Immobilization of Sertoli cells on islets of Langerhans
Naohiro
Takemoto
a,
Yuji
Teramura
bc and
Hiroo
Iwata
*a
aDepartment of Reparative Materials, Institute for Frontier Medical Sciences, Kyoto University, 53 Kawahara-Cho, Shogoin, Sakyo-Ku, Kyoto 606-8507, Japan. E-mail: iwata@frontier.kyoto-u.ac.jp; Tel: +81-75-751-4119
bRadioisotope Research Center, Kyoto University, Yoshida-Konoe-Cho, Sakyo-Ku, Kyoto 606-8501, Japan
cDepartment of Immunology, Genetics and Pathology (IGP), Uppsala University, Dag Hammarskjölds väg 20, 751 85, Uppsala, Sweden
First published on 28th November 2012
Abstract
Sertoli cells play a crucial role in creating the immunoprivileged environment of the testis. We examined the survival of islets of Langerhans after co-transplantation with Sertoli cells. Sertoli cells near islets should protect the graft from rejection. In this study, conjugates of single stranded oligonucleotides, poly(ethylene glycol) and phospholipids (ssDNA-PEG-DPPE) were used to immobilize Sertoli cells on islets. The 20-mer of deoxyadenylic acid (oligo(dA)20) and 20-mer of deoxythymidylic acid (oligo(dT)20) were presented as ssDNAs on the surfaces of Sertoli cells and islets, respectively, through the hydrophobic interaction between a lipid unit of the conjugates and the cell membrane. The Sertoli cells were immobilized on the islets through hybridization between oligo(dA)20 and oligo(dT)20. When Sertoli cell-immobilized islets were infused into the liver of mice through the portal vein, the Sertoli cells remained around the islets.
Introduction
In clinical islet transplantation, immunosuppressive therapy is required to control rejection reactions to the graft by the host immune system. Although steroid-free immunosuppressive therapy has been introduced in current islet transplantations,1 there are still risks of some side effects, such as opportunistic infections, malignant tumors and impairment of islet functions.2 Alternatives to administrating immunosuppressive drugs are needed to overcome these problems. One approach is the bioartificial pancreas, in which the islets are enclosed within a semi-permeable membrane.3,4 Other approaches, such as ultra-violet light radiation of islets,5 culture of islets under low temperature,6 and co-transplantation of islets with Sertoli cells,7,8 have been tested.Sertoli cells play a crucial role in creating the immunoprivileged environment of the testis9 by secreting immunoprotective factors10,11 and forming a barrier to inhibit infiltrating lymphocytes.10,12,13 Some groups reported long term survival of islets when co-transplanted with Sertoli cells7,8 into the subcapsular site of the kidney for co-localization. However, the co-localization of Sertoli cells with islets is difficult in the liver because for clinical islet transplantation islets and Sertoli cells are to be transfused into the portal vein together. The islets congregate in larger veins in the liver, but Sertoli cells are usually carried to more peripheral smaller veins by the blood flow.
It has been reported that conjugates of single stranded oligonucleotides (ssDNA) and long alkyl chains can be used to attach cells to other cells without deterioration of cell survival and functions.14–18 In our previous studies, we synthesized conjugates of ssDNA and poly(ethylene glycol)-conjugated phospholipid (ssDNA-PEG-lipid) and employed them to immobilize cells on an islet19 for improvement of graft survival.
In this study, ssDNA-PEG-lipid was applied to immobilize Sertoli cells on the islet surface, as shown in Scheme 1. The surface of Sertoli cells and islets were modified with 20-mer of deoxyadenylic acid (oligo(dA)20) and 20-mer of complementary deoxythymidylic acid (oligo(dT)20), respectively, conjugated to PEG-lipids. Sertoli cells were immobilized on islets through DNA hybridization between oligo(dA)20 and oligo(dT)20. The cell composites of Sertoli cells and islets were infused into the mouse portal vein to examine the co-localization of Sertoli cells with islets.
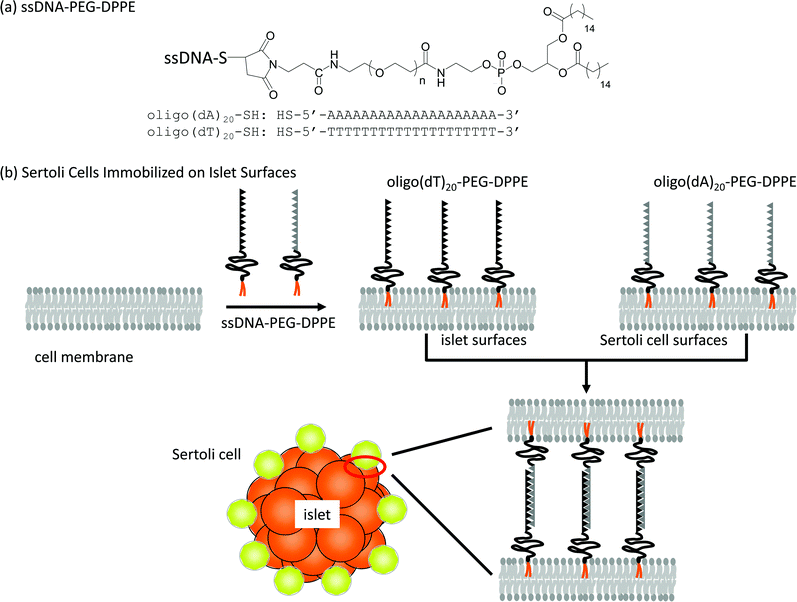 | ||
| Scheme 1 Immobilization of Sertoli cells onto an islet using ssDNA-PEG-DPPEs. (a) Oligo(dA)20 and oligo(dT)20 were used as ssDNA conjugated to PEG-DPPE. (b) A schematic illustration of the anchoring of ssDNA-PEG-DPPE to a lipid bilayer of the cell membrane and immobilization of Sertoli cells onto an islet through hybridization between oligo(dA)20 and oligo(dT)20. | ||
Materials and methods
Materials
The following were purchased from the indicated suppliers: α-N-hydroxysuccinimidyl-ω-maleimidyl poly(ethylene glycol) (NHS-PEG-Mal, Mw: 5000) and 1, 2-dipalmitoyl-sn-glycerol-3-phosphatidylethanolamine (DPPE) from NOF Corporation (Tokyo, Japan); oligo(dA)20 and oligo(dT)20, which carry a protected SH group at the 5′-end (oligo(dA)20-SH, oligo(dT)20-SH) or fluorescein isothiocyanate (FITC) at the 5′-end (FITC-oligo(dA)20, FITC-oligo(dT)20) from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA); dichloromethane, triethylamine, chloroform, diethyl ether, Blocking One, trypsin, penicillin–streptomycin mixed solution and heparin sodium salt from Nacalai Tesuque (Kyoto, Japan); ethylenediamine tetraacetic acid (EDTA) and Hoechst 33258 nuclear stain from Dojindo Laboratories (Kumamoto, Japan); Alexa 488-labeled rabbit anti-goat IgG, DNase, N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) buffer solution, Hanks’ balanced salt solution (HBSS), and Medium 199 from Invitrogen Co. (Carlsbad, CA, USA); fetal bovine serum (FBS) from Equitech-Bio, Inc. (Kerrville, TX, USA); phosphate-buffered saline (PBS) from Nissui Pharmaceutical Co. Ltd (Tokyo, Japan); enzyme-linked immunosorbent assay (ELISA) kits for the insulin assay from Shibayagi Co., Ltd (Gunma, Japan) and for Activin A from R&D Systems, Inc. (Minneapolis, MN, USA); polyclonal goat anti-mouse GATA-4 from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA); polyclonal guinea pig anti-swine insulin from Dako (Glostrup, Denmark); tetrarhodamine isothiocyanate (TRITC)-labeled rabbit anti-guinea pig IgG from Abcam plc. (Cambridge, UK); CellTracker™ Green CMFDA from Lonza Walkersville, Inc. (Walkersville, MD, USA); formalin solution, dithiothreitol (DTT), Tween 20, and Triton X-100 from Wako Pure Chemical (Osaka, Japan); Collagenase from Nitta Gelatin (Osaka, Japan); the NAP-5 column from GE healthcare (GE Healthcare UK Ltd, Buckinghamshire, UK); and Tissue-Tek® from Sakura Fine Technical Co., Ltd (Tokyo, Japan).Isolation of Sertoli cells and islets
All animal experiments were approved by the Kyoto University Animal Care Committee. Sertoli cells were isolated from BALB/c mice (7 day old males, Japan SLC, Inc., Shizuoka, Japan) by Korbutt's method.7 Briefly, testis were collected from mice, chopped into paste with scissors, and digested for 10 min at 37 °C in a collagenase solution (2.5 mg mL−1 in HBSS). The digest was washed with HBSS supplemented with 1 mM EDTA (HBSS/EDTA) and 0.5% bovine serum albumin, additionally digested for 10 min at 37 °C with trypsin (25 μg mL−1) and DNase (4 μg mL−1) in HBSS/EDTA, and washed with HBSS. The final cell pellet was resuspended in Medium 199 supplemented with 10% FBS, 100 units mL−1 penicillin and 100 μg mL−1 streptomycin. The cells were cultured in the medium for 3 days after isolation.To identify Sertoli cells, the isolated cells were immunohistochemically stained for GATA-4, which is a transcription factor and known to be abundantly expressed in the nuclei of Sertoli cells.20,21 Cells were fixed in 4% formalin solution at 37 °C for 15 min, permeabilized in 0.2% Triton X-100 in PBS at room temperature (rt) for 15 min, treated with Blocking One for 1 h to block nonspecific binding of antibodies and then washed with PBS. The cells were immersed in 2% polyclonal goat anti-mouse GATA-4 in Blocking One and left for 1 h at rt, followed by washing with PBS. The cells were incubated with fluorescent labeled secondary antibody, 0.2% Alexa 488-labeled rabbit anti-goat IgG in Blocking One at rt for 1 h, followed by washing with PBS containing 0.05% Tween 20. Cell nuclei were counterstained with Hoechst 33258. The stained cells were observed under a fluorescence microscope (IX71, Olympus Optical, Co., Ltd, Tokyo, Japan). Population of GATA-4 positive cells was elucidated from 300 cells counts.
Islets were isolated from BALB/c mice (6 week old males, Japan SLC, Inc., Shizuoka, Japan) by the collagenase digestion method.22 Briefly, the pancreas was collected from mice, chopped into paste with scissors, and digested for 18 min at 37 °C in a collagenase solution (0.5 mg mL−1 in HBSS). Islets were isolated from the digest by Ficoll density gradient purification, and cultured for 3 days in Medium 199 supplemented with 10% FBS, 8.8 mM HEPES buffer, 100 units mL−1 penicillin, 100 μg mL−1 streptomycin, and 8.8 units mL−1 heparin to remove cells damaged by the isolation procedure.
Immobilization of Sertoli cells onto islets
Oligo(dA)20- and oligo(dT)20-PEG-DPPEs, which were synthesized from Mal-PEG-DPPE and oligo(dA)20- or oligo(dT)20-SH as described previously,19 were used for surface modification of Sertoli cells and islets, respectively. A solution of oligo(dT)20-PEG-DPPE (50 μL, 500 μg mL−1 in PBS) was added to an islet suspension in serum-free medium, and the mixture was incubated at rt for 1.5 h. Oligo(dT)20-PEG-DPPE modified islets were washed three times with serum-free medium and then mixed with a solution of FITC-oligo(dA)20 (100 μL, 500 μg mL−1 in PBS), washed with serum-free medium and observed under a confocal laser scanning microscope (FLUOVIEW, FV500, Olympus Optical Co. Ltd, Tokyo, Japan) to examine the presence of oligo(dT)20 on islets and the hybridization with oligo(dA)20. The fluorophore was excited by an argon laser (488 nm) and the fluorescence was detected through a bandpass filter (510–550 nm).Sertoli cell pellets (2 × 106 cells) were collected by centrifugation (180g, 5 min at 25 °C) after treatment with trypsin. The cells were washed with HBSS to remove the medium. After the addition of oligo(dA)20-PEG-DPPE solution (100 μL, 500 μg mL−1 in PBS) to the cell pellet, the cell suspension was incubated for 30 min with gentle agitation at rt. After washing with HBSS, oligo(dA)20-PEG-DPPE-modified cells were obtained by centrifugation (180g, 5 min, 25 °C, twice). To examine the presence of oligo(dA)20 on Sertoli cells, the cells were mixed with a solution of FITC-oligo(dT)20 (100 μL, 500 μg mL−1 in PBS), washed with HBSS and observed under a confocal laser scanning microscope.
Oligo(dA)20-PEG-DPPE-modified Sertoli cells (2 × 106) and oligo(dT)20-PEG-DPPE-modified islets (200 islets) were mixed in serum-free medium (200 μL). The mixture was incubated for 60 min with gentle agitation at rt. The Sertoli cell-immobilized islets were obtained after washing with culture medium. The Sertoli cell-immobilized islets were immunohistochemically stained for GATA-4 and were observed under a confocal laser scanning microscope. In addition, Sertoli cells stained by CellTracker™ were also employed for easy finding of Sertoli cells after their transplantation.
Insulin and activin A secreted from Sertoli cell-immobilized islets
Sertoli cell-immobilized islets and naïve islets (50 islets each) were mixed with glucose solutions in Krebs–Ringer's buffer (KRB) at concentrations of 0.1 g dL−1, 0.3 g dL−1 and 0.1 g dL−1 glucose sequentially for intervals of 1 h in each solution at 37 °C. The supernatants were collected after each 1 h incubation and the insulin concentrations were determined by ELISA according to the manufacturer's instructions. Sertoli cell-immobilized islets and naïve islets (50 islets each) were incubated in serum-free medium at 37 °C for 24 h. The supernatants were collected and their Activin A concentrations were determined by ELISA according to the manufacturer's instructions.Intraportal transplantation of Sertoli cell-immobilized islets into the liver
BALB/c mice (6 week old males, Japan SLC, Inc.) were used as recipients. Sertoli cells were stained by CellTracker™ prior to immobilization. Sertoli cell-immobilized islets (500 islets/100 μL) were transplanted into the liver through a portal vein of the mice. Mice were anesthetized during surgery by mask inhalation of isoflurane using a specialized instrument (400 Anesthesia Unit; Univentor, Malta); the isoflurane concentration was 4.5% to 5.0% for induction and 2.0% for maintenance with an airflow rate of 200 mL min−1. Mice were sacrificed immediately after transplantation of Sertoli cell-immobilized islets. Livers were retrieved and then immersed in 4% formalin solution for 2 days at rt. The formalin solution was removed and the livers were then embedded in Tissue-Tek® for freezing. The frozen specimens were sliced into 6-μm thick sections for immunohistochemical staining. The sections were observed under a fluorescence microscope (BX51, Olympus Optical Co., Ltd, Tokyo, Japan).Sertoli cell-immobilized islets (500 islets/100 μL) without CellTracker™ staining were also used for transplantation. As a control experiment, 100 μL of a mixed suspension of Sertoli cells (5 × 106 cells) and islets (500 islets) was infused into the liver through a portal vein. Immediately after transplantation, mice were sacrificed and paraffin-embedded liver specimens were sliced into sections 4-μm thick. After deparaffinization, the sections were permeabilized with 0.2% Triton X-100 in PBS at rt for 15 min. The samples were then treated with Blocking One for 1 h, followed by washing with PBS. The sections were treated with 1% polyclonal guinea pig anti-swine insulin and 2% polyclonal goat anti-mouse GATA-4 in Blocking One for 1 h at rt. The sections were then incubated with fluorescent labeled secondary antibody, 2% TRITC-labeled rabbit anti-guinea pig IgG and 0.2% Alexa 488-labeled rabbit anti-goat IgG in Blocking One, at rt for 1 h followed by washing with PBS containing 0.05% Tween 20. The stained sections were observed under a fluorescence microscope.
Results
Immobilization of Sertoli cells onto islets
Amphiphilic polymers, ssDNA-PEG-DPPEs, were used to immobilize Sertoli cells onto islets through DNA hybridization between oligo(dA)20 and oligo(dT)20 as shown in Scheme 1. Sertoli cells were isolated from the testis of male BALB/c mice and seeded on a Petridish. Their purity was examined after 3 days culture by immunohistochemical staining for GATA-4, which is specifically expressed in the nuclei of Sertoli cells.20,21 About 90% of cells were GATA-4 positive, as shown in Fig. 1, and were used in the following experiments. Sertoli cells and islets were treated with oligo(dA)20-PEG-DPPE and oligo(dT)20-PEG-DPPE, respectively. We already reported that the surface modification with PEG-lipid did not affect the islet function.23 Viability of Sertoli cells after treatment with oligo(dA)20-PEG-DPPE was 75.2 ± 4.2%, as estimated by the trypan blue exclusion method.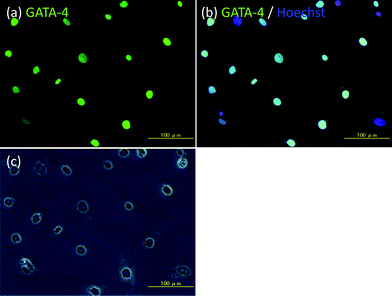 | ||
| Fig. 1 Immunohistochemical examination of Sertoli cells isolated from the testis of male BALB/c mice. (a) GATA-4 (green fluorescence), (b) GATA-4 (green fluorescence) and Hoechst 33258 (blue fluorescence), and (c) phase contrast microscopic image. Scale bar: 100 μm. | ||
After washing with HBSS and serum-free medium, Sertoli cells and islets were treated with oligo(dA)20-PEG-DPPE and oligo(dT)20-PEG-DPPE, respectively. Each was then mixed with a solution containing the corresponding complementary FITC-ssDNA. They were then observed under a confocal laser microscope (see Fig. 2(a-1) and (b-1)). Bright fluorescent rings were found at the periphery of the islets and the Sertoli cells. When the Sertoli cells and islets were mixed with mismatched ssDNA, that is, FITC-oligo(dA)20 and FITC-oligo(dT)20 solutions, respectively, no fluorescent rings were seen (see Fig. 2(a-2) and (b-2)). These results indicate that FITC was immobilized on Sertoli cell and islet surface through hybridization between oligo(dA)20 and oligo(dT)20. Oligo(dA)20- and oligo(dT)20-PEG-DPPEs were present on Sertoli cells and islets, respectively, and maintained the ability to hybridize with complementary ssDNA, that is, FITC-oligo(dT)20 and FITC-oligo(dA)20.
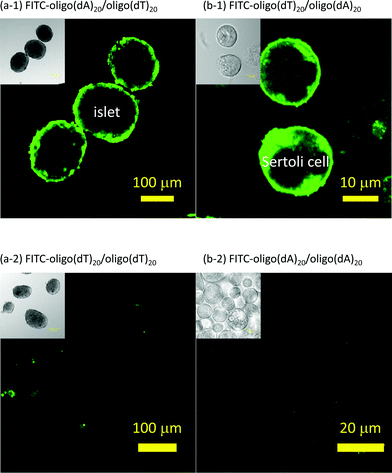 | ||
| Fig. 2 Surface modification of islets and Sertoli cells with ssDNA-PEG-DPPE. Sertoli cells and islets were observed under a confocal laser scanning fluorescent microscope. (a-1) FITC-oligo(dA)20 fixed on islets modified with oligo(dT)20-PEG-DPPE. (a-2) Islets were modified with oligo(dT)20-PEG-DPPE and then mixed with FITC-oligo(dT)20. (b-1) FITC-oligo(dT)20 fixed on Sertoli cells modified with oligo(dA)20-PEG-DPPE. (b-2) Sertoli cells were modified with oligo(dA)20-PEG-DPPE and then FITC-oligo(dA)20. | ||
Fig. 3 shows that Sertoli cells were immobilized on islets through hybridization between oligo(dA)20 and oligo(dT)20, and contains images of the aggregates observed under a confocal florescence microscope. Sertoli cells and islets were treated with oligo(dA)20-PEG-DPPE and oligo(dT)20-PEG-DPPE, respectively. After washing with HBSS and serum-free medium, suspensions of each were mixed together and left for 60 min. Existence of Sertoli cells was examined by immunohistochemical staining of GATA-4 which was specifically expressed in Sertoli cells.20,21 GATA-4 positive cells were found on the islet surface as shown in Fig. 3(a). Sertoli cells stained with CellTracker™ were also used to see location of Sertoli cells in the aggregates. Sertoli cells were clearly observed at the periphery of the islets immediately after the immobilization (see Fig. 3(b)). However, few cells were found at the periphery of the islets when oligo(dT)20-PEG-DPPE modified-islets were mixed with naïve Sertoli cells, that were observed under a phase contrast microscope (see Fig. 3(c)). When the aggregate prepared via hybridization were cultured for 4 days, some of Sertoli cells migrated into the inside of islets (see Fig. 3(d)). These results clearly demonstrate that Sertoli cells could be immobilized on islets through hybridization between oligo(dT)20 on the islets and oligo(dA)20 on the Sertoli cells.
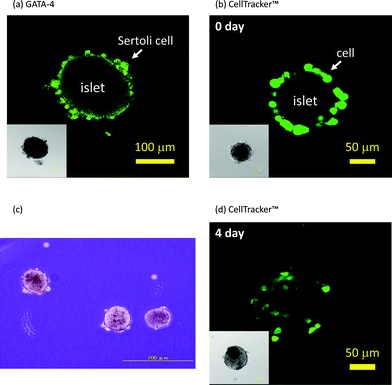 | ||
| Fig. 3 Immobilization of Sertoli cells onto islets. (a) Immunohistochemical staining of GATA-4 of Sertoli cells immobilized on an islet with the ssDNA-PEG-DPPE method. (b) Sertoli cells (stained with CellTracker™) immobilized on an islet with the ssDNA-PEG-DPPE method just after immobilization. (c) A phase contrast microscopic image of a mixture of oligo(dT)20-PEG-DPPE-modified islets and naïve Sertoli cells. (d) Sertoli cells (stained with CellTracker™) immobilized on an islet after 4 days of culture. Insets: phase contrast microscopic image. | ||
In vitro functional evaluation of the Sertoli cell and islet composite
Static glucose stimulation tests were performed on Sertoli cell-immobilized islets (50 islets) and islets without immobilization of Sertoli cells (naïve islets). As shown in Fig. 4(a), when the glucose concentration was increased from 0.1 to 0.3 g dL−1, both naïve and Sertoli cell-immobilized islets increased insulin release. When the glucose concentration was again reduced to 0.1 g dL−1, the insulin release returned to basal levels. No significant difference in insulin release was observed between the naïve and Sertoli cell-immobilized islets. These results indicate immobilization of Sertoli cells on the islet surface using ssDNA-PEG-DPPE did not impair the ability of islets to regulate insulin release.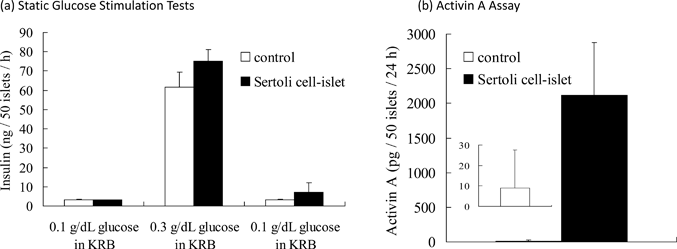 | ||
| Fig. 4 The functional evaluation of Sertoli cell-immobilized islets in vitro. (a) Glucose stimulation test of Sertoli cell-immobilized islets and naïve islets. Islets were sequentially exposed to 0.1, 0.3 and 0.1 g dL−1 glucose solutions in Krebs–Ringer buffer (KRB) at 37 °C for 1 h each. The amounts of released insulin are expressed as mean ± SD for n = 3. (b) Activin A release from Sertoli cell-immobilized islets and naïve islets. The islets were incubated in serum-free medium at 37 °C for 24 h. The amounts of released Activin A are expressed as mean ± SD for n = 5. | ||
Activin A is secreted by Sertoli cells24 and was examined in Sertoli cells on islets to determine if they were active. Activin A was secreted from Sertoli cell-immobilized islets; whereas, no secretion was seen from naïve islets (see Fig. 4(b)). These results indicate immobilization of Sertoli cells on the islet surface using ssDNA-PEG-DPPE did not affect the ability of Sertoli cells to secrete Activin A. Sertoli cells could be immobilized onto islets without loss of function of islets or Sertoli cells.
Co-localization of Sertoli cells around islets in the liver
Sertoli cell-immobilized islets were infused through the portal vein into the liver. Livers were retrieved from the recipient mice immediately after transplantation and thin sections prepared. When Sertoli cells were fluorescently stained with CellTracker™, fluorescently-stained cells were clearly observed at the periphery of the islet (see Fig. 5(a-1)). The cells were also immunohistochemically stained for GATA-4 to identify Sertoli cells not stained with CellTracker™. As seen in Fig. 5(b), the islet was surrounded by GATA-4 positive cells. As a control, a simple mixture of Sertoli cells and islets was infused into the liver (Fig. 5(c)). No GATA-4 positive cells could be found around these islets after the mixture was infused through the portal vein into the liver. These results clearly show ssDNA-PEG-DPPE is effective in localizing Sertoli cells on islets in the liver, even when they are exposed to blood flow.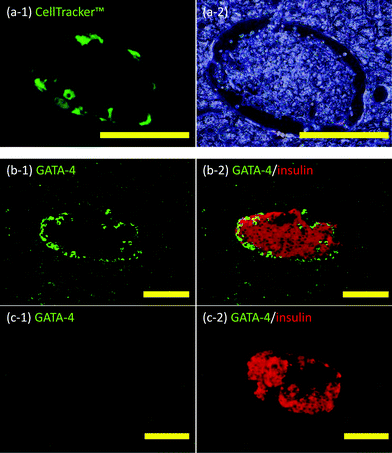 | ||
| Fig. 5 Histochemical analyses of Sertoli cells and islets in liver veins. (a) Sertoli cell-immobilized islets in the mouse liver: (a-1) CellTracker™ (green fluorescence) and (a-2) phase contrast microscopic image. (b) Sertoli cell-immobilized islets in the mouse liver: (b-1) GATA-4 (green fluorescence) and (b-2) Merged images of GATA-4 (green fluorescence) and insulin (red fluorescence). (c) Naïve islets in the mouse liver: (c-1) GATA-4 (green fluorescence) and (c-2) merged images of GATA-4 (green fluorescence) and insulin (red fluorescence). Scale bar: 100 μm. | ||
Discussion
Various bioactive substances, such as proteins,25 liposomes26 and cells,19 have been immobilized on the cell surface using hetero-bifunctional polymers. One end of these polymers interacts with the cell membrane and another functional group is used to introduce a bioactive substance on the cell surface.27,28 Several methods have been used to introduce polymers on the cell surface, including covalent conjugation of polymers carrying N-hydroxyl-succinimidyl esters to amino groups of membrane proteins,27,28 electrostatic interactions between cationic polymers and the negatively charged cell surface,27,28 and anchoring of long alkyl chains of amphiphilic polymers into the lipid bilayer of the cell membrane through hydrophobic interactions, as was done in this study. Each method has advantages and disadvantages. In the covalent conjugation methods, a stable bond forms between polymers and membrane proteins, but the functions of membrane proteins are often impaired.29 The electrostatic interaction using cationic polymers simply involves adding a polymer solution to a cell suspension, however, most cationic polymers are cytotoxic and most cells are deteriorated or killed after treatment with them.30 In the hydrophobic interaction method, cells can be modified by simply adding amphiphilic polymers carrying long alkyl chains to a cell suspension and no major or clear deterioration of cell functions or integrity have been observed.27,28 Thus, we employed amphiphilic polymers in this study.In this method, bioactive substances are immobilized on the cell through the polymers. The avidin–biotin complex is most often used.27,28 It can form stable bonding under physiological conditions and is suitable for in vitro examination of modified cells. Various avidins are commercially available, but are derived from albumen or bacteria. These avidins are hard to use for in vivo experiments, particularly in the clinical setting, because they are expected to induce unfavorable immune reactions. As an alternative, we employed DNA duplex formation. Oligo(dA)20 and oligo(dT)20 can form a stable complex under physiological conditions and the immunogenicity of DNA is much lower than that of avidin.
We were able to immobilize Sertoli cells on islets without loss of functions of either using amphiphilic ssDNA-PEG-DPPEs, as shown in Fig. 4. The aggregates of Sertoli cells and islets could be co-transplanted into the mouse liver though the portal vein without detachment of Sertoli cells from the islet surface as shown in Fig. 5. Further studies are necessary to determine if our method is clinically useful for the co-transplantation of Sertoli cells and islets in the liver. Factors to be examined include the stability of the cell composite, life span of Sertoli cells in the liver and the blood compatibility of Sertoli cells. However, cell immobilization using ssDNA-PEG-DPPEs is a versatile method to immobilize any kind of cells, such as regulatory T cells and endothelial cells, on islets without exerting harmful effects on the cells. We expect this method will increase the success rate of islet transplantations.
Although we have mainly focused on the application of ssDNA-PEG-lipid to prolong graft survival of islets, this technology has many potential applications in modification of surface of cells and tissues with bioactive substances, study of cell–cell interaction and three dimensional tissue fabrication from dissociated single cells as discussed previously.16,19
Conclusions
Sertoli cells, important for creating the immunoprivileged environment of the testis, can be immobilized on the surfaces of pancreatic islets through DNA hybridization using ssDNA-PEG-DPPE. The immobilizations did not influence islet morphology and islet ability to secrete insulin. Importantly, the function of Sertoli cells was maintained on the islet surface. After intraportal transplantation of Sertoli cell-immobilized islets, the Sertoli cells were attached to the islets surface in the liver. We expect this method will improve the success rate of islet transplantations.Acknowledgements
This study was supported in part by a Grant-in-Aid for Scientific Research (A) (No. 21240051), Challenging Exploratory Research (No. 21650118) and Japan Society for the Promotion of Science (JSPS) Fellows (No. 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3121) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and the Ministry of Health, Labor, and Welfare of Japan (H20-007).
3121) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and the Ministry of Health, Labor, and Welfare of Japan (H20-007).
References
- T. Froud, C. Ricordi, D. A. Baidal, M. M. Hafiz, G. Ponte, P. Cure, A. Pileggi, R. Poggioli, H. Ichii, A. Khan, J. V. Ferreira, A. Pugliese, V. V. Esquenazi, N. S. Kenyon and R. Alejandro, Am. J. Transplant., 2005, 5, 2037–2046 CrossRef.
- L. Luzi, J. Mol. Med., 1999, 77, 49–56 CrossRef CAS.
- H. Iwata, T. Takagi, H. Amemiya, H. Shimizu, K. Yamashita, K. Kobayashi and T. Akutsu, J. Biomed. Mater. Res., 1992, 26, 967–977 CrossRef CAS.
- F. Lim and A. M. Sun, Science, 1980, 210, 908–910 CAS.
- P. Y. Benhamou, E. Stein, C. Hober, M. Miyamoto, Y. Watanabe, Y. Nomura, P. C. Watt, T. Kenmochi, F. C. Brunicardi and Y. Mullen, Horm. Metab. Res., 1995, 27, 113–120 CrossRef CAS.
- C. Ricordi, P. E. Lacy, K. Sterbenz and J. M. Davie, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 8080–8084 CrossRef CAS.
- G. S. Korbutt, J. F. Elliott and R. V. Rajotte, Diabetes, 1997, 46, 317–322 CAS.
- H. P. Selawry and D. F. Cameron, Cell Transplant., 1993, 2, 123–129 CAS.
- J. W. Streilein, Science, 1995, 270, 1158–1159 CrossRef CAS.
- D. Bellgrau, D. Gold, H. Selawry, J. Moore, A. Franzusoff and R. C. Duke, Nature, 1995, 377, 630–632 CrossRef CAS.
- A. S. Cupp, G. Kim and M. K. Skinner, Biol. Reprod., 1999, 60, 1304–1313 CrossRef CAS.
- T. S. Grith, T. Brunner, S. M. Fletcher, D. R. Green and T. A. Ferguson, Science, 1995, 270, 1189–1192 Search PubMed.
- D. E. Jenne and J. Tschopp, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 7123–7127 CrossRef CAS.
- G. G. Borisenko, M. A. Zaitseva, A. N. Chuvilin and G. E. Pozmogova, Nucleic Acids Res., 2009, 37, e28 CrossRef.
- H. Liu, B. Kwong and D. J. Irvine, Angew. Chem., Int. Ed., 2011, 50, 7052–7055 CrossRef CAS.
- N. S. Selden, M. E. Todhunter, N. Y. Jee, J. S. Liu, K. E. Broaders and Z. J. Gartner, J. Am. Chem. Soc., 2012, 134, 765–768 CrossRef CAS.
- S. C. Hsiao, B. J. Shum, H. Onoe, E. S. Douglas, Z. J. Gartner, R. A. Mathies, C. R. Bertozzi and M. B. Francis, Langmuir, 2009, 25, 6985–6991 CrossRef CAS.
- Z. J. Gartner and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 4606–4610 CrossRef CAS.
- Y. Teramura, L. N. Minh, T. Kawamoto and H. Iwata, Bioconjugate Chem., 2010, 21, 792–796 CrossRef CAS.
- S. A. McCoard, D. D. Lunstra, T. H. Wise and J. J. Ford, Biol. Reprod., 2001, 64, 689–695 CrossRef CAS.
- R. S. Viger, C. Mertineit, J. M. Trasler and M. Nemer, Development, 1998, 125, 2665–2675 CAS.
- P. E. Lacy and M. Kostianovky, Diabetes, 1967, 16, 35–39 CAS.
- Y. Teramura and H. Iwata, Transplantation, 2011, 91, 271–278 CrossRef CAS.
- J. P. de Winter, H. M. Vanderstichele, M. A. Timmerman, L. J. Blok, A. P. Themmen and F. H. de Jong, Endocrinology, 1993, 132, 975–982 CrossRef CAS.
- N. Takemoto, Y. Teramura and H. Iwata, Bioconjugate Chem., 2010, 22, 673–678 CrossRef.
- H. Chen, Y. Teramura and H. Iwata, Biomaterials, 2011, 32, 7971–7977 CrossRef CAS.
- Y. Teramura and H. Iwata, Soft Matter, 2010, 6, 1081–1091 RSC.
- Y. Teramura and H. Iwata, Adv. Drug Delivery Rev., 2010, 62, 827–840 CrossRef CAS.
- D. Rabuka, M. B. Forstner, J. T. Groves and C. R. Bertozzi, J. Am. Chem. Soc., 2008, 130, 5947–5953 CrossRef CAS.
- D. Fischer, Y. Li, B. Ahlemeyer, J. Krieglstein and T. Kissel, Biomaterials, 2003, 24, 1121–1131 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2013 |
