Pancreatic cancer gene therapy using an siRNA-functionalized single walled carbon nanotubes (SWNTs) nanoplex
Tommy
Anderson
a,
Rui
Hu
a,
Chengbin
Yang
a,
Ho Sup
Yoon
b and
Ken-Tye
Yong
*a
aSchool of Electrical and Electronic Engineering, Nanyang Technological University, Singapore 639798, Singapore. E-mail: ktyong@ntu.edu.sg; Tel: +65-6790-5444
bDivision of Structural Biology & Biochemistry, School of Biological Sciences, Nanyang Technological University, Singapore 639798, Singapore
First published on 17th June 2014
Abstract
The discovery of RNA interference (RNAi) has created a new platform for cancer therapy applications. This approach utilizes small interfering RNA (siRNA) molecules to regulate the expression of a specific target gene and subsequently suppresses the growth of the cancer cells. However, the formulation of free siRNAs alone is incapable of transfecting cells as they are negatively charged and degrade in biological fluids. For successful siRNA transfection, a biocompatible and functional carrier is needed. In this contribution, we demonstrated the preparation of functionalized single walled carbon nanotubes (SWNTs) as efficient siRNA carriers and utilized the SWNTs/siRNA nanoplex for the in vitro gene therapy of pancreatic cancer. Through fluorescent imaging and quantitative flow cytometric analysis, we observed a high siRNA transfection efficiency mediated by the nanoplex formulation. We demonstrated the successful internalization of the nanoplex by the pancreatic cancer cell and the subsequent release of the siRNAs from the nanoplex, which resulted in a down-regulation of the target gene. In addition, the functionalized SWNTs proved to be highly biocompatible as assessed by cell viability tests. Our results suggest that in the near future the SWNTs may be able to serve as a multifunctional nanoplatform for the in vivo targeted gene therapy of pancreatic cancer.
Introduction
Cancer has remained one of the most fatal diseases in the 21st century. In the US, it was estimated that 1.6 million new cases and 580![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths from cancer were reported in 2013.1 Among various types of cancers, pancreatic cancer has been the 4th and 9th leading cause of cancer death in the US and worldwide, respectively.1,2 More importantly, pancreatic cancer patients possess a mere 4% chance of a 5-year survival rate.3 Due to these reasons, more effective treatment strategies for pancreatic cancer patients are urgently needed. Compared to conventional cancer therapies, RNA interference (RNAi) might provide a new way for the treatment of cancer. RNAi was first discovered in the late 1990s4 and has quickly received wide attention due to its potential application for gene therapy.5 The concept involves the use of sequence-specific small interfering RNAs (siRNAs), typically with a length of 20–25 base pairs, to mediate the degradation of the homologous messenger RNA and consequently to regulate the expression of the targeted gene.6–8 The high targeting specificity and efficiency of RNAi have initiated substantial clinical research trials in evaluating the potential of this approach for treating diseases,9,10 such as pancreatic cancer.11 Despite some success in these research trials, there is certainly more room for improvement in these studies. siRNAs are negatively charged and fragile in biological media.12 Thus, it is challenging to deliver bare siRNAs into cells as they generally result in a poor transfection efficiency and an ineffective gene therapy outcome. To overcome this challenge, several methods such as electroporation, calcium phosphate transfection, and microinjection have been developed13 for delivering the siRNA formulation to the target cells. However, these methods are complex and tedious. Moreover, they are not suitable for long-term use for in vivo applications. Such challenges can be overcome by using functionalized nanocarriers that aid the efficient delivery of siRNAs to the target cells.
000 deaths from cancer were reported in 2013.1 Among various types of cancers, pancreatic cancer has been the 4th and 9th leading cause of cancer death in the US and worldwide, respectively.1,2 More importantly, pancreatic cancer patients possess a mere 4% chance of a 5-year survival rate.3 Due to these reasons, more effective treatment strategies for pancreatic cancer patients are urgently needed. Compared to conventional cancer therapies, RNA interference (RNAi) might provide a new way for the treatment of cancer. RNAi was first discovered in the late 1990s4 and has quickly received wide attention due to its potential application for gene therapy.5 The concept involves the use of sequence-specific small interfering RNAs (siRNAs), typically with a length of 20–25 base pairs, to mediate the degradation of the homologous messenger RNA and consequently to regulate the expression of the targeted gene.6–8 The high targeting specificity and efficiency of RNAi have initiated substantial clinical research trials in evaluating the potential of this approach for treating diseases,9,10 such as pancreatic cancer.11 Despite some success in these research trials, there is certainly more room for improvement in these studies. siRNAs are negatively charged and fragile in biological media.12 Thus, it is challenging to deliver bare siRNAs into cells as they generally result in a poor transfection efficiency and an ineffective gene therapy outcome. To overcome this challenge, several methods such as electroporation, calcium phosphate transfection, and microinjection have been developed13 for delivering the siRNA formulation to the target cells. However, these methods are complex and tedious. Moreover, they are not suitable for long-term use for in vivo applications. Such challenges can be overcome by using functionalized nanocarriers that aid the efficient delivery of siRNAs to the target cells.
