Design, synthesis and characterization of a hexapeptide bio-inspired by acetylcholinesterase and its interaction with pesticide dichlorvos
Glauco Pilon dos
Santos
a,
Bianca Ferreira da
Silva
a,
Saulo Santesso
Garrido
a,
Marcello
Mascini
b and
Hideko
Yamanaka
*a
aUNESP – Univ Estadual Paulista, Institute of Chemistry, Rua Prof. Francisco Degni 55, 14800-900, Araraquara, SP, Brazil. E-mail: hidekoy@iq.unesp.br
bFaculty of Bioscience and Technology for Food, Agriculture and Environment, University of Teramo, 64023, Teramo, Italy
First published on 15th October 2013
Abstract
This paper describes the molecular modeling design, synthesis and characterization of a new bio-inspired hexapeptide of acetylcholinesterase enzyme and its interaction with the organophosphate pesticide dichlorvos monitored by UV-Vis spectroscopy and mass spectrometry. This strategy can contribute to the development of synthetic receptors to be coupled to biosensor transducers, avoiding the issues associated with proteins such as low stability under different pH and temperature conditions and high production cost. The resulting data of this work indicate a strong interaction between the pesticide dichlorvos and the hexapeptide (NH3+-Glu-His-Gly-Gly-Pro-Ser-COO−) with a binding constant of 4.10 × 105 M−1 and the formation of an adduct by covalent binding on the serine residue from the hexapeptide.
1. Introduction
Enzymes have been used in the construction of analytical devices to quantify substrates by catalytic reactions.1,2 Therefore, high price and low stability at higher temperatures and extremely high or low pH values generally result in complete loss of activity, which impedes widespread use of these biological molecules. Particularly in the biosensor area, the use of biological receptors still limits their commercialization as biotechnological devices mainly because of the limitations on the reproducibility, reliability and short lifetime of the device.3In this context, metallic complexes4,5 and molecularly imprinted polymers6,7 have been proposed to mimic enzymes. On the other hand, studies of new computational methodologies for designing artificial molecules as a mimic of enzyme active sites are considered to be attractive for the development of tailor-made receptors to be coupled to biosensor transducers.8 The use of recently developed molecular modeling software and techniques, synthetic methods, as well as the construction of new and improved synthetic receptors, offers a great help for the rational design of bio-inspired systems with high specificity and affinity.9
Although, the design of these new synthetic molecules requires a good knowledge of protein-folding predictions and surface-binding chemistries in order to obtain sufficiently detailed information to perform rational design of proteins.10 Helpful information on the key role of the structure–function of proteins usually involves research in biochemistry, bioinformatics, molecular biology, instrumental methods such as X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, and computational approaches.11
Laboratory-based research between simulated binding scores and experimental results has been reported, showing the ability of synthetic receptors such as aptamers or peptides to bind the target.12,13 These ligands are able to selectively interact with different molecules, similarly to enzymes or biological receptors. Especially the peptides, generally synthesized by solid-phase methodology, offer some advantages compared to other affinity approaches such as low cost, storage stability, simple and rapid synthesis.14
Mascini et al.15 computationally designed two different hexapeptides for binding cocaine, with the experimental screening done by solid phase extraction followed by liquid chromatography mass spectrometry in plasma samples. Rosca et al.16 developed a 20 amino acid biomimetic peptide derived from collagen IV with an anti-angiogenic activity in breast cancer. Cui et al.17 evaluated by molecular modeling a biomimetic peptide for the detection of trinitrotoluene, without the stability issues associated with proteins and with a preferred recognition molecule.
The approach chosen in this work relies on the design and development of an artificial hexapeptide inspired by the acetylcholinesterase enzyme (AChE) binding site. The highly selective properties of the biological receptor make it an ideal target for mimicking organophosphate (OP) and carbamate (CM) pesticide recognition.18 The high toxicity of both pesticide classes is ascribed to their ability to inhibit the AChE activity, which is essential to break down the neurotransmitter acetylcholine at cholinergic synapses. Inhibition of AChE activity allows acetylcholine to remain active in the synapse, thus leading to fatal consequences.19
The identification of the binding site of AChE and the target enzyme of CM and OP pesticides has led to the knowledge of the inhibition mechanism, given in the work by Millard et al.20 where structural and functional information on the amino acids involved in the pesticide binding is reported. Studying the three-dimensional structure of the complex between the CM/OP pesticides and the AChE, it was found to reproduce a very similar shape of the AChE binding site to artificial oligopeptides.21
In particular, the docking program FRED22 used in this research has already been proved to calculate binding scores in line with other software and with experimental data. The hexapeptide selected for experimental purposes (NH3+-Glu-His-Gly-Gly-Pro-Ser-COO−) derived from a previously tested tetrapeptide (NH3+-His-Glu-Pro-Ser-COO−)21 has the ability to bind the pesticide paraoxon with a binding constant of 5.06 × 102 M−1. The tetrapeptide sequence already studied was modified by swapping histidine with glutamic acid and adding two glycine amino acids in order to achieve a better simulation of the AChE binding site geometry.
