge focus mainly on the N7 or C8 of purines2–4 or the C5 position of pyrimidines5,6 and although labeling directly onto the sugar edge is a promising approach, such attachment points are so far restricted to the 2′-position of the ribose.7,8 Such derivatization is, however, known to heavily affect the sugar pucker and thus also stability and hybridization properties of the RNA duplex. In contrast, attachment of labels on the sugar edge at positions other than the 2′-position of the ribose has not yet been explored.
With the exception of the rare methyl or methylthio-modifications of adenine on position 2,9–11 guanine is the only nucleobase that carries naturally occurring modifications on the sugar edge. Since such modifications occur naturally in Watson–Crick helices12 this suggests guanosine as a promising candidate, where chemical labeling at this position may be expected to minimally alter its hybridization properties. Reports have been published with regard to the effect of methylation of the exocyclic N2 amine of guanosine on base pairing properties.
Altogether, this modification seems to have only a minimal impact and a significant difference is first observed with double methylation on position N2.7,12–14
We therefore decided to explore labeling of RNA at the guanosine sugar edge in an siRNA model system. Research associated with RNA interference,15 a process that moderates gene expression in cells, is often accompanied by fluorescent labeling of the siRNAs. This approach makes the intracellular tracing of the siRNAs possible.16 The 5′-end of the antisense strand is a particular target for positional labeling on the sugar edge. Because the 5'OH is phosphorylated in vivo, methylation of this OH-group has been reported to ablate RNAi activity.17,18
To investigate in detail the effect of 5′-siRNA labeling, we designed and synthesized a phosphoramidite building block for RNA synthesis, bearing a moiety whose alkyne functional group would enable CuAAC-type labeling with an azide, a method that has been used to great effect in bioconjugation chemistry between fluorescent dyes and RNA/DNA.21–27 Here, the exocyclic N2 of guanosine was our target for a diazotization reaction, followed by a nucleophilic substitution by a fluoride ion.14,19,20,28
The alkyne moiety was introduced via nucleophilic substitution of the incorporated fluoride by propargylamine. The subsequent transformation of this propargyl containing guanosine derivative into its phosphoramidite (Fig. 1a) was followed by its incorporation onto an siRNA antisense strand (5′-end) during solid phase oligonucleotide synthesis. The click product of the resulting strand with an Atto 590 azide dye was HPLC purified and hybridized to a commercially available sense strand (either unlabeled or containing an Atto 488 dye on its 3′-end), as is illustrated in Fig. 1b.
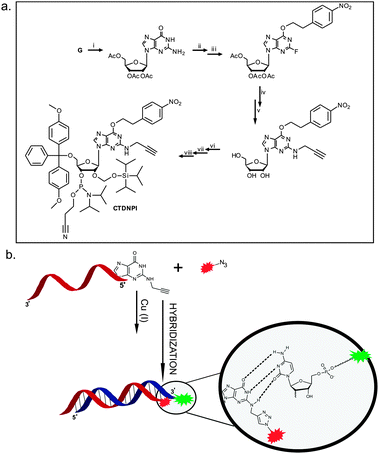 | ||
| Fig. 1 (a) Eight step synthetic route to the final alkyne functionalized phosphoramidite (CTDNPI): (i) peracetylation of guanosine, (ii) ether synthesis via the Mitsunobu reaction, (iii) diazotization of the aromatic amine and nucleophilic substitution by the fluoride ion,14,19,20 (iv) nucleophilic substitution of the fluoride ion by propargylamine and concomitant hydrolysis, (v) 5′-O-DMT protection, (vi) 2′-O-TOM protection and (vii) 3′-O-phosphitylation. (b) Click reaction between the Atto 590 azide and the alkyne functionalized oligonucleotide, followed by a hybridization step with the labeled sense strand, showing reserved base pairing between the modified G and the C of the sense strand. | ||
For validation of our labeling method, structural and functional data were acquired, starting with absorbance melting measurements of the RNA duplexes. These provided information on the effect of the labeling method on the thermal stability of the duplex. Fig. 2 shows that the temperature-dependent differential UV-absorption measurements of the siRNA double strands showed only a very slight difference in the melting temperatures of labeled strands, as compared to the unlabeled strands. A small destabilization, caused by the fluorescent labeling on the 5′-end of the RNA, resulted in a melting temperature difference of a mere 1 °C. In order to clarify whether this was due to the dye in general, or the labeling on the sugar edge in particular, a comparison was made between our construct and an antisense strand labeled via its 5′ end. Resulting similar Tm values of the two constructs indicate that labeling on the N2 of guanosine has the same little structural impact as has the labeling on the 5′-position, although the labels are attached at two completely different sites of the nucleotide. It can be concluded that the observed effects are due to the presence of the label on the 5′-nucleotide as such and are not associated specifically with our new labeling position.