Recent advancements in the field of nanotechnology have opened up new avenues in utilizing nanoscale materials for biomedical applications. Different nanomaterials such as quantum dots (QDs), gold nanorods (GNRs), and polymeric nanoparticles have been used in biomedical applications such as bioimaging,14,15 biosensing,16 and targeted drug delivery.17,18 The size and surface morphology of these nanoparticles have made them suitable for cell transfection usage. Among all of these nanomaterials, carbon nanotubes (CNTs) are of great interest and have received broad attention due to their rich surface chemistries and the ease of loading biomolecules onto the hollow tubes. CNTs are carbon macromolecules in the shape of long tube nanostructures. These nanotubes are either composed of single or multiple rolled graphene sheets that form seamless tubes of single walled nanotubes (SWNTs) or multi walled nanotubes (MWNTs). SWNTs and MWNTs are quite similar in their structures although SWNTs exhibit smaller diameters and a more perfect structure geometry. In general, CNTs have a high aspect ratio, a high surface area and excellent electrical and mechanical properties,19,20 and have attracted wide interest for their applications in bioelectronics and nanomedicine.21
It is known that pristine CNTs have inert and hydrophobic surfaces, and tend to aggregate and form bundles due to strong hydrophobic, van der Waals, and π–π stacking interactions.22,23 Because of the hydrophobicity of pristine CNTs, they cannot disperse adequately in any aqueous solution, which makes it very challenging to use them for the many applications that require a stable suspension. In particular, for biomedical applications, CNTs need to be dispersed in biological buffers for in vitro and in vivo studies. Both physical and chemical approaches have been used for preparing aqueous dispersions of CNTs in recent years. Physical methods such as sonication are used to make dispersions of CNTs but this approach results in the development of defects in the nanotubes24 and poor dispersibility in water.25 In comparison, chemical functionalization approaches result in the excellent colloidal stability of aqueous dispersions of CNTs through facile surface modification steps such as covalent or non-covalent functionalization methods. Covalent functionalization methods generally involve the addition of functional groups (e.g. carboxyl-, hydroxyl-, and amine-groups) on the CNTs surface that will help to improve their dispersibility in water.26–28 This method usually requires breaking the carbon bonds of the CNT walls and subsequently adding functional groups to the structure.28,29 For example, Ramanathan et al. have covalently functionalized SWNTs with amide and amine groups to enable the aqueous dispersibility of the nanotubes.30 On the other hand, non-covalent methods involve coating the surface of CNTs with surface ligands such as phospholipids and surfactant-like polymers through hydrophobic interactions. This functionalization approach creates water-dispersible CNTs where the hydrophobic part of the surface ligand interacts with the CNTs hydrophobic surface and the hydrophilic part of the ligand protrudes out from the surface of the CNTs and interacts favorably with water molecules. Moore et al. have explored a non-covalent method to prepare SWNT dispersions by using various surfactants.31 To date, functionalized CNTs have been extensively used for nanomedicine applications such as imaging,32,33 sensing,34,35 tissue engineering,36 disease therapy,32,37 drug discovery38 and delivery.39
Previous studies have shown that nanoparticles are efficient functional nanocarriers for transporting biomolecules into cells. This is particularly beneficial for delivering target-specific fragile biomolecules to targeted cellular components, especially those that are prone to degradation after exposure to biological fluids. A good example is the delivery of siRNA molecules with nanocarriers such as chitosan nanoparticles,40 magnetic nanoparticles,41 polymer nanoparticles,10,42–45 gold nanoparticles,46,47 silica nanoparticles,48 polymer coated calcium phosphate nanoparticles,49 quantum dots50 and CNTs. As compared to other nanoparticles, CNTs have unique properties including their non-metal origin, high surface-to-volume ratio, high aspect ratio and unique chemical structure. These properties are extremely useful for enabling CNTs to be used as nanocarriers as they can be loaded with a high amount of functional groups to increase their biocompatibility and with various types of cargo molecules. Furthermore, due to their high aspect ratio, CNTs have been observed to be able to enter cells by non-endocytotic pathways.51,52 Although there are contradictory results which need further investigations,53 this is an interesting finding which could be beneficial for cellular delivery.
The application of CNTs for the delivery of siRNA into cells has been explored using various functionalization methods.54 For example, Dai's group have used CNTs functionalized with disulfide bonds for controlled release in cells for the delivery of thiol-modified siRNAs.55,56 This method was shown to induce a higher knockdown efficiency on human T cells than that of liposomes. CNTs have also been covalently functionalized with amine groups to deliver siRNAs in vitro and in vivo.57 Several groups have also explored the covalent functionalization of CNTs with cationic polymers,58 PEI,59 or dendrimer60 for siRNA attachment via electrostatic interaction. More importantly, studies have shown that the CNT-polymer conjugates exhibit lower cytotoxicity than polymers alone.58,61 In this study, we demonstrated the use of non-covalently functionalized SWNTs as transfection nanocarriers for delivering mutant K-Ras siRNAs for gene silencing of pancreatic cancer cells. Specifically, we first functionalized SWNTs with amphiphilic and cationic polymers for aqueous dispersion. The functionalized SWNTs were further complexed with siRNAs to form an SWNTs/siRNA nanoplex for gene therapy applications. Our results demonstrated that siRNAs can be efficiently delivered into the pancreatic cancer cells using the synthesized nanoplex formulation and induce gene silencing in the PANC-1 cells. In addition, the cytotoxicity study showed that the functionalized SWNTs are biocompatible over a wide range of concentrations. More importantly, the modification was performed using cationic poly(allylamine hydrochloride), which has a lower cytotoxicity than poly(diallyldiamine hydrochloride) or poly(ethylenimine).62 Due to the non-covalent nature of the functionalization, defects and breakage of the pristine CNT structure is minimized. This preserves the electronic properties of the CNTs and would be useful for applying functions such as Raman imaging and photothermal therapy simultaneously.
Experimental methods
Functionalization of carbon nanotubes
SWNTs were made water dispersible following a modified version of the procedure of Kam et al.55 Briefly, 160 mg of raw HiPCo Carbon Nanotube powder (containing 12.3 wt% of pure SWNT, Nanointegris) was mixed with mPEG-DSPE-5000 (Laysan Bio Inc.) at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 (SWNT
2.5 (SWNT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mPEG-DSPE-5000). To the above mixture, 20 mL of mini-Q DI water was added and sonicated for 4 hours in a water bath, resulting in a uniformly dispersed black solution. Insoluble impurities and bundled SWNTs were removed by centrifugation steps. Centrifugation was performed at 6000g for 30 minutes. The supernatant liquid was collected and centrifuged at 13
mPEG-DSPE-5000). To the above mixture, 20 mL of mini-Q DI water was added and sonicated for 4 hours in a water bath, resulting in a uniformly dispersed black solution. Insoluble impurities and bundled SWNTs were removed by centrifugation steps. Centrifugation was performed at 6000g for 30 minutes. The supernatant liquid was collected and centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500g for 30 minutes, before final centrifugation at 21
500g for 30 minutes, before final centrifugation at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200g for 1 hour. The supernatant liquid from the last stage was filtered through a 100 kDa filter (Corning) and washed several times to remove any excess mPEG-DSPE. The SWNTs were re-suspended in water for characterization. To further functionalize the SWNTs, 200 μL of 1 mg mL−1 poly(allylamine hydrochloride) (PAH, Sigma-Aldrich) was added dropwise into 800 μL of SWNT solution (Abs = 0.461, at 808 nm) under vigorous stirring. The solution was then centrifuged at 9400g for 10 minutes to remove the aggregates.
200g for 1 hour. The supernatant liquid from the last stage was filtered through a 100 kDa filter (Corning) and washed several times to remove any excess mPEG-DSPE. The SWNTs were re-suspended in water for characterization. To further functionalize the SWNTs, 200 μL of 1 mg mL−1 poly(allylamine hydrochloride) (PAH, Sigma-Aldrich) was added dropwise into 800 μL of SWNT solution (Abs = 0.461, at 808 nm) under vigorous stirring. The solution was then centrifuged at 9400g for 10 minutes to remove the aggregates.
Characterization of SWNT solution
The absorption spectra of SWNTs were measured using a Shimadzu UC-2450 UV-VIS Spectrophotometer. Samples were loaded into a standard 10 mm quartz cuvette and measured against water as the reference. Hydrodynamic size measurement based on dynamic light scattering (DLS) and zeta potential measurement by phase analysis light scattering (PALS) were carried out using a Brookhaven 90Plus Particle Size Analyzer. For Raman spectra measurement and AFM imaging, SWNTs samples weres prepared on a Si wafer. Raman spectroscopy was carried out using a WITec Raman system with an excitation laser at 532 nm. AFM images were obtained with a Shimadzu SPM-9500J2 instrument in the tapping mode.Cell culture and transfection
The human pancreatic cancer cell PANC-1 (CRL-1469, American Type Culture Collection) was maintained in culture with Dulbecco's Modified Eagle's Medium (DMEM, Hyclone), supplemented with 10% fetal bovine serum (FBS, Hyclone), 100 μg mL−1 penicillin (Gibco) and 100 μg mL−1 streptomycin (Gibco). Cells were cultured at 37 °C in a humidified atmosphere with 5% CO2. The day prior to transfection, cells were seeded onto 6-well plates to give 30%–50% confluence at the time of transfection. Before transfection, the cell medium was replaced with 1.1 mL of OPTI-MEM (Invitrogen) Serum Free medium. Mutant K-Ras siRNA–FAM (K-Ras siRNAFAM, sense strand: 5′-FAM-GUUGGAGCUGAUGGCGUAGUU-3′; Antisense: 5′-CUACGCCAUCAGCUCCAACUU-3, from Shanghai GenePharma, China) was solubilized in DEPC treated water at a density of 10 μM. Functionalized SWNT-PAH solution (10 μL) was mixed with siRNAFAM (20 μL, 10 μM) with gentle vortex mixing and left undisturbed for 20 minutes. The SWNT-PAH-siRNAFAM conjugates were then added to the PANC-1 cell culture and the cells were continuously cultured. Free siRNAFAM was used in a parallel experiment at the same dosage level. As a positive control, a commercially available transfection reagent Oligofectamine™ (Invitrogen)-coupled siRNAFAM was used according to the vendor's instructions. The gene expression was monitored at 48 hours post-transfection. For transfection efficiency examination, flow cytometry and fluorescence imaging were performed at 4 hours post-treatment.Fluorescence microscopy
In vitro fluorescence microscopy images were obtained using a fluorescence microscope (Eclipse-Ti, Nikon). The cells were washed with phosphate-buffered saline (PBS), fixed with 4% formaldehyde and mounted onto glass slides with DAPI loaded mounting media (Fluoroshield™, Sigma-Aldrich) before imaging. To image the cells, filter sets for DAPI (two band pass filters 350/50 nm and 460/50 nm, and a 400 nm long pass filter as the dichroic mirror) and FITC (two band pass filters 480/30 nm and 540/50 nm, and a 505 nm long pass filter as the dichroic mirror) were applied for DAPI and FAM signals, respectively.Flow cytometry
Transfected cells were washed with PBS before collection with trypsin, and then washed again afterwards. The FAM served as the luminescent marker (filter set for FITC was applied) to determine the transfection efficiency quantitatively. Measurements were performed using a FACSCalibur flow cytometer (Becton Dickinson, Mississauga, CA).Gene expression analysis
RNA was extracted from PANC-1 cells using TRIzol reagent (Invitrogen) 48 hours post transfection. The amount was quantified using a spectrophotometer (Nano- Drop ND-1000). A reverse transcriptase kit from Promega was used to reverse transcribe this total RNA (2 μg) to cDNA following the vendor’s instructions. The K-Ras mRNA expression level was determined by semi-quantitative PCR (Polymerase Chain Reaction), and compared to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is a housekeeping gene used for the normalization of gene expression.63 Electrophoresis of the PCR result was carried out on 1.2% ethidium bromide-stained agarose gel and observed under a UV transilluminator (Bio-Rad). Primers: K-Ras 5′-AGAGTGCCTTGACGATACAGC-3′ (sense), 5′-ACAAAGAAAGCCCTCCCCAGT-3′ (antisense); GAPDH 5′-ACCACAGTCCATGCCATCAC-3′ (sense), GAPDH 5′-TCCACCACCCTGTTGCTGTA-3′ (antisense).Cell viability study
Cell viability was measured by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, Sigma) assay. For this, cells were seeded in a 96-well plate at a density of 5 × 103 cells per well and incubated with a range of concentrations of SWNT and SWNT-PAH for 24 hours. After 24 hours, MTT (5 mg mL−1, 20 μL) in PBS was added and the cells were incubated for 4 hours at 37 °C with 5% CO2. Dimethylsulfoxide (DMSO, 150 μL, Sigma) was then added to solubilize the precipitate with 5 minutes of gentle shaking. Absorbance was measured with a microplate reader (Bio-Rad) at a wavelength of 490 nm. The cell viability was determined by normalizing the absorbance of the sample well against that from the control well and was expressed as a percentage, assigning the viability of the non-treated cells as 100%.Results and discussion
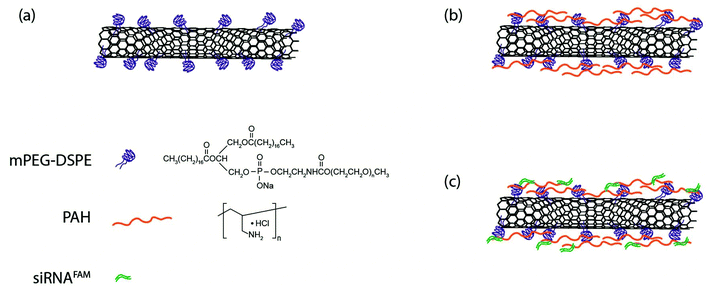
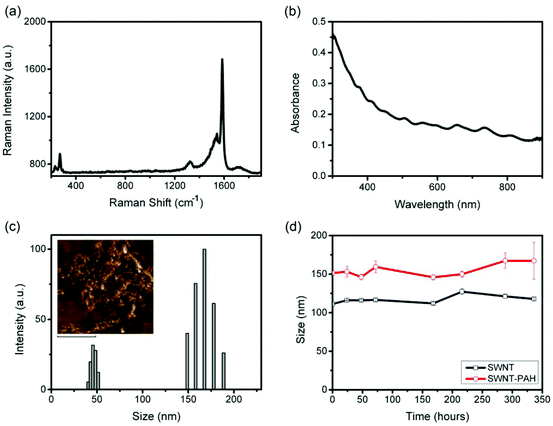