This work reports the evaluation by computational design of this new hexapeptide bio-inspired by the active site of acetylcholinesterase enzyme. The hexapeptide sequence was synthesized by the solid phase methodology and characterized by high performance liquid chromatography and mass spectrometry. The binding constant between the pesticide dichlorvos and the hexapeptide beyond investigations on the adduct formation was also determined by UV-Vis spectroscopy and mass spectrometry, respectively. The resulting data of this work were presented and discussed.
2. Materials and methods
2.1 Chemicals
The polymeric support Fmoc-Ser(tBu)-WangResin, with a substitution of 0.63 mmol g−1, all the Fmoc-amino acids (Fmoc-Ser(tBu), Fmoc-Pro, Fmoc-Gly, Fmoc-His(trt), Fmoc-Glu(tBu)) and N-hydroxybenzotriazole (HOBt) were obtained from Novabiochem. N,N′-Diisopropylcarbodiimide (DIC) and trifluoroacetic acid (TFA) were acquired from Fluka. Triisopropylsilane (TIS) and 1,2-ethanedithiol (EDT) were purchased from Acros. Solvents for chromatographic procedures were of high performance liquid chromatography (HPLC) grade. Dichloromethane (DCM), dimethylformamide (DMF), glacial acetic acid, dichlorvos, disodium hydrogen phosphate (Na2HPO4) and sodium dihydrogen phosphate (NaH2PO4) were obtained from Sigma-Aldrich. Acetonitrile and piperidine solution 20% (v/v) in DMF were acquired from Hexis. The pesticide stock solution was prepared in acetonitrile and diluted with phosphate buffer solution 0.01 M at pH 7.0 to the desired concentration. All the aqueous solutions were prepared with ultrapure water (resistance >18 MΩ cm, Millipore Inc., USA) of Milli-Q quality.2.2 Instrumentation
Absorbance measurements were performed using a Hewlett-Packard UV-Vis Spectrophotometer Model 8453 controlled with Chemstation software, using a quartz cell with a light path of 10 mm and a capacity of 500 μL (Hellma Analytics).The chromatographic analysis of the synthesized peptide was carried out using a Shimadzu model LC-10 A/C-47A using a reverse phase Nucleosil C18 column (25 × 0.46 cm; 5 μm; 300 Å).
A hybrid triple quadrupole linear ion trap mass spectrometer (LC-ESI-MS/MS 3200Q TRAP, Applied Biosystems/MDS Sciex) equipped with a Turbo Ion Spray ion source was used to analyze the hexapeptide sequence and its interaction with the pesticide dichlorvos.
2.3 Computational design
Molecular modeling calculations were performed using a desktop PC with a 3.4 GHz Intel Core I7-2600 processor having 8 GBytes DDR3 RAM with 1333 MHz bus, running Microsoft Windows 7 Professional 64 bits.All structures were designed and cleaned up with Hyperchem 8.0.5. The hexapeptide was designed in zwitterionic mode. An OpenEye Scientific Software package under academic license was used. The energy minimization process was carried out using SZYBKI 1.5.1 in its default parameterization.23 OMEGA 2.4.3 was used to generate conformers to both dichlorvos and hexapeptide.24 To consider the hexapeptide flexibility, 10 conformers were required to make a reliable computed binding score, as reported in other work.13 The program was used with MMFF as the force field. The docking software FRED 2.2.522 was used to conduct the virtual screening applying default parameters. VIDA 4.1.1 was used for visualization, post-calculation analysis and representation.25
2.4 Solid phase peptide synthesis
The peptide synthesis was manually performed using the solid phase methodology26 in order to reach the desired sequence: NH3+-Glu-His-Gly-Gly-Pro-Ser-COO−. Specifically in this study, the first amino acid residue was already coupled to the resin through an ester bond with the α-carboxyl group (Fmoc-Ser(tBu)-WangResin). On the coupling of each amino acid was used a molar excess of 3 equivalents of the Fmoc-amino acid and 3 equivalents of the condensing agents (DIC/HOBt) in a mixture of DCM–DMF (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) for 2 hours. After coupling, the qualitative ninhydrin test was performed to evaluate the effectiveness of the reaction; the recoupling procedure was done when the ninhydrin test was positive. The removal of the Fmoc group after the coupling of each amino acid was done by adding piperidine solution 20% (v/v) in DMF for 20 minutes. All the steps of coupling and deprotection are followed by alternating washing steps with DCM and DMF.
50) for 2 hours. After coupling, the qualitative ninhydrin test was performed to evaluate the effectiveness of the reaction; the recoupling procedure was done when the ninhydrin test was positive. The removal of the Fmoc group after the coupling of each amino acid was done by adding piperidine solution 20% (v/v) in DMF for 20 minutes. All the steps of coupling and deprotection are followed by alternating washing steps with DCM and DMF.
The cleavage of resin and removal of the side chain protecting groups were performed with a mixture containing 95% TFA, 2.5% ultrapure water and 2.5% triisopropylsilane at 25 °C for 2 hours. Then, the hexapeptide was precipitated, washed with cold ethyl ether and extracted in a mixture of water–acetic acid (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5). Then, the hexapeptide was separated by filtration, concentrated and lyophilized.
5). Then, the hexapeptide was separated by filtration, concentrated and lyophilized.
The purification of the crude hexapeptide sequence was done by semipreparative HPLC using a reverse phase Nucleosil C18 column (25 × 2.5 cm; 15 μm; 300 Å) on a linear gradient, from 0 to 25% of ACN containing 0.036% TFA, in 90 minutes. The characterization of the purified peptide was done by analytical HPLC using a reverse phase Nucleosil C18 column (25 × 0.46 cm; 5 μm; 300 Å) on a linear gradient, from 5 to 95% of ACN containing 0.036% of TFA, in 30 minutes, with a flow rate of 1 mL min−1 and detection at 220 nm. The peptide was also analyzed using electrospray mass spectrometry.
2.5 Spectrophotometric evaluation of the interaction between the hexapeptide and the pesticide
Absorbance measurements for the evaluation of the interaction between the hexapeptide and the pesticide dichlorvos were performed by keeping the concentration of the peptide at 8 × 10−4 mol L−1 and varying the concentration of dichlorvos in the range of 1 × 10−8 to 1 × 10−3 mol L−1. All the solutions were prepared in 0.01 M phosphate buffer solution at pH 7.0 and incubated for 2 minutes before the measurement. Changes in the absorption at λmax = 250 nm, related to the complex formation, was considered on the calculation of the binding constant, by comparing its extinction coefficient with the value obtained for the free hexapeptide, adapting the mathematic model applied by Zhang et al.272.6 Mass spectrometry measurements
The characterization of the hexapeptide sequence after the synthesis was done by direct injection through electrospray ionization mass spectrometry (ESI-MS) at the following settings: positive ionization mode, syringe flow = 10 μL min−1, curtain gas = 10, ion spray voltage = 4500 V, ion source gas 1 = 50, declustering potential = 90 V, ion source gas nitrogen at 20 L min−1, entrance potential = 10 V, ion energy = 1.0 V, and a detector channel electron multiplier at 2100 V.The preparation of a non-stoichiometrically mixed solution containing the peptide and an excess of pesticide was carried out in order to identify the formation of the complex by mass spectrometry. The concentration of both analytes used for the direct injection was 1 ppm, prepared in phosphate buffer solution 0.01 M at pH 7.0. The adduct formation was evaluated using the same settings as described above, except for ion source gas 1 = 13 and a detector channel electron multiplier at 2200 V.