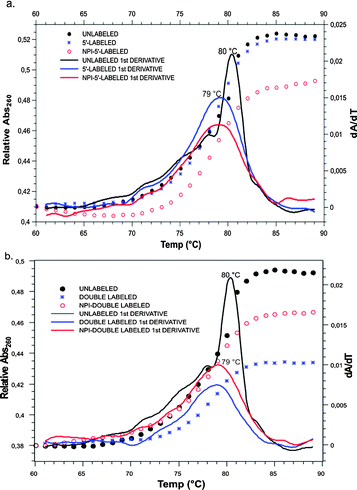 | ||
| Fig. 2 Comparison of the UV melting curves of the unlabeled siRNA duplex (black) with those of the (a) single labeled and (b) double labeled constructs. | ||
In addition, we added an Atto 488 label on the 3′-end of the opposing strand, in order to determine if fluorescent labeling at this particular domain of siRNA is affecting melting in general (Fig. 2b). Once again, the Tm did not change upon introduction of the label. As a result, labeled constructs thus obtained are suitable for in vivo tracking of intact siRNA during live cell imaging.29 Imaging of siRNA labeled on the sugar edge (Fig. S6, ESI§) shows that transfection, cellular uptake and intracellular distribution are unimpeded.
We applied our new labeling scheme to investigate the effect of labeling on RNAi activity. Literature on the effect of 5′-labeling of the antisense strand is controversial,30,31 with assessments ranging from “no effect”31 to “complete loss of RNAi activity”.32–34 Because this type of labeling is important for a FRET-based imaging technique of intact siRNA developed in our lab,29 we decided to quantitatively address this question by determining IC50-values in an established reporter gene assay, using the eGFP reporter gene.15,16,29 The expression of the enhanced green fluorescent protein (eGFP) in HEK cells was monitored by FACS at varying concentrations of the different siRNA double strands. As shown in Fig. 3a, attachment of dye to the 3′ end of the sense strand did not alter the IC50. However, attachment to the 5′ end of the antisense strand of various dyes, such as TAMRA, Atto 590 and Atto 647N, via the phosphate (chemical structure shown in Fig. S7, ESI§) did increase the IC50 by ∼5-, 10- and 12-fold, respectively, thereby roughly correlating with the molecular weight of the dye. Especially the only 5-fold decrease due to the TAMRA-label shows that labeling at the 5′ end is mildly detrimental, but does by no means completely abolish RNAi activity. As mentioned above, this labeling site was considered critical because methylation of the 5'OH ablated RNAi activity.17,18 The question then arises, if the observed mild impediment results from the labeling chemistry, i.e. if the attachment of the spacer onto the 5′-phosphate makes a major contribution to the increased IC50. To answer this question, we used our new phosphoramidite. As shown in Fig. 3b, the corresponding siRNA, sugar edge-labeled with Atto 590 surprisingly displays an IC50, which is increased by a factor of 50 compared to the reference and by a factor of 5 compared to the phosphate labeled with the same dye. Thus, avoiding the 5'OH of the ribose for labeling is not necessarily advantageous. Both the structure of the dye as well as the attachment site have an effect on RNAi activity, with the latter effect more pronounced.
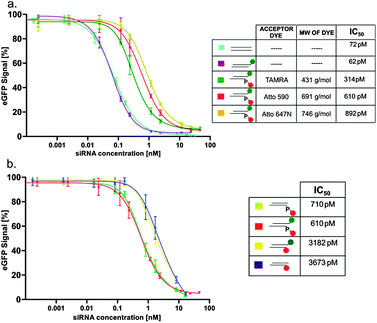 | ||
Fig. 3 Comparison of the concentration dependent knockdown efficiencies of various constructs on the eGFP signal: (a) ( ) unlabeled, ( ) unlabeled, ( ) only 3′-labeled and ( ) only 3′-labeled and ( ) double labeled, with different acceptor dyes on the 5′-end; (b) ( ) double labeled, with different acceptor dyes on the 5′-end; (b) ( ) only 5′-labeled via phosphate linkage, ( ) only 5′-labeled via phosphate linkage, ( ) double labeled via phosphate linkages, ( ) double labeled via phosphate linkages, ( ) double labeled, with the 5′-label on the guanosine sugar edge and ( ) double labeled, with the 5′-label on the guanosine sugar edge and ( ) only 5′-labeled via the guanosine sugar edge. ) only 5′-labeled via the guanosine sugar edge. | ||
In summary, we have explored the sugar edge of the guanine nucleobase for derivatization by synthesizing CTDNPI, a phosphoramidite, which, after incorporation into RNA via solid phase synthesis, offers an alkynyl moiety for functionalization via CuAAC. This functionalization is, however, otherwise inert. The building block was used in structure–function studies, probing a new labeling site at the 5′-end of the antisense strand in siRNA, offering a new technique for dual labeling in FRET-based experiments. Our quantitative assessment of the effect of 5′-labeling on RNAi activity shows that all used 5′-labeled siRNAs are still highly potent despite the labels. Our new phosphoramidite has allowed us to determine that the mild increase in IC50 caused by 5′-labeling of the siRNA antisense strand does not depend on the attachment site, but rather is caused by the presence of the dye in the approximate vicinity of the 5′-nucleotide. Since larger dyes impede RNAi activity more strongly, the effects are likely the result of a steric clash of the dye with protein factors of the RNAi pathway.
Support by Dr Malte Paulsen and Dr Andreas Vonderheit of the IMB Core Facilities Cytometry and Microscopy, respectively, is gratefully acknowledged. The authors also wish to acknowledge Professor Andres Jäschke and Mr Heiko Rudi at the IPMB (University of Heidelberg) for the MALDI-TOF measurements.
Notes and references
- L. M. Hall, M. Gerowska and T. Brown, Nucleic Acids Res., 2012, 40, e108 CrossRef CAS.
- N. M. Thomsen, V. Vongsutilers and P. M. Gannett, Crit. Rev. Eukaryotic Gene Expression, 2011, 21, 155–176 CrossRef CAS.
- F. Jordan and H. Niv, Biochim. Biophys. Acta, 1977, 476, 265–271 CrossRef CAS.
- A. St Clair, G. Xiang and L. W. McLaughlin, Nucleosides Nucleotides, 1998, 17, 925–937 CAS.
- W. A. Cristofoli, L. I. Wiebe, E. De Clercq, G. Andrei, R. Snoeck, J. Balzarini and E. E. Knaus, J. Med. Chem., 2007, 50, 2851–2857 CrossRef CAS.
- D. J. Hurley, S. E. Seaman, J. C. Mazura and Y. Tor, Org. Lett., 2002, 4, 2305–2308 CrossRef CAS.
- C. Höbartner, H. Mittendorfer, K. Breuker and R. Micura, Angew. Chem., 2004, 43, 3922–3925 Search PubMed.
- C. Kreutz, H. Kahlig, R. Konrat and R. Micura, J. Am. Chem. Soc., 2005, 127, 11558–11559 CrossRef CAS.
- W. J. Burrows, D. J. Armstrong, F. Skoog, S. M. Hecht, J. T. Boyle, N. J. Leonard and J. Occolowitz, Science, 1968, 161, 691–693 CAS.
- S. M. Hecht, N. J. Leonard, W. J. Burrows, D. J. Armstrong, F. Skoog and J. Occolowitz, Science, 1969, 166, 1272–1274 CAS.
- D. M. Reddy, P. F. Crain, C. G. Edmonds, R. Gupta, T. Hashizume, K. O. Stetter, F. Widdel and J. A. McCloskey, Nucleic Acids Res., 1992, 20, 5607–5615 CrossRef CAS.
- A. Y. Kobitski, M. Hengesbach, S. Seidu-Larry, K. Dammertz, C. S. Chow, A. van Aerschot, G. U. Nienhaus and M. Helm, Chem. Biol., 2011, 18, 928–936 CrossRef CAS.
- P. S. Pallan, C. Kreutz, S. Bosio, R. Micura and M. Egli, RNA, 2008, 14, 2125–2135 CrossRef CAS.
- J. P. Rife, C. S. Cheng, P. B. Moore and S. A. Strobel, Nucleic Acids Res., 1998, 26, 3640–3644 CrossRef CAS.
- T. N. Campbell and F. Y. Choy, Genet. Mol. Res., 2004, 3, 282–287 CAS.
- A. Järve, J. Muller, I. H. Kim, K. Rohr, C. MacLean, G. Fricker, U. Massing, F. Eberle, A. Dalpke, R. Fischer, M. F. Trendelenburg and M. Helm, Nucleic Acids Res., 2007, 35, e124 CrossRef.
- P. Y. Chen, L. Weinmann, D. Gaidatzis, Y. Pei, M. Zavolan, T. Tuschl and G. Meister, RNA, 2008, 14, 263–274 CrossRef CAS.
- D. S. Schwarz, G. Hutvagner, B. Haley and P. D. Zamore, Mol. Cell, 2002, 10, 537–548 CrossRef CAS.
- J. F. Gerster and R. K. Robins, J. Am. Chem. Soc., 1965, 87, 3752–3759 CrossRef CAS.
- R. Micura, C. Hobartner, R. Rieder, C. Kreutz, B. Puffer, K. Lang and H. Moroder, Current protocols in nucleic acid chemistry, ed. S. L. Beaucage, et al., 2007, ch. 1, Unit 1 15 Search PubMed.
- A. H. El-Sagheer and T. Brown, Chem. Soc. Rev., 2010, 39, 1388–1405 RSC.
- K. Fauster, M. Hartl, T. Santner, M. Aigner, C. Kreutz, K. Bister, E. Ennifar and R. Micura, ACS Chem. Biol., 2012, 7, 581–589 CrossRef CAS.
- C. Y. Jao and A. Salic, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15779–15784 CrossRef CAS.
- F. Seela, V. R. Sirivolu and P. Chittepu, Bioconjugate Chem., 2008, 19, 211–224 CrossRef CAS.
- T. S. Seo, Z. Li, H. Ruparel and J. Ju, J. Org. Chem., 2003, 68, 609–612 CrossRef CAS.
- Q. Shen, S. Tang, W. Li, Z. Nie, Z. Liu, Y. Huang and S. Yao, Chem. Commun., 2012, 48, 281–283 RSC.
- P. M. Gramlich, S. Warncke, J. Gierlich and T. Carell, Angew. Chem., 2008, 47, 3442–3444 CAS.
- J. F. Gerster and R. K. Robins, J. Org. Chem., 1966, 31, 3258–3262 CrossRef CAS.
- M. Hirsch, D. Strand and M. Helm, Biol. Chem., 2012, 393, 23–35 CrossRef CAS.
- Y. Dorsett and T. Tuschl, Nat. Rev. Drug Discovery, 2004, 3, 318–329 CrossRef CAS.
- J. Harborth, S. M. Elbashir, K. Vandenburgh, H. Manninga, S. A. Scaringe, K. Weber and T. Tuschl, Antisense Nucleic Acid Drug Dev., 2003, 13, 83–105 CrossRef CAS.
- Y. L. Chiu and T. M. Rana, Mol. Cell, 2002, 10, 549–561 CrossRef CAS.
- J. Martinez, A. Patkaniowska, H. Urlaub, R. Luhrmann and T. Tuschl, Cell, 2002, 110, 563–574 CrossRef CAS.
- A. Nykänen, B. Haley and P. D. Zamore, Cell, 2001, 107, 309–321 CrossRef.
Footnotes |
| † In memory of Professor H. Goni Khorana. |
| ‡ This article forms part of the ‘Nucleic acids: new life, new materials’ web themed issue. |
| § Electronic supplementary information (ESI) available. See DOI: 10.1039/c2cc34015a |
| This journal is © The Royal Society of Chemistry 2012 |