Fig. 1 shows a schematic illustration of the functionalization process for preparing SWNTs/siRNA nanoplexes for the gene therapy of pancreatic cancer cells. The carbon nanotubes were made water-dispersible by functionalizing them with PEGylated phospholipids (mPEG-DSPE). Homogenous SWNTs dispersion was obtained by further removing the amorphous carbons and other impurities from the dispersion by using filtration and centrifugation. The phospholipid-functionalized SWNTs exhibited negatively charged surfaces and subsequently they were coated with a positively charged polymer poly(allylamine hydrochloride) (PAH) through electrostatic interaction forces. PAH is a cationic polymer with repeating amine groups connected along the carbon chain backbone. PAH generally exhibits a much lower cytotoxicity as compared with other cationic polymers such as poly(dimethyl diallyl ammonium chloride) (PDDAC) and polyethylenimine (PEI).62 Thus, we have chosen PAH to modify the SWNTs surface. The addition of PAH onto the negatively-charged SWNTs surface creates a layer of PAH coating thereby altering the SWNTs surface charge to become positive. These positively-charged SWNTs were used to complex with the negatively-charged siRNA molecules through electrostatic interaction. The resulting SWNT/siRNA nanoplexes have a net positively charged surface that is useful for the intracellular delivery process.64The prepared SWNTs dispersion was systematically characterized by Raman spectrometry, UV-Vis spectrophotometry, and dynamic light scattering as shown in Fig. 2. Raman spectroscopy identified several unique features of the SWNTs and the spectra confirmed the structure of the nanotubes in the solution. In particular, as shown in Fig. 2a, a large peak is observed at 1587 cm−1, which is attributable to the graphitic G-Band of the SWNTs.65 A smaller D-Band peak is observable around 1300 cm−1, indicating that there are some defects and impurities within the SWNTs.65 This may be a result of the long sonication process in the preparation process which breaks and introduces defect sites to the SWNTs.66 Finally, the Radial Breathing Mode (RBM) of the SWNTs was also detectable at the lower region of 150–250 cm−1, which is related to the radial mode and diameter of the SWNTs.65 These SWNTs dispersed in water were then characterized by UV-Vis spectrophotometry, and their absorption spectrum is shown in Fig. 2b. According to Kam et al., the molar extinction coefficient of SWNTs was estimated to be 7.9 × 106 M−1 cm−1 at a wavelength of 808 nm.37 Although this value is dependent on the average length of the SWNTs, we adopted it as a useful reference and guideline to estimate the concentration of the nanotubes applied in our experiments.
The size of the functionalized SWNTs can be measured by different methods. For example, imaging modalities such scanning or transmission electron microscopy (SEM or TEM) can be used to determine the diameter, length and shape of 1-D SWNTs.22 However, such measurements are carried out on dehydrated or unsolvated SWNTs samples and the obtained values do not reflect the actual colloidal particle behavior. In our experiment, the size of the mPEG-DSPE functionalized SWNTs is represented in terms of the effective hydrodynamic diameter and the values were determined by the dynamic light scattering (DLS) technique. Measurement of the hydrodynamic diameter takes into account the effect of the surfactant size in solution media, Brownian motion of the nanoparticles, the aqueous solvation layer and diffusion of the nanoparticle.67 Hence, it is particularly important to consider the hydrodynamic diameter for in vivo applications.68,69 Despite the fact that the SWNTs are not spherical in their inherited form, our measured hydrodynamic size value for SWNTs is still very useful as a guide for understanding the colloidal stability of the formulation in various biological buffers. In addition, such measurements will be useful for studying cellular interaction with nanoparticles. For example, in cellular uptake studies, the approximate effective diameter of SWNTs is useful for comparison with other nanoparticles formulations.70 The effective hydrodynamic diameter of our SWNTs was estimated to be 110 nm (Fig. 2c), which is comparable to polymeric nanoparticles45 and mesoporous silica nanoparticles,48 and hence they are suitable for application as nanocarriers for intracellular delivery. To further verify the structure and morphology of the SWNTs, further investigation of the SWNTs was performed using atomic force microscopy (AFM) in our study. Our AFM analysis showed that the dispersed SWNTs display a silk-like structure (Fig. 2c, inset). We have monitored the hydrodynamic size over different time points and the results indicated that the SWNTs dispersion exhibited excellent colloidal stability and no changes in the size were observed over two weeks (Fig. 2d). The zeta potential value of these SWNTs was measured to be −25 mV, indicating a negatively charged surface for our prepared formulation. After coating the SWNTs surface with PAH, an increase in the hydrodynamic size from 110 to 150 nm was observed and a surface charge reversal was detected with a zeta potential value of +40 mV. The colloidal stability of the functionalized SWNT/PAH was also monitored and they were shown to have good colloidal stability comparable to that prior to PAH coating (Fig. 2d).
Prior to the use of nanomaterials for biomedical applications, the cytotoxicity issue of materials needs to be addressed. CNTs, in general, possess low cytotoxicity yet it is dependent on the type and degree of functionalization of CNTs.71 The cytotoxicity of the prepared SWNTs formulations was evaluated by carrying out MTT assays. PANC-1 cells were treated with phospholipid functionalized SWNTs, before and after PAH coating, at various concentrations. As shown in Fig. 3, the viability of the treated cells was maintained at over 80% for a concentration as high as 600 pM for 24 hours post treatment. Moreover, we did not observe any signs of morphological damage to the treated cells. This result indicates that the functionalized SWNTs are highly biocompatible and that they can be safely applied for biomedical applications within the concentration range reported here. It is also worth mentioning that, although the PAH coating changed the surface potential of the SWNTs from negative to positive, it did not affect the biocompatibility and colloidal stability of the prepared formulation.
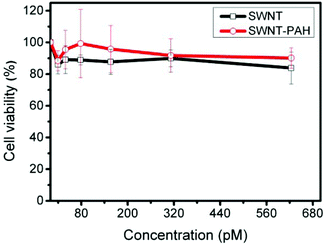 | ||
| Fig. 3 Cytotoxicity study of SWNT and SWNT/PAH by MTT assay. PANC-1 cancer cells were treated with various concentrations of SWNT and SWNT/PAH for 24 hours. Values shown are means ± SD, n = 5. | ||
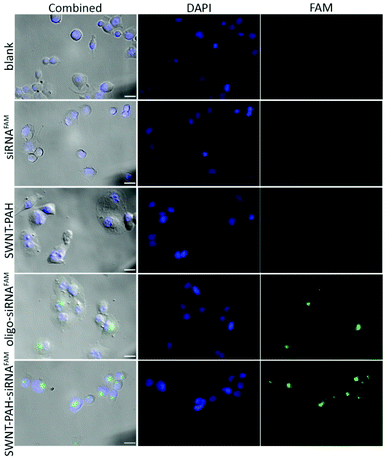
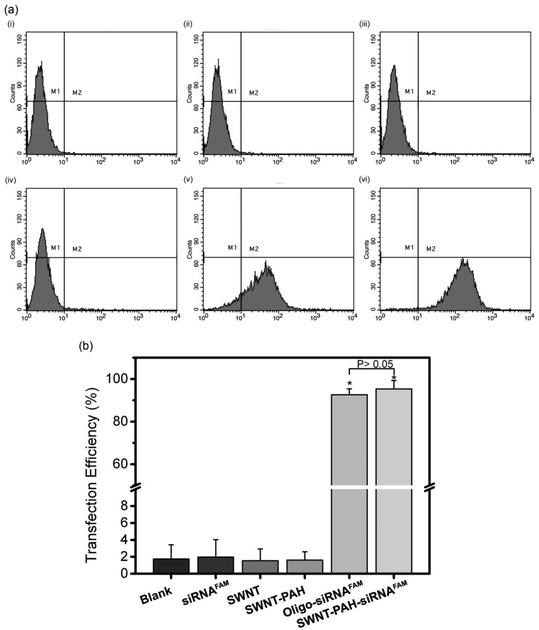
To demonstrate the potential therapeutic application of the SWNTs/PAH nanoformulation for gene therapy, the nanoparticles were complexed with siRNAs (SWNTs/PAH/siRNA nanoplexes) by electrostatic interaction between SWNT/PAH and siRNA molecules. These freshly made nanoplexes were then used for gene transfection against the mutant K-Ras gene in PANC-1 pancreatic cells. It is worth mentioning that the hydrodynamic size of the nanoplexes was monitored to be stable in aqueous solution for up to 72 hours. The K-Ras oncogene is a member of the Ras gene family which encodes important proteins that regulate normal cellular activities such as cell proliferation, differentiation and survival of the cell.72,73 Previous studies have shown that the mutational activation of the K-Ras gene occurs in almost all pancreatic cancers and causes abnormalities in cellular functions.74 These K-Ras siRNAs were labelled with fluorescent FAM (siRNAFAM) for visualization and were transfected to PANC-1 cells with different formulations, as shown in Fig. 4. The positive control group was transfected with a commercial transfection reagent Oligofectamine™ loaded with siRNAFAM (Oligo/siRNAFAM). For comparison, results from the non-transfected cells, cells transfected with solely siRNAFAM or with SWNT/PAH dispersion are also presented in Fig. 4. As shown in the figure, comparably high FAM fluorescent signals were observed in cells treated with the SWNTs/PAH/siRNAFAM nanoplexes and the Oligo/siRNAFAM formulation. This indicates that the siRNAs can be successfully delivered into the PANC-1 cells with the assistance of SWNTs/PAH nanocarrier. On the contrary, the FAM signal is not observable in the cells transfected with only siRNAFAM, due to the inability of bare siRNAFAM to enter the PANC-1 cells. Quantitative assessment of the transfection efficiency of siRNAFAMs by SWNTs/PAH/siRNAFAM nanoplexes formulation was performed by flow cytometric analysis. In the experiments, the FAM fluorescent signals from each cell were recorded and a threshold value was determined using the negative control group (cells treated with SWNT/PAH only, Fig. 5a). The transfection efficiency was measured as the fraction of the cell count with fluorescent signals above the threshold (Fig. 5b). The results of the flow cytometric analysis were in good agreement with the observations of the fluorescence microscopy analysis study. The cells treated with naked siRNAFAMs exhibit a negligible signal which is comparable with the background from the blank cells and the cells treated with SWNTs/PAH formulation. In contrast, significantly high values of (92.6 ± 2.1%) and (95.3 ± 4%) were observed for cells treated with the Oligo/siRNAFAM and SWNTs/PAH/siRNAFAM nanoplex, respectively. These results confirm that these functionalized SWNTs are efficient siRNA carriers for cell transfection.
After the successful delivery of the SWNTs/PAH/siRNA nanoplex into the cell cytoplasm, the siRNAs were released from the nanoplex and started to initiate the RNAi process. To validate the successful gene transfection, we examined down regulation of the targeted mRNA in the cells by PCR. Fig. 6 shows the relative expression levels of the mutant K-Ras mRNA in PANC-1 cells after treatment with different formulations for 48 hours. Similar expression levels were observed in the untreated cells (100 ± 12.2%) and the cells treated with SWNT (109.7 ± 5.73%) or SWNT/PAH (113.1 ± 4.11%) formulations. Cells treated with only siRNAs also exhibited high mutant K-Ras gene expression (106 ± 8.8%). In contrast, a notable knockdown was observed for the cells treated with the SWNTs/PAH/siRNA nanoplex formulation as the expression level of the targeted mutant K-Ras mRNA was suppressed to (66.88 ± 5.14%), which is comparable to that of the Oligo/siRNA formulation (56.92 ± 1.27%). Interestingly, covalently functionalized SWNTs electrostatically attached to siRNAs also reached similar knockdown efficiency as compared to the positive control.59,60 However, the knockdown efficiency was lower as compared to SWNTs attached to siRNA via disulfide bonds.56 It is worth noting that measurement of the knockdown efficiency could be affected by the duration of transfection, especially for nanocarriers with electrostatically loaded siRNA,40,46 as the nanocarrier conjugate might be more stable such that it releases siRNAs slower than the transfection agent in the positive control group.60 Nevertheless, this result suggests that the prepared SWNTs/PAH/siRNA nanoplex is effective as a transfection agent and the nanoplex is able to release the siRNA molecules within the cell and subsequently initiate the RNAi process for specific gene knockdown.
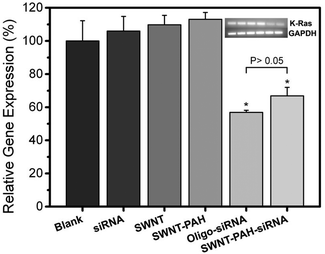 | ||
| Fig. 6 Relative gene expression levels of mutant K-Ras mRNA in PANC-1 cancer cells of different treatment groups. Values shown are means ± SD, n = 3 (*, P < 0.001 compared to negative control). | ||
The use of nanoparticles as carriers for gene transfection has received much attention recently. Compared to viral vectors, nanoparticles present a much lower risk for gene delivery applications. However, the potential toxicity of nanoparticles, especially for long-term settings, is still a major concern and certainly extensive studies are needed to understand their long term impact in the body. Nevertheless, some polymer based nanoparticle formulations have shown quite promising results for gene delivery and some of these formulations have been used for conducting clinical phase I human trials10 and such clinical trials will certainly shed some light on the continuous development of a RNAi based therapeutic strategy for human diseases. In this work, we demonstrated an alternative approach of using SWNTs for pancreatic cancer gene therapy. Our results have shown that after non-covalent functionalization with a biocompatible polymer, these one dimensional wire-like SWNTs are effective in loading siRNAs molecules. Moreover, we have demonstrated that the SWNTs/PAH/siRNA nanoplex displayes a high gene transfection efficiency with effective knockdown of the targeted gene in the pancreatic cancer cells. Most importantly, the MTT assay has shown that the formulation has a low toxicity over a wide range of concentrations. Previous in vivo studies have also demonstrated that CNTs are safe for use in animals.75,76 In the near future, we foresee that multifunctional and multimodal CNTs formulations could be engineered for advanced healthcare applications since the extremely large surface area of nanotubes makes them suitable for integration with drug molecules, imaging contrast agents, and targeting ligands. Furthermore, the multifunctionality of CNTs would be beneficial for applications in theranostics. The integration of all these factors into CNTs will be essential for future cancer therapy applications.
Conclusion
In summary, we have engineered SWNTs as a nanocarrier for siRNA delivery into pancreatic cancer cells. Non-covalent functionalization was performed on the SWNTs, which minimizes the creation of defects on the SWNT surface. An aqueous dispersion of the SWNTs was obtained by functionalization with PEGylated phospholipids through hydrophobic interaction. Further modification with a cationic polymer poly(allylamine hydrochloride) was performed on the SWNTs to alter their surface potential. The prepared SWNTs dispersion possessed a high biocompatibility over a wide range of concentrations as shown in a cell viability study and so was ready to be used for applications in a biological environment. The positively charged SWNTs were complexed with siRNAs for targeting the mutant K-Ras gene in PANC-1 cells by electrostatic interaction, thereby promoting gene therapy. A high gene transfection efficiency mediated by the SWNTs/PAH/siRNAFAM nanoplex was observed by using fluorescence microscopy and flow cytometric analysis. The knockdown of the targeted gene proved the successful internalization of the nanoplex by the cell. The siRNA molecules were released from the nanoplex and initiated the RNAi process. Based on these results we envision that with further developments the functionalized SWNTs might be able to provide a multifunctional platform for targeted cancer therapy with high efficiency.Acknowledgements
This study was supported by the Start-up grant (M4080141.040) from Nanyang Technological University, Tier 1 Academic Research Funds (M4010360.040 RG29/10) from Singapore Ministry of Education and partially from the Singapore Ministry of Education under the Tier 2 Research Grant MOE2010-T2-2-010 (4020020.040 ARC2/11).Notes and references
- R. Siegel, D. Naishadham and A. Jemal, CA Cancer J. Clin., 2013, 63, 11–30 CrossRef PubMed.
- J. Ferlay, H.-R. Shin, F. Bray, D. Forman, C. Mathers and D. M. Parkin, Int. J. Cancer, 2010, 127, 2893–2917 CrossRef CAS PubMed.
- A. Vincent, J. Herman, R. Schulick, R. H. Hruban and M. Goggins, Lancet, 2011, 378, 607–620 CrossRef.
- A. Fire, S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Nature, 1998, 391, 806–811 CrossRef CAS PubMed.
- S. M. Elbashir, J. Harborth, W. Lendeckel, A. Yalcin, K. Weber and T. Tuschl, Nature, 2001, 411, 494–498 CrossRef CAS PubMed.
- S. M. Hammond, E. Bernstein, D. Beach and G. J. Hannon, Nature, 2000, 404, 293–296 CrossRef CAS PubMed.
- A. J. Hamilton and D. C. Baulcombe, Science, 1999, 286, 950–952 CrossRef CAS.
- P. D. Zamore, T. Tuschl, P. A. Sharp and D. P. Bartel, Cell, 2000, 101, 25–33 CrossRef CAS.
- J. Tabernero, G. I. Shapiro, P. M. LoRusso, A. Cervantes, G. K. Schwartz, G. J. Weiss, L. Paz-Ares, D. C. Cho, J. R. Infante, M. Alsina, M. M. Gounder, R. Falzone, J. Harrop, A. C. S. White, I. Toudjarska, D. Bumcrot, R. E. Meyers, G. Hinkle, N. Svrzikapa, R. M. Hutabarat, V. A. Clausen, J. Cehelsky, S. V. Nochur, C. Gamba-Vitalo, A. K. Vaishnaw, D. W. Y. Sah, J. A. Gollob and H. A. Burris, Cancer Discovery, 2013, 3, 406–417 CrossRef CAS PubMed.
- M. E. Davis, J. E. Zuckerman, C. H. J. Choi, D. Seligson, A. Tolcher, C. A. Alabi, Y. Yen, J. D. Heidel and A. Ribas, Nature, 2010, 464, 1067–1070 CrossRef CAS PubMed.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Nat. Mater., 2013, 12, 967–977 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS PubMed.
- I. R. Gilmore, S. P. Fox, A. J. Hollins and S. Akhtar, Curr. Drug. Deliv., 2006, 3, 147–155 CrossRef CAS.
- R. Hu, W.-C. Law, G. Lin, L. Ye, J. Liu, J. Liu, J. L. Reynolds and K.-T. Yong, Theranostics, 2012, 2, 723 CrossRef CAS PubMed.
- K.-T. Yong, H. Ding, I. Roy, W.-C. Law, E. J. Bergey, A. Maitra and P. N. Prasad, ACS Nano, 2009, 3, 502–510 CrossRef CAS PubMed.
- C. Yu and J. Irudayaraj, Anal. Chem., 2006, 79, 572–579 CrossRef PubMed.
- R. Savla, O. Taratula, O. Garbuzenko and T. Minko, J. Control. Release, 2011, 153, 16–22 CrossRef CAS PubMed.
- J. Wang, W. Liu, Q. Tu, J. Wang, N. Song, Y. Zhang, N. Nie and J. Wang, Biomacromolecules, 2010, 12, 228–234 CrossRef PubMed.
- R. H. Baughman, A. A. Zakhidov and W. A. de Heer, Science, 2002, 297, 787–792 CrossRef CAS PubMed.
- Carbon nanotubes: synthesis, structure, properties, and applications, ed. M. S. Dresselhaus, G. Dresselhaus and P. Avouris, Springer, Berlin Heidelberg, 2001 Search PubMed.
- K. Kostarelos, A. Bianco and M. Prato, Nat. Nanotechnol., 2009, 4, 627–633 CrossRef CAS PubMed.
- M. Foldvari and M. Bagonluri, Nanomedicine, 2008, 4, 173–182 CrossRef CAS PubMed.
- A. Ortiz-Acevedo, H. Xie, V. Zorbas, W. M. Sampson, A. B. Dalton, R. H. Baughman, R. K. Draper, I. H. Musselman and G. R. Dieckmann, J. Am. Chem. Soc., 2005, 127, 9512–9517 CrossRef CAS PubMed.
- K. L. Lu, R. M. Lago, Y. K. Chen, M. L. H. Green, P. J. F. Harris and S. C. Tsang, Carbon, 1996, 34, 814–816 CrossRef CAS.
- H. T. Ham, Y. S. Choi and I. J. Chung, J. Colloid Interface Sci., 2005, 286, 216–223 CrossRef CAS PubMed.
- N. Karousis, N. Tagmatarchis and D. Tasis, Chem. Rev., 2010, 110, 5366–5397 CrossRef CAS PubMed.
- D. Tasis, N. Tagmatarchis, A. Bianco and M. Prato, Chem. Rev., 2006, 106, 1105–1136 CrossRef CAS PubMed.
- A. Hirsch and O. Vostrowsky, in Functional Molecular Nanostructures, ed. A. D. Schlüter, Springer, Berlin Heidelberg, 2005, vol. 245, pp. 193–237 Search PubMed.
- A. Hirsch, Angew. Chem., Int. Ed., 2002, 41, 1853–1859 CrossRef CAS.
- T. Ramanathan, F. T. Fisher, R. S. Ruoff and L. C. Brinson, Chem. Mater., 2005, 17, 1290–1295 CrossRef CAS.
- V. C. Moore, M. S. Strano, E. H. Haroz, R. H. Hauge, R. E. Smalley, J. Schmidt and Y. Talmon, Nano Lett., 2003, 3, 1379–1382 CrossRef CAS.
- A. L. Antaris, J. T. Robinson, O. K. Yaghi, G. Hong, S. Diao, R. Luong and H. Dai, ACS Nano, 2013, 7, 3644–3652 CrossRef CAS PubMed.
- A. De La Zerda, C. Zavaleta, S. Keren, S. Vaithilingam, S. Bodapati, Z. Liu, J. Levi, B. R. Smith, T.-J. Ma, O. Oralkan, Z. Cheng, X. Chen, H. Dai, B. T. Khuri-Yakub and S. S. Gambhir, Nat. Nanotechnol., 2008, 3, 557–562 CrossRef CAS PubMed.
- P. W. Barone, S. Baik, D. A. Heller and M. S. Strano, Nat. Mater., 2004, 4, 86–92 CrossRef PubMed.
- R. J. Chen, S. Bangsaruntip, K. A. Drouvalakis, N. Wong Shi Kam, M. Shim, Y. Li, W. Kim, P. J. Utz and H. Dai, Proc. Natl. Acad. Sci. U.S.A., 2003, 100, 4984–4989 CrossRef CAS PubMed.
- B. S. Harrison and A. Atala, Biomaterials, 2007, 28, 344–353 CrossRef CAS PubMed.
- N. W. S. Kam, M. O'Connell, J. A. Wisdom and H. Dai, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11600–11605 CrossRef CAS PubMed.
- M. Prato, K. Kostarelos and A. Bianco, Acc. Chem. Res., 2007, 41, 60–68 CrossRef PubMed.
- Z. Liu, X. Sun, N. Nakayama-Ratchford and H. Dai, ACS Nano, 2007, 1, 50–56 CrossRef CAS PubMed.
- J. Malmo, H. Sørgård, K. M. Vårum and S. P. Strand, J. Controlled Release, 2012, 158, 261–268 CrossRef CAS PubMed.
- C. Boyer, P. Priyanto, T. P. Davis, D. Pissuwan, V. Bulmus, M. Kavallaris, W. Y. Teoh, R. Amal, M. Carroll, R. Woodward and T. St Pierre, J. Mater. Chem., 2010, 20, 255–265 RSC.
- G. Lin, R. Hu, W.-C. Law, C.-K. Chen, Y. Wang, H. Li Chin, Q. T. Nguyen, C. K. Lai, H. S. Yoon, X. Wang, G. Xu, L. Ye, C. Cheng and K.-T. Yong, Small, 2013, 9, 2757–2763 CrossRef CAS PubMed.
- Y. Patil and J. Panyam, Int. J. Pharm., 2009, 367, 195–203 CrossRef CAS PubMed.
- M. E. Davis, Mol. Pharm., 2009, 6, 659–668 CrossRef CAS PubMed.
- T.-M. Sun, J.-Z. Du, L.-F. Yan, H.-Q. Mao and J. Wang, Biomaterials, 2008, 29, 4348–4355 CrossRef CAS PubMed.
- A. C. Bonoiu, S. D. Mahajan, H. Ding, I. Roy, K.-T. Yong, R. Kumar, R. Hu, E. J. Bergey, S. A. Schwartz and P. N. Prasad, Proc. Natl. Acad. Sci. U.S.A., 2009, 106, 5546–5550 CrossRef CAS PubMed.
- A. Elbakry, A. Zaky, R. Liebl, R. Rachel, A. Goepferich and M. Breunig, Nano Lett., 2009, 9, 2059–2064 CrossRef CAS PubMed.
- H. Meng, M. Liong, T. Xia, Z. Li, Z. Ji, J. I. Zink and A. E. Nel, ACS Nano, 2010, 4, 4539–4550 CrossRef CAS PubMed.
- F. Pittella, M. Zhang, Y. Lee, H. J. Kim, T. Tockary, K. Osada, T. Ishii, K. Miyata, N. Nishiyama and K. Kataoka, Biomaterials, 2011, 32, 3106–3114 CrossRef CAS PubMed.
- J. Jung, A. Solanki, K. A. Memoli, K.-I. Kamei, H. Kim, M. A. Drahl, L. J. Williams, H.-R. Tseng and K. Lee, Angew. Chem., 2010, 122, 107–111 CrossRef.
- K. Kostarelos, L. Lacerda, G. Pastorin, W. Wu, W. Sebastien, J. Luangsivilay, S. Godefroy, D. Pantarotto, J.-P. Briand, S. Muller, M. Prato and A. Bianco, Nat. Nanotechnol., 2007, 2, 108–113 CrossRef CAS PubMed.
- D. Pantarotto, R. Singh, D. McCarthy, M. Erhardt, J.-P. Briand, M. Prato, K. Kostarelos and A. Bianco, Angew. Chem., Int. Ed., 2004, 43, 5242–5246 CrossRef CAS PubMed.
- N. W. S. Kam, Z. Liu and H. Dai, Angew. Chem., 2006, 118, 591–595 CrossRef.
- K. Bates and K. Kostarelos, Adv. Drug Delivery Rev., 2013, 65, 2023–2033 CrossRef CAS PubMed.
- N. W. S. Kam, Z. Liu and H. Dai, J. Am. Chem. Soc., 2005, 127, 12492–12493 CrossRef CAS PubMed.
- Z. Liu, M. Winters, M. Holodniy and H. Dai, Angew. Chem., Int.Ed., 2007, 46, 2023–2027 CrossRef CAS PubMed.
- J. E. Podesta, K. T. Al-Jamal, M. A. Herrero, B. Tian, H. Ali-Boucetta, V. Hegde, A. Bianco, M. Prato and K. Kostarelos, Small, 2009, 5, 1176–1185 CrossRef CAS.
- R. Krajcik, A. Jung, A. Hirsch, W. Neuhuber and O. Zolk, Biochem. Biophys. Res. Commun., 2008, 369, 595–602 CrossRef CAS PubMed.
- S. Foillard, G. Zuber and E. Doris, Nanoscale, 2011, 3, 1461–1464 RSC.
- A. Battigelli, J. T.-W. Wang, J. Russier, T. Da Ros, K. Kostarelos, K. T. Al-Jamal, M. Prato and A. Bianco, Small, 2013, 9, 3610–3619 CrossRef CAS PubMed.
- M. Liu, B. Chen, Y. Xue, J. Huang, L. Zhang, S. Huang, Q. Li and Z. Zhang, Bioconjugate Chem., 2011, 22, 2237–2243 CrossRef CAS PubMed.
- E. Zhao, Z. Zhao, J. Wang, C. Yang, C. Chen, L. Gao, Q. Feng, W. Hou, M. Gao and Q. Zhang, Nanoscale, 2012, 4, 5102–5109 RSC.
- O. Thellin, W. Zorzi, B. Lakaye, B. De Borman, B. Coumans, G. Hennen, T. Grisar, A. Igout and E. Heinen, J. Biotechnol., 1999, 75, 291–295 CrossRef CAS.
- A. Verma and F. Stellacci, Small, 2010, 6, 12–21 CrossRef CAS PubMed.
- R. Saito, C. Fantini and J. Jiang, in Carbon Nanotubes: Advanced Topics in the Synthesis, Structure, Properties and Applications, ed. A. Jorio, G. Dresselhaus and M. S. Dresselhaus, Springer, Berlin Heidelberg, 2008, vol. 111, pp. 251–286 Search PubMed.
- K. Welsher, Z. Liu, S. P. Sherlock, J. T. Robinson, Z. Chen, D. Daranciang and H. Dai, Nat. Nanotechnol., 2009, 4, 773–780 CrossRef CAS PubMed.
- T. Pons, H. T. Uyeda, I. L. Medintz and H. Mattoussi, J. Phys. Chem. B, 2006, 110, 20308–20316 CrossRef CAS PubMed.
- H. S. Choi, W. Liu, P. Misra, E. Tanaka, J. P. Zimmer, B. Itty Ipe, M. G. Bawendi and J. V. Frangioni, Nat. Biotechnol., 2007, 25, 1165–1170 CrossRef CAS PubMed.
- M. Longmire, P. L. Choyke and H. Kobayashi, Nanomedicine, 2008, 3, 703–717 CrossRef CAS PubMed.
- H. Jin, D. A. Heller, R. Sharma and M. S. Strano, ACS Nano, 2009, 3, 149–158 CrossRef CAS PubMed.
- C. M. Sayes, F. Liang, J. L. Hudson, J. Mendez, W. Guo, J. M. Beach, V. C. Moore, C. D. Doyle, J. L. West, W. E. Billups, K. D. Ausman and V. L. Colvin, Toxicol. Lett., 2006, 161, 135–142 CrossRef CAS PubMed.
- S. L. Campbell, R. Khosravi-Far, K. L. Rossman, G. J. Clark and C. J. Der, Oncogene, 1998, 17, 1395–1413 CAS.
- M. Malumbres and M. Barbacid, Nat. Rev. Cancer, 2003, 3, 459–465 CrossRef CAS PubMed.
- E. Rozenblum, M. Schutte, M. Goggins, S. A. Hahn, S. Panzer, M. Zahurak, S. N. Goodman, T. A. Sohn, R. H. Hruban, C. J. Yeo and S. E. Kern, Cancer Res., 1997, 57, 1731–1734 CAS.
- Z. Liu, C. Davis, W. Cai, L. He, X. Chen and H. Dai, Proc. Natl. Acad. Sci. U.S.A., 2008, 105, 1410–1415 CrossRef CAS PubMed.
- Z. Liu, W. Cai, L. He, N. Nakayama, K. Chen, X. Sun, X. Chen and H. Dai, Nat. Nanotechnol., 2007, 2, 47–52 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |