Cationic, helical polypeptide-based gene delivery for IMR-90 fibroblasts and human embryonic stem cells
Jonathan Yena, Yanfeng Zhangb, Nathan P. Gabrielsonb, Lichen Yinb, Linna Guana, Isthier Chaudhuryb, Hua Luc, Fei Wang*ade and Jianjun Cheng*ab
aDepartment of Bioengineering, University of Illinois at Urbana–Champaign, Urbana, IL 61801, USA
bDepartment of Materials Science and Engineering, University of Illinois at Urbana–Champaign, Urbana, IL 61801, USA. E-mail: jianjunc@illinois.edu
cDepartment of Chemistry, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA
dDepartment of Cell and Developmental Biology, University of Illinois at Urbana–Champaign, Urbana, IL 61801, USA. E-mail: feiwang@life.illinois.edu
eInstitute of Genomic Biology, University of Illinois at Urbana–Champaign, Urbana, IL 61801, USA
First published on 8th April 2013
Abstract
Diblock copolymers consisting of poly(ethylene glycol)-block-poly(γ-4-(((2-(piperidin-1-yl)ethyl)amino)methyl)benzyl-L-glutamate) (PEG-b-PVBLG-8) were synthesized and evaluated for their ability to mediate gene delivery in hard-to-transfect cells like IMR-90 human fetal lung fibroblasts and human embryonic stem cells (hESCs). The PEG-b-PVBLG-8 contained a membrane-disruptive, cationic, helical polypeptide block (PVBLG-8) for complexing with DNA and a hydrophilic PEG block to improve the biocompatibility of the gene delivery vehicle. The incorporation of PEG effectively reduced the toxicity of the helical PVBLG-8 block without dramatically compromising the polymer's ability to destabilize membranes or form complexes with DNA. PEG-b-PVBLG-8 copolymers with low (n = 76) and high (n = 287) degrees of polymerization (n) of the PVBLG-8 block were synthesized and evaluated for gene delivery. PEG-b-PVBLG-8 diblock polymers with a high degree of polymerization have a greater transfection efficiency and lower toxicity in IMR-90 cells than the commercial reagent Lipofectamine 2000. The usefulness of PEG-b-PVBLG-8 was further demonstrated via the successful transfection of hESCs without a measured loss in cell pluripotency markers.
1. Introduction
Both viral and non-viral gene delivery have been explored extensively for basic research as well as therapeutic applications. Viral gene delivery is typically more efficient than its non-viral counterpart but poses increased risks of immunogenicity, insertional mutagenesis and viral integration into the host genome.1 While the integration event results in permanent transgene expression—which may not be desirable for all applications—it also provides a route for the prolonged expression of undesired viral components in host cells. As non-viral gene delivery relies on synthetic polymers or lipids and DNA that is explicitly free from viral components, it is generally considered a safer alternative than viral gene therapy. Therefore, there is a push to develop non-viral gene delivery materials to match the efficiency of viral vectors.2–11In the pursuit of regenerative medicine, non-viral gene delivery is an important tool to manipulate and control cell fate. One of the latest applications of gene delivery is the reprogramming of terminally differentiated cells into induced pluripotent stem cells (iPSCs) or, more recently, induced neurons, blood progenitors and cardiomyocytes.12–14 In the pioneering work of Yu et al., a recombinant lentivirus carrying the genes for OCT4, SOX2, NANOG and LIN28 was used to reprogram IMR-90 fetal lung fibroblasts into iPSCs.15 While the resulting iPSCs were characteristically and functionally pluripotent, they nonetheless retained remnants of viral DNA as a result of lentiviral integration. This could have serious implications if tissue derived from such iPSCs was used therapeutically. Non-viral approaches to achieve safe, virus-free reprogramming of IMR-90 cells using synthetic polymers and lipids, however, have been fruitless as existing gene delivery materials have proven to be inefficient at mediating effective gene delivery in these cells.
The genetic manipulation of human embryonic stem cells is an important tool in regenerative medicine. The control and over-expression of specific genes afforded by gene delivery are valuable not only in efforts to control stem cell fate, but also to study cell behavior in differentiation and gene targeting studies.16 Unfortunately, recent efforts to develop new polymeric materials for the non-viral transfection of human embryonic stem cell colonies has resulted in only slightly increased efficiency compared to existing commercial products.17
Polypeptides were among the first set of materials examined as non-viral gene delivery agents.18–20 Given the simplicity in synthesis and formulation with anionic DNA, cationic poly(L-lysine) (PLL) was one of the most intensively studied gene delivery polypeptides. However, as a DNA delivery vector, unmodified PLL suffered from low transfection efficiency. Even following modification with functional moieties like saccharide,21,22 imidazole23 and guanidinium groups,24 PLL has proven to be a largely ineffective gene delivery vector. Nonetheless, there have also been numerous attempts to create novel gene delivery vehicles with modified polypeptides, like poly(glycoamidoamine)s25 and HPMA-oligolysines.26
Many biologically active peptides share facially amphipathic helical domains as a common structural motif.27,28 Peptides which possess this structure are often able to interact with and destabilize the lipid bilayers of cell membranes. In terms of gene delivery, cell membrane destabilization can facilitate cell internalization and escape from endocytic vesicles.29,30 PLL and modified PLL, however, adopt random coil structures because strong intramolecular side-chain charge repulsion prohibits α-helix formation. As such, PLL functions as a conventional polyelectrolyte in gene delivery studies and exhibits limited membrane activity.
Recently, we reported a new cationic polypeptide possessing a pH-, ionic- and temperature-stable cationic helical structure.31 Traditionally, charged polypeptides adopt random coil orientations because strong intramolecular side-chain charge repulsion prohibits α-helix formation. In our reported polypeptide, termed PVBLGn-8 where n is the degree of polymerization, the helical structure is stabilized by increasing the distance between the charged side chain groups and the polypeptide backbone. This has the net effect of both minimizing the effect of charge repulsion while simultaneously stabilizing the helix through hydrophobic interaction between the pendent side chain groups. Our prior characterization of PVBLGn-8 peptides as non-viral gene delivery materials revealed that the helical structure facilitates strong, disruptive interactions with cell membranes which aid the endosomal escape of the endocytosed complexes of polypeptide and DNA.32 Although gene delivery with PVBLGn-8 proved to be effective in COS-7 and HEK 293 cells with comparable performance to the commercial transfection agent Lipofectamine 2000,32 it was largely ineffective in IMR-90 fibroblasts due to its substantial toxicity in these cells. Here, we report the design and synthesis of a diblock copolymer incorporating polyethylene glycol (PEG) to the PVBLGn-8 polypeptide. We demonstrate that the addition of PEG to PVBLGn-8 markedly decreases the toxicity of PVBLGn-8 yet preserves the biological activity and gene delivery efficiency of the polypeptide. Moreover, with its reduced toxicity, PEG-b-PVBLG-8 is able to effectively transfect IMR-90 fibroblasts and human embryonic stem cells (Scheme 1), which suggests that it may be possible to ultimately achieve higher reprogramming efficiency of fibroblasts with a safer non-viral vector.
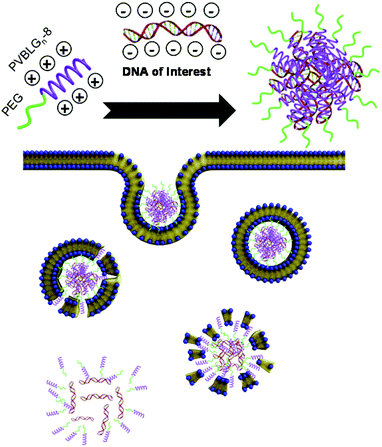 | ||
| Scheme 1 Plasmid DNA condensation by PEG-b-PVBLG-8 and the uptake and release of DNA inside the cell. | ||
2. Materials and methods
2.1 General
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received unless otherwise specified. Anhydrous dimethylformamide (DMF) was dried by a column packed with 4 Å molecular sieves and stored in a glovebox. Tetrahydrofuran (THF) and hexane were dried by a column packed with alumina and stored in a glove box. OptiMEM and Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA, USA). pEGFP-N1 was obtained from Elim Biopharmaceuticals (Hayward, CA, USA). The human embryonic stem cell line H1 was cultured in mTeSR 1 medium from Stem Cell Technologies (Vancouver, Canada). Milli-Mark™ Anti-SSEA-4-PE were purchased from EMD Millipore (Billerica, MA, USA). IMR-90 fetal lung fibroblast cells were purchased from ATCC (Manassas, VA, USA) and cultured in MEM containing Earle's balanced salt solution supplemented with 10% fetal bovine serum. L-Glutamic acid copper(II) complex, copper(II) salt, tetrahydrate33 and γ-(4-vinylbenzyl)-L-glutamate N-carboxyanhydride (VB-Glu-NCA)31,34 were prepared by following previously reported procedures.2.2 Instrumentation
NMR spectra were recorded on a Varian UI400 MHz, a UI500NB MHz or a VXR-500 MHz spectrometer. Tandem gel permeation chromatography (GPC) experiments were performed on a system equipped with an isocratic pump (Model 1100, Agilent Technology, Santa Clara, CA, USA), a DAWN HELEOS 18-angle laser light scattering detector (also known as a multi-angle laser light scattering (MALLS) detector, Wyatt Technology, Santa Barbara, CA, USA) and an Optilab rEX refractive index detector (Wyatt Technology, Santa Barbara, CA, USA). The detection wavelength of HELEOS was set at 658 nm. Separations were performed using serially connected size exclusion columns (100, 500, 103 and 104 Å Phenogel columns, 5 μm, 300 × 7.8 mm, Phenomenex, Torrance, CA, USA) at 60 °C using DMF containing 0.1 M LiBr as the mobile phase. The MALLS detector was calibrated using pure toluene with no need for external polymer standards and was used for the determination of the absolute molecular weights. The molecular weights (MWs) of all polymers were determined based on the dn/dc value of each sample calculated offline by using the internal calibration processed by ASTRA V software (version 5.1.7.3, Wyatt Technology, Santa Barbara, CA, USA). Infrared spectra were recorded on a PerkinElmer 100 serial FTIR spectrophotometer equipped with universal attenuated total reflectance (ATR), which enabled the analysis of polymer samples in powder form. Circular dichroism (CD) measurements were carried out on a JASCO J-700 or a JASCO J-720 CD Spectrometer. Ozone was produced by an OZV-8S ozone generator manufactured by Ozone Solutions Inc. (Hull, IA, USA). Lyophilization was performed on a FreeZone lyophilizer (Labconco, Kansas City, MO, USA). Flow cytometry analysis was conducted on a BD FACSCanto 6 color flow cytometry analyzer (Becton Dickinson, Franklin Lakes, NJ, USA). Cells were visualized with a Zeiss Axiovert 40 CFL fluorescence microscope equipped with a 20× objective (Thornwood, NY, USA). Zeta potential and particle size were analyzed with a Malvern Zetasizer (Worcestershire, UK).2.3 General procedure for the polymerization of VB-Glu-NCA
We followed a previously established procedure to synthesize and polymerize NCAs to prepare polypeptides.35–38 In a glovebox, VB-Glu-NCA (56 mg, 0.2 mmol) was dissolved in DMF (1 mL) followed by the addition of PEG-amine and 1,5,7-triazabicyclo[4.4.0]dec-5-ene (TBD) at various monomer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amine
amine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TBD ratios (Table 1). The polymerization solutions were stirred at room temperature for 24–60 h untill VB-Glu-NCA was consumed. Aliquots of the polymerization solutions were diluted to 10 mg polymer mL−1 using DMF containing 0.1 M LiBr and analyzed by GPC. The real-time concentration of NCA was quantified by measuring the intensity of the anhydride band at 1784 cm−1 by FTIR. The conversion of VB-Glu-NCA was determined by comparing the VB-Glu-NCA concentration in the polymerization solution versus the initial VB-Glu-NCA concentration. When the polymerization was complete, the majority of the DMF was removed under vacuum and the polymer was precipitated with ether (15 mL). The resulting PEG-b-PVBLG polymer was sonicated in ether for 5 min and centrifuged to remove the remaining solvent. After the sonication–centrifugation steps were repeated two more times, PEG-b-PVBLG was collected and dried under vacuum (44 mg and 75% yield, and 35 mg and 68% yield for PEG113-b-PVBLG76 and PEG113-b-PVBLG287, respectively). 1H NMR (TFA-d, 500 MHz): δ 7.53 (d, 2H, J = 7.0 Hz, ArH), 7.39 (d, 2H, J = 7.0 Hz, ArH), 6.84 (dd, 1H, J1 = 11.0 Hz, J2 = 18.0 Hz C6H4CH
TBD ratios (Table 1). The polymerization solutions were stirred at room temperature for 24–60 h untill VB-Glu-NCA was consumed. Aliquots of the polymerization solutions were diluted to 10 mg polymer mL−1 using DMF containing 0.1 M LiBr and analyzed by GPC. The real-time concentration of NCA was quantified by measuring the intensity of the anhydride band at 1784 cm−1 by FTIR. The conversion of VB-Glu-NCA was determined by comparing the VB-Glu-NCA concentration in the polymerization solution versus the initial VB-Glu-NCA concentration. When the polymerization was complete, the majority of the DMF was removed under vacuum and the polymer was precipitated with ether (15 mL). The resulting PEG-b-PVBLG polymer was sonicated in ether for 5 min and centrifuged to remove the remaining solvent. After the sonication–centrifugation steps were repeated two more times, PEG-b-PVBLG was collected and dried under vacuum (44 mg and 75% yield, and 35 mg and 68% yield for PEG113-b-PVBLG76 and PEG113-b-PVBLG287, respectively). 1H NMR (TFA-d, 500 MHz): δ 7.53 (d, 2H, J = 7.0 Hz, ArH), 7.39 (d, 2H, J = 7.0 Hz, ArH), 6.84 (dd, 1H, J1 = 11.0 Hz, J2 = 18.0 Hz C6H4CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.91 (d, 1H, J = 18.0 Hz, C6H4CH
CH2), 5.91 (d, 1H, J = 18.0 Hz, C6H4CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.43 (d, 1H, J = 11.0 Hz, C6H4CH
CH2), 5.43 (d, 1H, J = 11.0 Hz, C6H4CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.26 (m, 2H, ArCH2), 4.80 (m, 1H, CHCH2CH2COOCH2), 4.13 (m, –OCH2CH2– in PEG), 2.68 (m, 2H, CHCH2CH2COO), 2.30 (m, 1H, CHCH2CH2COO), 2.12 (m, 1H, CHCH2CH2COO).
CH2), 5.26 (m, 2H, ArCH2), 4.80 (m, 1H, CHCH2CH2COOCH2), 4.13 (m, –OCH2CH2– in PEG), 2.68 (m, 2H, CHCH2CH2COO), 2.30 (m, 1H, CHCH2CH2COO), 2.12 (m, 1H, CHCH2CH2COO).
2.4 General procedure for the synthesis of poly(ethylene glycol)-block-poly(γ-(4-aldehydebenzyl)-L-glutamate) (PEG-b-PABLG)
PEG-b-PVBLG (40 mg) was dissolved in chloroform (30 mL) at −78 °C. Oxygen was then bubbled into the solution for 1 min followed by the bubbling of ozone until the solution became blue. The ozone was then replaced by oxygen, which was bubbled into the solution for 2 min until the solution became colorless. The solution was degassed and back filled with nitrogen. Dimethyl sulfide (1 mL) was then added and the solution was stirred at room temperature overnight. Afterwards, the solvent was removed under vacuum and the resulting PEG113-b-PABLG76 was purified by sonicating the polymer in methanol (3 × 15 mL) and collected by centrifugation. PEG113-b-PABLG76 was dried under vacuum (33 mg, 82% yield). PEG113-b-PABLG287 was synthesized from PEG113-b-PVBLG287 by following a similar procedure for the synthesis of PEG113-b-PABLG76 with 86% yield. 1H NMR (TFA-d, 500 MHz): δ 10.31 (1H, CHOC6H4), 8.40 (d, 2H, J = 7.0 Hz, ArH), 7.96 (d, 2H, J = 7.0 Hz, ArH), 5.71 (2H, CHOC6H4CH2), 5.21 (1H, CHCH2CH2CO2CH2), 4.10 (m, –OCH2CH2– in PEG), 3.12 (2H, CHCH2CH2), 2.75 (1H, CHCH2CH2), 2.56 (1H, CHCH2CH2).2.5 General procedure for the preparation of PEG-b-PVBLG-8
PEG-b-PABLG (20 mg), N-(2-aminoethyl)piperidine (5 molar equiv. relative to the Glu repeating unit of PEG-b-PABLG) and borane–pyridine complex (5 molar equiv.) were mixed in DMF (3 mL) and stirred at 50 °C for 48 h (Table 2). The mixture was poured into 3 M HCl (3 mL) and dialyzed against water for 48 h. The resulting PEG-b-PVBLG-8 was lyophilized. The yields of PEG-b-PVBLG-8 copolymers were between 60 and 70%, with grafting efficiencies greater than 95%, which was determined as previously reported.32| Starting polymer | Product | Grafting eff.b (%) | −[θ]222![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c (10−3 degree cm2/dmol) c (10−3 degree cm2/dmol) | Helical contentd (%) |
|---|---|---|---|---|
a Reducing reagent (5 molar equiv.) was used. Reaction was carried out for 48 h at 50 °C.b The grafting efficiency was determined by 1H NMR analysis.c The mean residue molar ellipticity was calculated by following literature-reported formulas: ellipticity ([θ]222 nm in cm2 degree dmol−1) = (millidegrees × mean residue weight)/(path length in millimeters × concentration of polypeptide in mg mL−1).d The α-helix contents of the polypeptides were calculated using the following equation: % α-helix = (−[θ]222 + 3000)/39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.40 000.40 | ||||
| PEG113-b-PABLG76 | PEG113-b-PVBLG76-8 (PEV-L) | >95 | 32.8 | 91.8 |
| PEG113-b-PABLG287 | PEG113-b-PVBLG287-8 (PEV-H) | >95 | 34.6 | 96.4 |
2.6 General procedure for the analysis of polypeptide conformations by circular dichroism (CD)
Circular dichroism studies were performed on JASCO J-700 and J-720 CD spectrometers. Samples were prepared at polymer concentrations of 0.01–0.1 mg mL−1 unless otherwise specified. In a representative experiment, the sample solution was placed in a quartz cell with a path length of 0.5 cm and the mean residue molar ellipticity of the polymer was calculated based on the measured apparent ellipticity according to the equation: Ellipticity ([θ] in degree cm2 dmol−1) = (millidegrees × mean residue weight)/(path length in millimeters × concentration of polypeptide in mg mL−1).39 For helix-temperature dependency studies, the temperature of the sample chamber containing the quartz cell was varied from 4 to 70 °C using a water bath. A minimum of 10 min was allowed for sample temperature equilibration prior to collecting CD measurements. The α-helix contents of the polypeptides were calculated using the following equation: % α-helix = (−[θ]222 + 3000)/39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.40
000.402.7 Agarose gel retardation
A solution of DNA (0.5 μg) was prepared in OptiMEM (50 μL). Separately, a solution of polypeptide in OptiMEM (50 μL) was prepared to achieve the desired polypeptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA weight ratio. Following mixing of the two solutions, complexes were incubated at room temperature for 20 min, after which an aliquot (20 μL) was withdrawn and a loading dye (4 μL) was added. The mixture was then run on a 2% agarose gel (100 V, 60 min). DNA was stained with ethidium bromide and visualized on a Gel Doc imaging system (Biorad, Herclues, CA, USA).
DNA weight ratio. Following mixing of the two solutions, complexes were incubated at room temperature for 20 min, after which an aliquot (20 μL) was withdrawn and a loading dye (4 μL) was added. The mixture was then run on a 2% agarose gel (100 V, 60 min). DNA was stained with ethidium bromide and visualized on a Gel Doc imaging system (Biorad, Herclues, CA, USA).2.8 Characterization of polymer–DNA complex with zeta potential and dynamic light scattering
Solutions of DNA (25 μg) were prepared in OptiMEM (400 μL). Separately, a solution of polypeptide (1 mg) was prepared in OptiMEM (400 μL). A solution of Lipofectamine 2000 (50 μL, 1 mg mL−1) in OptiMEM (400 μL) was also prepared as a control. The DNA solution was then mixed with either the polypeptide or Lipofectamine 2000 solution and allowed to incubate at rt for 20 min. The size and surface charge of the resulting polyplexes were analyzed by dynamic light scattering (DLS) and zeta potential.2.9 Transfection of IMR-90 with Lipofectamine 2000 and PVBLG-8 polymers
IMR-90 cells were seeded at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 24-well plates one day prior to transfection. On the day of transfection, plasmid pEGFP-N1 DNA (1 μL, 1 mg mL−1) was diluted with OptiMEM (50 μL). Separately, Lipofectamine 2000 (2 μL, 1 mg μL−1) or the polymer solution (10–80 μL, 1 mg mL−1) was diluted with OptiMEM (50 μL). The individual solutions were then mixed gently and allowed to incubate for 5 min at rt, after which they were combined and allowed to incubate at rt for another 20 min. The cell medium was then aspirated and replaced with pre-warmed (37 °C) OptiMEM (500 μL). The complex solution was added dropwise to the cells. The cells were then incubated at 37 °C with 5% CO2 for 4 h, after which the cell medium was replaced with normal culture medium (500 μL). After incubation for a total of 48 h at 37 °C with 5% CO2, the cells were imaged with a fluorescent microscopy. The EGFP transfection efficiency was quantified by flow cytometry.
000 cells per well in 24-well plates one day prior to transfection. On the day of transfection, plasmid pEGFP-N1 DNA (1 μL, 1 mg mL−1) was diluted with OptiMEM (50 μL). Separately, Lipofectamine 2000 (2 μL, 1 mg μL−1) or the polymer solution (10–80 μL, 1 mg mL−1) was diluted with OptiMEM (50 μL). The individual solutions were then mixed gently and allowed to incubate for 5 min at rt, after which they were combined and allowed to incubate at rt for another 20 min. The cell medium was then aspirated and replaced with pre-warmed (37 °C) OptiMEM (500 μL). The complex solution was added dropwise to the cells. The cells were then incubated at 37 °C with 5% CO2 for 4 h, after which the cell medium was replaced with normal culture medium (500 μL). After incubation for a total of 48 h at 37 °C with 5% CO2, the cells were imaged with a fluorescent microscopy. The EGFP transfection efficiency was quantified by flow cytometry.2.10 Sample preparation and flow cytometry analysis
Prior to analysis by flow cytometry, transfected cells on the 24-well plate were washed with 1× PBS (500 μL for each well) to remove any residual serum, dead cells and debris. Next, trypsin (100 μL) was added and incubated for 5–10 min to detach the cells from the plate. PBS (100 μL) was then added and pipetted up and down to break up cell clumps. A solution of 4% paraformaldehyde (100 μL) was added to fix the cells. Samples were kept in covered flow cytometry tubes until analysis (BD FACSCanto, Franklin Lakes, NJ, USA).2.11 MTT assay of polymers
For MTT assays, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded in each well of a 96-well plate one day before transfection. The cells were then transfected as described above, save for an 80% reduction in volume and reagent quantity to accommodate the reduced well volume. The cells were incubated for 4 h at 37 °C in the transfection mix before being returned to a fresh growth medium. After 48 h, the cells were washed with PBS and an MTT solution was added. Following a 4 h incubation at 37 °C, an MTT solubilization solution (10% Triton X-100 in acidic (0.1 M HCl) isopropanol) was added to the cells and the absorbance of 570 nm light was quantified on a PerkinElmer plate reader (Waltham, MA, USA).
000 cells were seeded in each well of a 96-well plate one day before transfection. The cells were then transfected as described above, save for an 80% reduction in volume and reagent quantity to accommodate the reduced well volume. The cells were incubated for 4 h at 37 °C in the transfection mix before being returned to a fresh growth medium. After 48 h, the cells were washed with PBS and an MTT solution was added. Following a 4 h incubation at 37 °C, an MTT solubilization solution (10% Triton X-100 in acidic (0.1 M HCl) isopropanol) was added to the cells and the absorbance of 570 nm light was quantified on a PerkinElmer plate reader (Waltham, MA, USA).2.12 hESC transfection
hESCs were seeded in Matrigel-coated 24-well plates. Plasmid DNA (1 μL, 1 mg mL−1) was diluted in OptiMEM (50 μL). The polymer solution (10–40 μL, 1 mg mL−1) was diluted with OptiMEM (50 μL). The two solutions were then vortexed gently and allowed to incubate for 5 min at rt, after which they were combined and allowed to incubate for another 20 min at rt. Next, the mixtures were added to the cells dropwise and allowed to incubate at 37 °C for 4 h. The medium was then aspirated and fresh medium was added. After 48 h, the cells were stained with DAPI (250 μL, 3 nM) and SSEA-4-PE (250 μL, 0.02 mg mL−1), a pluripotency cell marker, for 30 min at 37 °C.3. Results
3.1 Synthesis and characterization of PEG-b-PVBLG-8 (PEV)
γ-(4-Vinylbenzyl)-L-glutamate N-carboxyanhydride (VB-Glu-NCA) was prepared by following previously reported methods.31,32,34 The ring-opening polymerization of VB-Glu-NCA with PEG-amine as the macroinitiator yielded PEG-block-poly(γ-(4-vinylbenzyl)-L-glutamate) (PEG-b-PVBLG) with controlled molecular weights (MWs) and narrow molecular-weight distributions (Scheme 2). At the VB-Glu-NCA–PEG-amine ratio of 100, the obtained Mn of 20.7 × 103 g mol−1 agreed well with the theoretical Mn of 24.6 × 103 g mol−1 and had a narrow molecular weight distribution of 1.21 (entry 1, Table 1). Two PEG-b-PVBLG copolymers were prepared with degrees of polymerization (DP) of 76 (PEG-b-PVBLG76) and 287 (PEG-b-PVBLG287) of the PVBLG block (Table 1). The ozonation of PEG-b-PVBLG yielded PEG-b-poly(γ-(4-aldehydebenzyl-L-glutamate) (PEG-b-PABLG), which served as the reactive intermediate that, through subsequent hydroamination and reduction with N-(2-aminoethyl)piperidine, yielded the desired PEG-b-PVBLG76-8 (PEV-L) and PEG-b-PVBLG287-8 (PEV-H). Grafting efficiencies greater than 95% were achieved for both PEV-L and PEV-H (Table 2).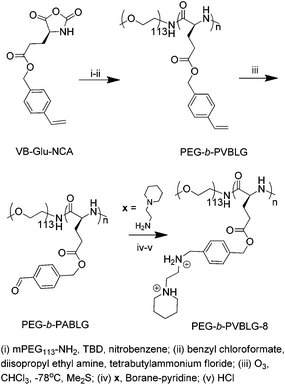 | ||
| Scheme 2 The chemical route for the preparation of PEG-b-PVBLG-8 from VB-Glu-NCA. | ||
Both PEV-L and PEV-H are highly soluble in water at pH 1–10 (>50 mg mL−1), which is drastically different from the corresponding parental (PEG-b-PVBLG) and intermediate polymers (PEG-b-PABLG) that are insoluble in water. The excellent water solubility of PEV-L and PEV-H is clearly related to their charged side groups, which make it possible for the application of PEV at physiological pH. Both PEV-L and PEV-H showed the characteristic CD spectra of an α-helix with two minima at 208 and 222 nm (Fig. 1a), consistent with our previously reported α-helical conformation of PVBLG-8.32,41 Helical contents of greater than 90% were observed for both PEV-L and PEV-H at pH 3 when the side chain amine groups are protonated (Table 2). As expected, the charge repulsion of the side groups had minimal effect on helix stability because the charged amine groups were placed far away from the polypeptide backbone. Furthermore, the helicity—as measured by the value of −[θ]222—was shown to be stable against pH and salt changes in the surrounding environment. For example, the −[θ]222 values of PEV-L and PEV-H remained unchanged when the solution pH was increased from 1 to 9 (Fig. 1b). The helices of PEV-L and PEV-H were also fairly stable in concentrated denaturing conditions, such as in 1 M NaCl (Fig. 1d) and 2 M urea (Fig. 1d) aqueous solutions. These observations suggested that PEVs would maintain their helical conformation in various extracellular and intracellular environments with well-preserved properties throughout the gene transfection processes.
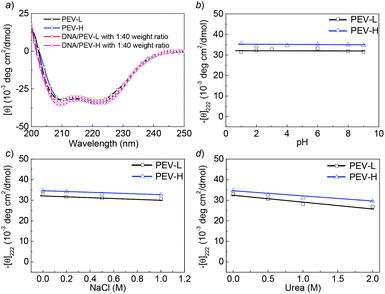 | ||
Fig. 1 (a) CD spectra in water of PEV-L, PEV-H, DNA–PEV-L, and DNA–PEV-H at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 weight ratio at pH 3. (b) The pH dependence of the residue molar ellipticity at 222 nm for PEV-L and PEV-H at 0.05 mg mL−1. (c) Salt dependence of residue ellipticity at 222 nm for PEV-L and PEV-H at pH 3 and 0.05 mg mL−1. (d) The helical stabilities of PEV-L and PEV-H at pH 3 and 0.05 mg mL−1 in the presence of urea. 40 weight ratio at pH 3. (b) The pH dependence of the residue molar ellipticity at 222 nm for PEV-L and PEV-H at 0.05 mg mL−1. (c) Salt dependence of residue ellipticity at 222 nm for PEV-L and PEV-H at pH 3 and 0.05 mg mL−1. (d) The helical stabilities of PEV-L and PEV-H at pH 3 and 0.05 mg mL−1 in the presence of urea. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer weight ratios between 1
polymer weight ratios between 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 and run on an agarose gel. The results for PEV-L can be seen in Fig. 2a. The addition of polymer in excess of 1
60 and run on an agarose gel. The results for PEV-L can be seen in Fig. 2a. The addition of polymer in excess of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (DNA
2 (DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer weight ratio) resulted in the formation of stable complexes which prohibited the migration of DNA under an electrophoretic force. Interestingly, at a 1
polymer weight ratio) resulted in the formation of stable complexes which prohibited the migration of DNA under an electrophoretic force. Interestingly, at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 DNA
2 DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer weight ratio, the DNA could still be seen in the loading well, indicating incomplete condensation. However, the DNA was no longer visible when sufficient polymer was added to achieve a 1
polymer weight ratio, the DNA could still be seen in the loading well, indicating incomplete condensation. However, the DNA was no longer visible when sufficient polymer was added to achieve a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 DNA
10 DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer weight ratio, indicating complete condensation. As can be seen in Fig. 1a, DNA binding does not affect the α-helicity of the peptide.
polymer weight ratio, indicating complete condensation. As can be seen in Fig. 1a, DNA binding does not affect the α-helicity of the peptide.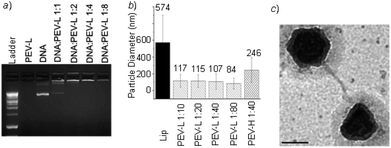 | ||
| Fig. 2 DNA–polymer complex analysis: (a) Gel retardation of the PEV-L at different DNA to polymer weight ratios. (b) Particle size analysis of the PEV-L copolymer and the DNA complex with different PVBLG-8 chain lengths and DNA to polymer weight ratios. (c) TEM image of PEV-L with the DNA complex. Scale bar: 200 nm. | ||
3.2 Dynamic light scattering
Dynamic light scattering revealed complexes formed between DNA and Lipofectamine 2000 at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 DNA
2 DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Lipofectamine 2000 weight ratio to be approximately 574 nm in diameter. Meanwhile, the hydrodynamic diameters of complexes of PEV-L and PEV-H at a 1
Lipofectamine 2000 weight ratio to be approximately 574 nm in diameter. Meanwhile, the hydrodynamic diameters of complexes of PEV-L and PEV-H at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 DNA
40 DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer weight ratio were substantially smaller—about 107 nm and 246 nm for PEV-L and PEV-H, respectively (Fig. 2b). Testing of DNA
polymer weight ratio were substantially smaller—about 107 nm and 246 nm for PEV-L and PEV-H, respectively (Fig. 2b). Testing of DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer weight ratios less than and greater than 1
polymer weight ratios less than and greater than 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 did not dramatically change the measured diameter—provided that a minimum amount of polymer was added to achieve complexation. For example, the diameters of complexes made with DNA
40 did not dramatically change the measured diameter—provided that a minimum amount of polymer was added to achieve complexation. For example, the diameters of complexes made with DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEV-L weight ratios of 1
PEV-L weight ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 1
20, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 and 1
40 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 were 117 nm, 115 nm, 107 nm, and 84 nm, respectively. This suggests that once a minimum amount of polypeptide is present—presumably enough to achieve a 1
80 were 117 nm, 115 nm, 107 nm, and 84 nm, respectively. This suggests that once a minimum amount of polypeptide is present—presumably enough to achieve a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 DNA
10 DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer weight ratio based on Fig. 2a—the excess polymer is not incorporated into the complexes and exists freely in the solution. Furthermore, despite the rod-like structure of the helical polypeptide, complexes formed between DNA and PEG-PVBLG-8 possessed a globular structure under TEM (Fig. 2c). Zeta potential measurements were performed to ensure that the complex formed between PEV-L (or PEV-H) and DNA were overall positively charged. PEV–DNA complexes formed possessed zeta potentials between 2 and 10 mV, while PVBLG-8/DNA complexes possessed zeta potentials between 20 and 30 mV (data not shown). The addition of the PEG block to the polypeptide shielded the cationic charge of the PVBLG-8 block.
polymer weight ratio based on Fig. 2a—the excess polymer is not incorporated into the complexes and exists freely in the solution. Furthermore, despite the rod-like structure of the helical polypeptide, complexes formed between DNA and PEG-PVBLG-8 possessed a globular structure under TEM (Fig. 2c). Zeta potential measurements were performed to ensure that the complex formed between PEV-L (or PEV-H) and DNA were overall positively charged. PEV–DNA complexes formed possessed zeta potentials between 2 and 10 mV, while PVBLG-8/DNA complexes possessed zeta potentials between 20 and 30 mV (data not shown). The addition of the PEG block to the polypeptide shielded the cationic charge of the PVBLG-8 block.3.3 Toxicity
The toxicity of the PEV-H was compared with PVBLG-8 homopolymer (P0) and Lipofectamine 2000 via MTT assays in IMR-90 cells. PVBLG-8 (P0) was found to be notably toxic to IMR-90 cells. At 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 DNA
40 DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer weight ratio, a viability of 6% was observed. The new diblock polymers of both low (PEV-L) and high (PEV-H) molecular weight showed substantially reduced toxicity to IMR-90 cells (Fig. 3a). Increasing the DNA
polymer weight ratio, a viability of 6% was observed. The new diblock polymers of both low (PEV-L) and high (PEV-H) molecular weight showed substantially reduced toxicity to IMR-90 cells (Fig. 3a). Increasing the DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer ratio resulted in increased toxicity for both PEV-L and PEV-H polymers. PEV-H–DNA complexes showed slightly reduced toxicity compared to the PEV-L–DNA complex, but both PEV-L- and PEV-H polymers were less toxic than Lipofectamine 2000 under similar conditions.
polymer ratio resulted in increased toxicity for both PEV-L and PEV-H polymers. PEV-H–DNA complexes showed slightly reduced toxicity compared to the PEV-L–DNA complex, but both PEV-L- and PEV-H polymers were less toxic than Lipofectamine 2000 under similar conditions.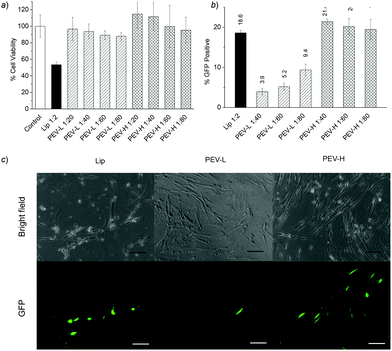 | ||
| Fig. 3 In vitro analysis in IMR-90 cells. (a) MTT cell viability assay of the DNA–polymer complex with different polymers and at different DNA to polymer weight ratios in IMR-90. The amount of DNA was fixed at 1 μg. (b) Initial testing for PEV-L along with Lipofectamine 2000 (Lip) and PVBLG-8 (P0) with varying pEGFP-N1 plasmid and polymer amounts. Transfection efficiency was analysed 48 h post-transfection with flow cytometry. (c) Fluorescent images of the transfection using Lip and PEV-H. Scale bars = 0.25 mm. | ||
3.4 Transfection
Transfection experiments were performed with the diblock co-polymers to determine whether their reduced toxicity impacted their ability to effectively deliver genes to IMR-90 cells. As shown in Fig. 3b, the commercial reagent Lipofectamine 2000 resulted in approximately 19% of treated cells expressing the delivered GFP transgene. Unmodified PVBLG-8 (P0), on the other hand, had a transfection efficiency of only 1.4% with low cell viability. While PVBLG-8 performed comparable to Lipofectamine 2000 in COS-7 and HeLa cells in previous studies, its poor performance as shown in Fig. 3b is likely due to the excessive toxicity of the polypeptide in IMR-90 cells. Reducing the toxicity of PVBLG-8 through the addition of PEG blocks resulted in an increase in IMR-90 transfection efficiency of 9.4% and 21.4% for PEV-L and PEV-H, respectively. Because of the reduced toxicity of PEV-L and PEV-H, the DNA dosage could be increased from 1 μg to 2 μg, which resulted in slightly higher transfection efficiencies, 13% and 27%, for PEV-L and PEV-H formulations, respectively (data not shown). Although this increase in efficiency was modest, it should be highlighted that the diblock polymers appeared to have reduced toxicity. For example, even though both Lipofectamine 2000- and PEV-H-transfected cells expressed similar amounts of GFP in Fig. 3c, the cells transfected with PEV-H possessed an overall healthier phenotype with flat and elongated shapes as opposed to cells transfected with Lipofectamine 2000 that appeared to be sparse and rounded.To further demonstrate the application of PEG-b-PVBLG-8, we next evaluated the gene delivery efficiency to H1 hESC. PEV-H transfection in separated H1 cells resulted in higher transfection efficiency than in cells plated as colonies (Fig. 4a and b). Moreover, the polymer was shown to have no impact on hESC pluripotency 48 h post-transfection. This was evidenced by the similar expression of stage-specific embryonic antigen-4 (SSEA-4) both before and after transfection in colonies and single cells (Fig. 4c). Further evidence of pluripotency was shown through a Western blot of the OCT4 pluripotency transcription factor before and after transfection with PEV-H (Fig. 4d). Combined with its efficiency, the mild cell impact of the PEV-H made it a promising reagent to manipulate the gene expression of human stem cells.
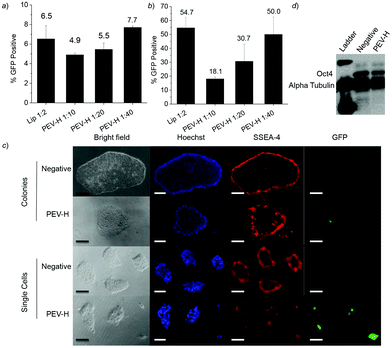 | ||
| Fig. 4 EGFP plasmid transfection efficiency using Lip and PEV-H of H1 hESC as (a) small colonies and (b) single cells as analysed by flow cytometry. (c) Bright field and fluorescence imaging of PEV-H transfection of EGFP plasmid into hESC as colonies and single cells. Colonies were stained with Hoechst and pluripotency marker SSEA-4 antibody conjugated with PE. Scale bar: 250 μm. (d) Western blot of cells isolated 72 h post-transfection demonstrating the protein expression of OCT4 in hESC H1 cells. | ||
4. Discussion
Transfection efficiency is often limited due to the toxicity of vectors. Generally, the more efficient the transfection material, the greater the impact it may have on cell health. This is largely due to the requirements of effective gene delivery—namely, a high polycationic charge to condense DNA and enable cell surface binding and membrane-lytic properties to facilitate intracellular escape from endocytic vesicles. While good for the delivery of nucleic acids, highly cationic materials can also bind and interfere with the function of necessary proteins within the cell. Moreover, the membrane lytic effect of materials can be unspecific and may act in a desirable as well as an undesirable manner. Effective gene delivery materials are able to balance their positive and negative tendencies so that they are efficient enough to allow macromolecules like DNA to enter the cells but safe enough that the cells are not irreparably damaged during the process. In this paper, we focus on the previously described PVBLG-8 materials and try to reduce their overall toxicity while maintaining their effective gene delivery performance.Previous characterizations of the helical polypeptide PVBLG-8 revealed that it can operate as an effective delivery vector for both DNA and siRNA.41 In both cases, its performance was demonstrated to be tied to its ability to form stable helices that cause pore formation within membranes. In the case of DNA delivery, the membrane lytic potential was essential to the escape of DNA–polypeptide complexes from endocytic vesicles. In the case of siRNA delivery, the helical PVBLG-8 caused pore formation within cell membranes to allow the non-endocytic diffusion of siRNA into the cell cytosol. Therefore, in our desire to reduce the toxicity of the PVBLG-8 materials, it was also essential to retain their helical structure. Unfortunately, just as helicity makes the materials effective delivery agents, it is also a contributing factor to overall toxicity. As such, we sought a strategy to append charge shielding materials to the helical PVBLG-8. By shielding the positive charge, we hoped to minimize the toxicity of the material by reducing its electrostatic attraction with negatively charged cell membranes. At the same time, since the helicity of the PVBLG-8 block was maintained (Fig. 1a), we hoped that the material would retain its ability to effectively escape the endosome and mediate effective gene delivery (Scheme 1).
To test the feasibility of incorporation of charge shielding groups, PEG was covalently conjugated with PVBLG-8 to yield the diblock polymer PEG-b-PVBLG-8. The PEG used had a MW of 5000 Da (DP of 113) and was conjugated to helical PVBLG-8 with DPs of 76 and 287 to yield the diblock materials PEV-L and PEV-H, respectively (Scheme 2). Circular dichroism experiments revealed that the incorporation of PEG did not alter the presence and stability of the helices even after the DNA is bound and condensed by the peptide (Fig. 1a). Moreover, the materials were also able to bind and condense plasmid DNA into spherical particles with diameters of the order of 100 nm (Fig. 2a–c). Despite the inclusion of PEG, these particles were demonstrated to retain an overall positive surface charge similar to commercial materials like Lipofectamine 2000 by analyzing their surface zeta potential (data not shown).
Toxicity measurements with the diblock materials revealed that the inclusion of a PEG block substantially increased the biocompatibility of the materials in IMR-90 cells. For example, treatment with the unmodified PVBLG-8 left only approximately 5% of IMR-90 cells viable. However, with the addition of a PEG block, cell viability increased dramatically and was less toxic than Lipofectamine 2000 (Fig. 3a). With its improved toxicity profile, PEG-b-PVBLG-8 was able to mediate effective transfection in IMR-90 cells. Previously, the toxicity of the materials was so extreme that only 1.4% of cells were successfully transfected. With the reduced toxicity, that number was increased to approximately 20%—an increase of approximately 14-fold. With its improved safety profile, PEG-b-PVBLG-8 was mild enough to transfect H1 human embryonic stem cells without affecting the expression of cell pluripotency markers (Fig. 4).
5. Conclusions
We prepared diblock copolymers consisting of poly(ethylene glycol)-block-poly(γ-4-(((2-(piperidin-1-yl)ethyl)amino)methyl)-benzyl-L-glutamate) (PEG-b-PVBLG-8) and evaluated their capability to mediate gene delivery in IMR-90 human fetal lung fibroblasts and human embryonic stem cells (hESCs). The PEG-b-PVBLG-8 contained a membrane-disruptive, cationic, helical polypeptide block (PVBLG-8) for complexing with DNA and a hydrophilic PEG block to improve the biocompatibility of the gene delivery vehicle. PEG-b-PVBLG-8 copolymers with low (n = 76) and high (n = 287) degrees of polymerization (n) of the PVBLG-8 block were synthesized. We found that the incorporation of PEG effectively reduced the toxicity of the helical PVBLG-8 block without dramatically compromising the complexation of copolymers with DNA and their transfection efficiencies. PEG-b-PVBLG-8 diblock polymer with a high degree of polymerization had a greater transfection efficiency and lower toxicity in IMR-90 cells than the commercial reagent Lipofectamine 2000. The usefulness of PEG-b-PVBLG-8 was further demonstrated via the successful transfection of hESCs without a measured loss in cell pluripotency markers. In contrast to many other polymer- and lipid-based transfection systems which utilize the proton sponge mechanism (e.g. polyethylenimine) or lipid mixing (e.g. DOTAP, DOPE, etc.) to facilitate endosomal escape, the peptides described here make use of a novel pore formation mechanism. As endocytic escape is generally considered one of the most challenging aspects of gene delivery, this alternative endosomolytic mechanism may prove useful when working with cell lines not readily amenable to transfection by current methods (i.e. IMR-90 and hES cells). As both IMR-90 and hESCs are important cell lines that are commonly used in the field of regenerative medicine, effective non-viral gene delivery to these cells is an important step in bringing regenerative medicine closer to clinical applications.Acknowledgements
This work was supported by NIH (Director's New Innovator Award 1DP2OD007246 and 1R21EB013379 for J.C) and NSF (CHE 1153122). J.Y. was funded at UIUC by National Science Foundation (NSF) Grant 0965918 IGERT: Training the Next Generation of Researchers in Cellular and Molecular Mechanics and BioNanotechnology.References
- U. P. Davé, N. A. Jenkins and N. G. Copeland, Gene therapy insertional mutagenesis insights, Science, 2004, 303, 333 CrossRef.
- K. Leong, H. Mao, K. Roy, S. Walsh and J. August, DNA-polycation nanospheres as non-viral gene delivery vehicles, J. Controlled Release, 1998, 53, 183 CrossRef CAS.
- K. Kizjakina, J. M. Bryson, G. Grandinetti and T. M. Reineke, Cationic glycopolymers for the delivery of pDNA to human dermal fibroblasts and rat mesenchymal stem cells, Biomaterials, 2011, 33, 1851–1862 CrossRef.
- P. M. McLendon, K. M. Fichter and T. M. Reineke, Poly (glycoamidoamine) vehicles promote pDNA uptake through multiple routes and efficient gene expression via caveolae-mediated endocytosis, Mol. Pharm., 2010, 7, 738–750 CrossRef CAS.
- S. Srinivasachari and T. M. Reineke, Versatile supramolecular pDNA vehicles via “click polymerization” of β-cyclodextrin with oligoethyleneamines, Biomaterials, 2009, 30, 928–938 CrossRef CAS.
- F. Yang, S. W. Cho, S. M. Son, S. R. Bogatyrev, D. Singh, J. J. Green, Y. Mei, S. Park, S. H. Bhang and B. S. Kim, Genetic engineering of human stem cells for enhanced angiogenesis using biodegradable polymeric nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 3317–3322 CrossRef CAS.
- F. Yang, J. Green, T. Dinio, L. Keung, S. Cho, H. Park, R. Langer and D. Anderson, Gene delivery to human adult and embryonic cell-derived stem cells using biodegradable nanoparticulate polymeric vectors, Gene Ther., 2009, 16, 533–546 CrossRef CAS.
- M. S. Shim and Y. J. Kwon, Controlled delivery of plasmid DNA and siRNA to intracellular targets using ketalized polyethylenimine, Biomacromolecules, 2008, 9, 444–455 CrossRef CAS.
- M. S. Shim and Y. J. Kwon, Acid-transforming polypeptide micelles for targeted nonviral gene delivery, Biomaterials, 2010, 31, 3404–3413 CrossRef CAS.
- M. S. Shim and Y. J. Kwon, Dual mode polyspermine with tunable degradability for plasmid DNA and siRNA delivery, Biomaterials, 2011, 32, 4009–4020 CrossRef CAS.
- X. Jiang, Y. Zheng, H. H. Chen, K. W. Leong, T. H. Wang and H. Q. Mao, Dual-sensitive micellar nanoparticles regulate DNA unpacking and enhance gene-delivery efficiency, Adv. Mater., 2010, 22, 2556–2560 CrossRef CAS.
- U. Pfisterer, A. Kirkeby, O. Torper, J. Wood, J. Nelander, A. Dufour, A. Björklund, O. Lindvall, J. Jakobsson and M. Parmar, Direct conversion of human fibroblasts to dopaminergic neurons, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 10343–10348 CrossRef CAS.
- R. Ambasudhan, M. Talantova, R. Coleman, X. Yuan, S. Zhu, S. A. Lipton and S. Ding, Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions, Cell Stem Cell, 2011, 113–118 CrossRef CAS.
- E. Szabo, S. Rampalli, R. M. Risueño, A. Schnerch, R. Mitchell, A. Fiebig-Comyn, M. Levadoux-Martin and M. Bhatia, Direct conversion of human fibroblasts to multilineage blood progenitors, Nature, 2010, 468, 521–526 CrossRef CAS.
- J. Yu, M. A. Vodyanik, K. Smuga-Otto, J. Antosiewicz-Bourget, J. L. Frane, S. Tian, J. Nie, G. A. Jonsdottir, V. Ruotti and R. Stewart, Induced pluripotent stem cell lines derived from human somatic cells, Science, 2007, 318, 1917–1920 CrossRef CAS.
- J. Zou, M. L. Maeder, P. Mali, S. M. Pruett-Miller, S. Thibodeau-Beganny, B. K. Chou, G. Chen, Z. Ye, I. H. Park and G. Q. Daley, Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells, Cell Stem Cell, 2009, 5, 97–110 CrossRef CAS.
- J. J. Green, B. Y. Zhou, M. M. Mitalipova, C. Beard, R. Langer, R. Jaenisch and D. G. Anderson, Nanoparticles for gene transfer to human embryonic stem cell colonies, Nano Lett., 2008, 8, 3126–3130 CrossRef CAS.
- M. Monsigny, A. C. Roche, P. Midoux and R. Mayer, Glycoconjugates as carriers for specific delivery of therapeutic drugs and genes, Adv. Drug Delivery Rev., 1994, 14, 1–24 CrossRef CAS.
- E. Wagner, D. Curiel and M. Cotten, Delivery of drugs, proteins and genes into cells using transferrin as a ligand for receptor-mediated endocytosis, Adv. Drug Delivery Rev., 1994, 14, 113–135 CrossRef CAS.
- S. K. Cho and Y. J. Kwon, Polyamine/DNA polyplexes with acid-degradable polymeric shell as structurally and functionally virus-mimicking nonviral vectors, J. Controlled Release, 2011, 150, 287–297 CrossRef CAS.
- T. Ferkol, J. C. Perales, F. Mularo and R. W. Hanson, Receptor-mediated gene transfer into macrophages, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 101–105 CrossRef CAS.
- P. Erbacher, M. T. Bousser, J. Raimond, M. Monsigny, P. Midoux and A. C. Roche, Gene transfer by DNA/glycosylated polylysine complexes into human blood monocyte-derived macrophages, Hum. Gene Ther., 1996, 7, 721–729 CrossRef CAS.
- D. Putnam, C. A. Gentry, D. W. Pack and R. Langer, Polymer-based gene delivery with low cytotoxicity by a unique balance of side-chain termini, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 1200–1205 CrossRef CAS.
- T. Okuda, A. Sugiyama, T. Niidome and H. Aoyagi, Characters of dendritic poly (-lysine) analogues with the terminal lysines replaced with arginines and histidines as gene carriers in vitro, Biomaterials, 2004, 25, 537–544 CrossRef CAS.
- N. P. Ingle, B. Malone and T. M. Reineke, Poly (glycoamidoamine)s: a broad class of carbohydrate-containing polycations for nucleic acid delivery, Trends Biotechnol., 2011, 443–453 CrossRef CAS.
- R. N. Johnson, D. S. H. Chu, J. Shi, J. G. Schellinger, P. M. Carlson and S. H. Pun, HPMA-oligolysine copolymers for gene delivery: optimization of peptide length and polymer molecular weight, J. Controlled Release, 2011, 303–311 CrossRef CAS.
- D. E. Robertson, R. S. Farid, C. C. Moser, J. L. Urbauer, S. E. Mulholland, R. Pidikiti, J. D. Lear, A. J. Wand, W. F. DeGrado and P. L. Dutton, Design and synthesis of multi-haem proteins, Nature, 1994, 425–432 CrossRef CAS.
- V. Muñoz and L. Serrano, Elucidating the folding problem of helical peptides using empirical parameters, Nat. Struct. Mol. Biol., 1994, 1, 399–409 Search PubMed.
- K. M. Stewart, K. L. Horton and S. O. Kelley, Cell-penetrating peptides as delivery vehicles for biology and medicine, Org. Biomol. Chem., 2008, 6, 2242–2255 CAS.
- S. Deshayes, M. Morris, G. Divita and F. Heitz, Cell-penetrating peptides: tools for intracellular delivery of therapeutics, Cell. Mol. Life Sci., 2005, 62, 1839–1849 CrossRef CAS.
- H. Lu, J. Wang, Y. G. Bai, J. W. Lang, S. Y. Liu, Y. Lin and J. Cheng, Ionic polypeptides with unusual helical stability, Nat. Commun., 2011, 2, 206 CrossRef.
- N. P. Gabrielson, H. Lu, L. C. Yin, D. Li, F. Wang and J. Cheng, Reactive and bioactive cationic α-helical polypeptide template for nonviral gene delivery, Angew. Chem., Int. Ed., 2012, 51, 1143–1147 CrossRef CAS.
- W. A. R. Vanheeswijk, M. J. D. Eenink and J. Feijen, An improved method for the preparation of gamma-esters of glutamic-acid and beta-esters of aspartic-acid, Synthesis-Stuttgart, 1982, 744–747 CrossRef CAS.
- H. Lu, Y. G. Bai, J. Wang, N. P. Gabrielson, F. Wang, Y. Lin and J. Cheng, Ring-opening polymerization of gamma-(4-vinylbenzyl)-L-glutamate N-carboxyanhydride for the synthesis of functional polypeptides, Macromolecules, 2011, 44, 6237–6240 CrossRef CAS.
- H. Lu, J. Wang, Y. Lin and J. Cheng, One-pot synthesis of brush-like polymers via integrated ring-opening metathesis polymerization and polymerization of amino acid N-carboxyanhydrides, J. Am. Chem. Soc., 2009, 131, 13582–13583 CrossRef CAS.
- H. Lu and J. Cheng, N-Trimethylsilyl amines for controlled ring-opening polymerization of amino acid N-carboxyanhydrides and facile end group functionalization of polypeptides, J. Am. Chem. Soc., 2008, 130, 12562–12563 CrossRef CAS.
- H. Lu and J. Cheng, Hexamethyldisilazane-mediated controlled polymerization of alpha-amino acid N-carboxyanhydrides, J. Am. Chem. Soc., 2007, 129, 14114–14115 CrossRef CAS.
- Y. G. Bai, H. Lu, E. Ponnusamy and J. Cheng, Synthesis of hybrid block copolymers via integrated ring-opening metathesis polymerization and polymerization of NCA, Chem. Commun., 2011, 47, 10830–10832 RSC.
- N. J. Greenfield, Using circular dichroism spectra to estimate protein secondary structure, Nat. Protoc., 2006, 1, 2876–2890 CrossRef CAS.
- J. A. Morrow, M. L. Segall, S. Lund-Katz, M. C. Phillips, M. Knapp, B. Rupp and K. H. Weisgraber, Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain, Biochemistry, 2000, 39, 11657–11666 CrossRef CAS.
- N. P. Gabrielson, H. Lu, L. Yin, K. H. Kim and J. Cheng, A cell-penetrating helical polymer for siRNA delivery to mammalian cells, Mol. Ther., 2012, 20, 1599–1609 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2013 |