3. Results and discussion
The hexapeptide was built taking into account the amino acid residues of the AChE binding pocket, but reducing the size to control the possible shape during the computational simulation. To achieve a better simulation of the AChE binding site geometry, the tetrapeptide NH3+-His-Glu-Pro-Ser-COO−, already tested to be able to bind pesticides,21 was mutated by swapping histidine with glutamic acid in the amino-terminal position and adding two glycine residues in the third and the fourth position, respectively, obtaining the hexapeptide NH3+-Glu-His-Gly-Gly-Pro-Ser-COO−. As reported in Table 1, the geometries of the amino acids involved in the pesticide binding of the AChE (the catalytic triad Ser-200, His-440, Glu-327), in the minimized tetrapeptide previously studied and in the new minimized hexapeptide proposed, were compared using as parameters the angles and distances of the Cα of each amino acid. The angles were calculated wherever the position in the peptides or AChE sequence using the Cα of the central amino acid (in boldface in Table 1). Considering the distances between the Cα of each amino acid involved in the binding, the AChE and hexapeptide serine distances from both histidine and glutamic acid were comparable and they were twice that calculated using amino acids from the tetrapeptide. Looking at the angles, the hexapeptide matched the geometry of the catalytic triad, showing minor differences. Instead, the tetrapeptide angles were quite different from the original spatial position in the enzyme AChE, especially in the angle formed by the amino acids His-Ser-Glu.| Angle Cα-Cα (deg) | |||
|---|---|---|---|
| AChE | EHGGPS | HEPS | |
| Ser-Glu-His | 55.7 | 47.2 | 50.6 |
| Glu-His-Ser | 98.1 | 114.7 | 85.6 |
| His-Ser-Glu | 26.3 | 18.8 | 43.8 |
| Distances Cα-Cα (Å) | ||||
|---|---|---|---|---|
| Ser | Glu | His | ||
| Ser | AChE | 0 | 10.00 | 8.34 |
| EHGGPS | 0 | 10.78 | 8.66 | |
| HEPS | 0 | 5.63 | 4.36 | |
| Glu | AChE | 10.00 | 0 | 4.47 |
| EHGGPS | 10.78 | 0 | 3.80 | |
| HEPS | 5.63 | 0 | 3.91 | |
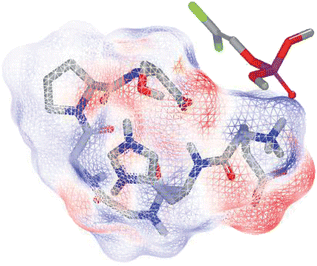
The binding score and the three-dimensional hexapeptide structure in docking dichlorvos were calculated by FRED. The hexapeptide 10 conformers' average binding score was –4.28 ± 0.53 having a 3-fold increase in binding compared to the previously tested tetrapeptide with a FRED 10 conformers' average binding score equal to –1.30 ± 0.18. As reported in Fig. 1, dichlorvos was in the center of the simulated hexapeptide electrostatic molecular surface. It should be noted that the in vivo interaction can be due to a synergic cooperation of the amino acid residues that have a certain amount of freedom to move around the carbon backbone (much larger than in proteins), increasing the probability of interaction with the target, especially when the target is found in the center of the complex.
Chemical hexapeptide synthesis started from the Fmoc-Ser(tBu)-WangResin, with a substitution of 0.63 mmol g−1. Using DIC/HOBt as the condensing agents, the six amino acids of the sequence were coupled by the activation of the amino group from an amino acid, generating reactive species in order to produce the amide bond with the other amino acid. After the coupling steps, the hexapeptide was precipitated, extracted in a mixture of water–acetic acid (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
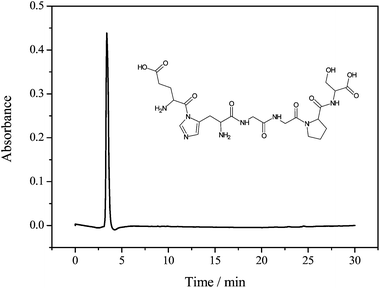
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5), separated by filtration, concentrated and lyophilized. The crude hexapeptide was purified by semipreparative HPLC. Fig. 2 shows the characterization of the desired peptide sequence by analytical HPLC after the process of purification. As can be seen, a retention time of 3.4 min was observed and a purity degree above 95% was obtained, enabling its application in the spectroscopic and spectrometric measurements.
5), separated by filtration, concentrated and lyophilized. The crude hexapeptide was purified by semipreparative HPLC. Fig. 2 shows the characterization of the desired peptide sequence by analytical HPLC after the process of purification. As can be seen, a retention time of 3.4 min was observed and a purity degree above 95% was obtained, enabling its application in the spectroscopic and spectrometric measurements.
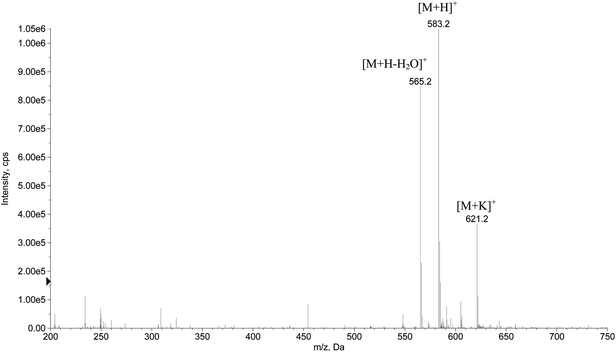
The purified hexapeptide was also analyzed in positive ionization mode using ESI-MS for the confirmation and identification of the ion related to the theoretical mass of the peptide (582.5 g mol−1). As can be seen in Fig. 3, the ratio m/z 583.2 is related to the ion [M + H]+. The corresponding loss of water [M + H − 18]+ at m/z 565.2 and the potassium adduct [M + K]+ at m/z 621.2 were also identified, proving that the desired hexapeptide was obtained by the solid phase synthesis.
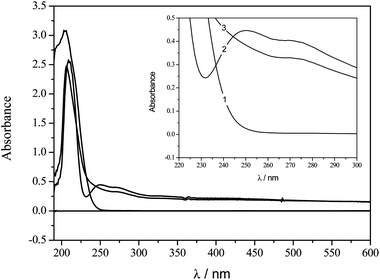
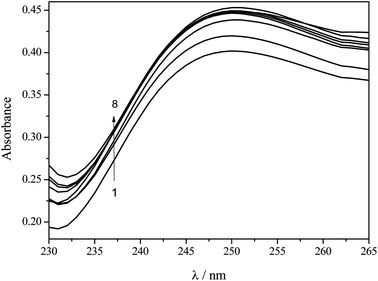
UV-Vis molecular absorption spectroscopy is an important technique employed for studying the affinity of compounds.28–30 As can be seen in Fig. 4, the pesticide dichlorvos presents a band with a maximum of absorption at λmax = 205 nm while the peptide presents bands at λmax = 210 and 275 nm. By the addition of a known amount of pesticide to a solution containing the same concentration of hexapeptide another band at λmax = 250 nm was observed, indicating the formation of a hexapeptide–pesticide complex.
The formation of the dichlorvos adduct with the hexapeptide proposed in this work was investigated by ESI-MS in positive ionization mode. As can be seen in Fig. 5, the ratio m/z 690.8 is related to the hexapeptide with an added mass of +108 after its complexation with the pesticide dichlorvos. Another ion of m/z 581.4 is related to a source fragmentation of the complex structure ([M + H − C2H7O3P]+). The toxicological mechanism involving the covalent binding of dichlorvos on the serine residue from the hexapeptide and its source fragmentation is proposed in Fig. 6. Li et al.31 also proposed that the organophosphorus and carbamate pesticides form a covalent bond with the serine residue of the enzyme binding site, identifying the adducts on Ser 198 of butyrylcholinesterase (BChE) with the pesticides dichlorvos, chlorpyrifos oxon, aldicarb and Baygon.
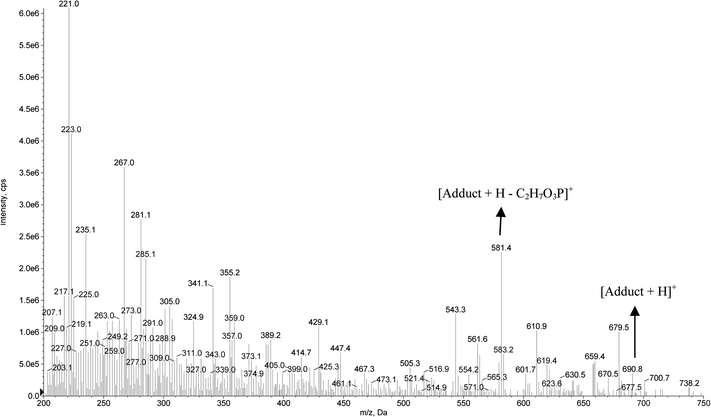 | ||
| Fig. 5 Full scan mass spectrum for the adduct formation between the hexapeptide and the pesticide dichlorvos. | ||
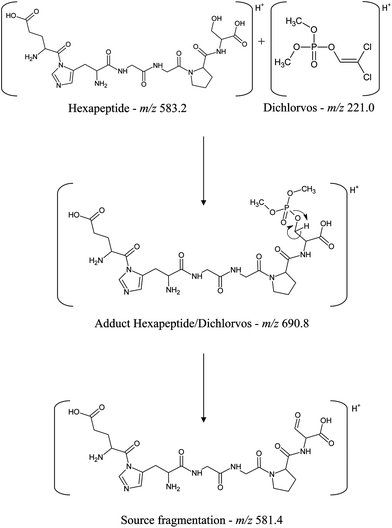 | ||
| Fig. 6 Scheme of the covalent binding of dichlorvos on the serine residue from the hexapeptide and its source fragmentation. | ||
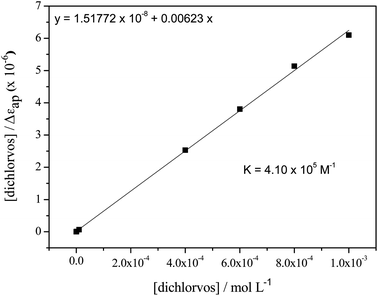
The binding constant (K) related to the formation of the hexapeptide–dichlorvos complex was evaluated by varying the pesticide concentration from 1 × 10−8 to 1 × 10−3 mol L−1 and keeping the hexapeptide concentration at 8 × 10−4 mol L−1. As can be seen in Fig. 7, increasing the concentration of dichlorvos leads to the formation of more products and consequently increases the absorption intensity at λmax = 250 nm.
The procedure for the calculation of K was carried out by adapting the mathematical model applied by Zhang et al.,27 where the half-reciprocal plot of [dichlorvos]/Δεapversus [dichlorvos] (Fig. 8), with Δεap = |εa − εf|, εa = Acomplex/[hexapeptide] and εf the extinction coefficient for the free hexapeptide (εf = 402.34 L mol−1 cm−1, obtained from the calibration curve of the hexapeptide at λmax = 275 nm in the range of 5 × 10−5 and 1 × 10−3 mol L−1), will have a slope of 1/Δε and an intercept of 1/(Δε × K). K is then given by the ratio of the slope to the intercept, resulting in a value of 4.10 × 105 M−1, which indicates a strong interaction between the hexapeptide and the pesticide dichlorvos.
The main mechanism of insecticide action is inhibition of enzyme cholinesterase; the proposed hexapeptide also interacted with acephate and methamidophos (organophosphate insecticide) but no interaction with the herbicide clomazone was detected.
4. Conclusions
The computational simulation data of the peptide conformers' average binding score were found to be in agreement with experimental laboratory results, supporting the methodology proposed in this work. The strong interaction between the pesticide dichlorvos and the hexapeptide, with a binding constant of 4.10 × 105 M−1 and the formation of an adduct by covalent binding on the serine residue, demonstrates that this synthetic receptor shows a promising future for coupling to biosensor transducers, avoiding the issues associated with proteins.Acknowledgements
The authors would like to thank FAPESP (Proc. 2010/04663-6), CNPq (Proc. 313307/2009-1) and FP7-PEOPLE-IRSES project BIOMIMIC 230849 for the financial support.References
- H.-W. Yanga, M.-Y. Hua, S. L. Chen and R.-Y. Tsai, Biosens. Bioelectron., 2013, 41, 172–179 CrossRef PubMed.
- P. Sun and Y. Wu, Sens. Actuators, B, 2013, 178, 113–118 CrossRef CAS PubMed.
- J. A. Berberich, L. W. Yang, I. Bahar and A. J. Russell, Acta Biomater., 2005, 1, 183–191 CrossRef PubMed.
- M. L. Carvalho, M. Santhiago, R. Peralta, A. Neves, G. A. Micke and I. C. Vieira, Talanta, 2008, 77, 394–399 CrossRef PubMed.
- A. C. Boni, M. D. P. T. Sotomayor, M. R. V. Lanza, S. M. C. N. Tanaka and A. A. Tanaka, J. Braz. Chem. Soc., 2010, 21, 1377–1383 CrossRef CAS PubMed.
- G. Wulff and J. Liu, Acc. Chem. Res., 2012, 45, 239–247 CrossRef CAS PubMed.
- G. Díaz-Díaz, D. Antuña-Jiménez, M. C. Blanco-López, M. J. Lobo-Castañón, A. J. Miranda-Ordieres and P. Tuñón-Blanco, TrAC, Trends Anal. Chem., 2012, 33, 68–80 CrossRef PubMed.
- M. Mascini, M. Del Carlo, D. Compagnone, G. Perez, L. A. Montero-Cabrera, S. Gonzalez and H. Yamanaka, Sensors and Microsystems. Lecture Notes in Electrical Engineering, 2011, 91, 403–407 CrossRef.
- S. Friedle, E. Reisner and S. J. Lippard, Chem. Soc. Rev., 2010, 39, 2768–2779 RSC.
- M. Sarikaya, C. Tamerler, A. K. Y. Jen, K. Schulten and F. Baneyx, Nat. Mater., 2003, 2, 577–585 CrossRef CAS PubMed.
- H.-L. Liu and J.-P. Hsu, Proteomics, 2005, 5, 2056–2068 CrossRef CAS PubMed.
- N. Dave and J. Liu, Chem. Commun., 2012, 48, 3718–3720 RSC.
- G. Perez, M. Mascini, M. Sergi, M. Del Carlo, R. Curini, L. A. Montero-Cabrera and D. Compagnone, J. Proteomics Bioinf., 2013, 6, 15–22 CAS.
- A. S. Galanis, F. Albericio and M. Grotli, Org. Lett., 2009, 11, 4488–4491 CrossRef CAS PubMed.
- M. Mascini, C. Montesano, M. Sergi, G. Perez, M. Cicco, R. Curini and D. Compagnone, Anal. Chim. Acta, 2013, 772, 40–46 CrossRef CAS PubMed.
- E. V. Rosca, J. E. Koskimaki, N. B. Pandey, A. C. Wolff and A. S. Popel, Cancer Biol. Ther., 2011, 12, 808–817 CrossRef CAS PubMed.
- Y. Cui, S. N. Kim, R. R. Naik and M. C. McAlpine, Acc. Chem. Res., 2012, 45, 696–704 CrossRef CAS PubMed.
- K. Mwila, M. H. Burton, J. S. Van Dyk and B. I. Pletchke, Environ. Monit. Assess., 2013, 185, 2315–2327 CrossRef CAS PubMed.
- D. Liu, W. Chen, J. Wei, X. Li, Z. Wang and X. Jiang, Anal. Chem., 2012, 84, 4185–4191 CrossRef CAS PubMed.
- C. B. Millard, G. Kryger, A. Ordentlich, H. M. Greenblatt, M. Harel, M. L. Raves, Y. Segall, D. Barak, A. Shafferman, I. Silman and J. L. Sussman, Biochemistry, 1999, 38, 7032–7039 CrossRef CAS PubMed.
- M. Mascini, M. Sergi, D. Monti, M. Del Carlo and D. Compagnone, Anal. Chem., 2008, 80, 9150–9156 CrossRef CAS PubMed.
- OpenEye Scientific Software, Santa Fe, New Mexico, FRED version 2.2.5 Search PubMed.
- OpenEye Scientific Software, Santa Fe, New Mexico, SZYBKI version 1.5.1 Search PubMed.
- OpenEye Scientific Software, Santa Fe, New Mexico, OMEGA version 2.4.3 Search PubMed.
- OpenEye Scientific Software, Santa Fe, New Mexico, VIDA version 4.1.1 Search PubMed.
- R. B. Merrifield, J. Am. Chem. Soc., 1963, 85, 2149–2154 CrossRef CAS.
- L. Z. Zhang and G.-Q. Tang, J. Photochem. Photobiol., B, 2004, 74, 119–125 CrossRef CAS PubMed.
- C. V. Uliana, G. S. Garbellini and H. Yamanaka, Sens. Actuators, B, 2013, 178, 627–635 CrossRef CAS PubMed.
- C. V. Uliana, G. S. Garbellini and H. Yamanaka, J. Braz. Chem. Soc., 2012, 23, 1469–1475 CrossRef CAS PubMed.
- Y. Liu and R. Liu, Food Chem. Toxicol., 2012, 50, 3298–3305 CrossRef CAS PubMed.
- B. Li, I. Ricordel, L. M. Schopfer, F. Baud, B. Megarbane, P. Masson and O. Lockridge, J. Appl. Toxicol., 2010, 30, 559–565 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |