Changing ligand number and type within nanocylindrical domains through kinetically constrained self-assembly – impacts of ligand ‘redundancy’ on human mesenchymal stem cell adhesion and morphology
Haiqing
Li†
a and
Justin J.
Cooper-White
*abc
aTissue Engineering and Microfluidic Laboratory, Australian Institute for Bioengineering and Nanotechnology, The University of Queensland, Cnr Cooper and College Rd, 4072, Queensland, Australia. E-mail: j.cooperwhite@uq.edu.au
bSchool of Chemical Engineering, The University of Queensland, College Rd, Brisbane, 4072, Queensland, Australia
cCSIRO, Division of Materials Science and Engineering, Clayton, 3169, Victoria, Australia
First published on 13th August 2014
Abstract
In this paper, we firstly describe a facile method by which sequential attachment of different adhesion peptides to a nanotopographical, self-assembled block copolymer cell culture surface is made possible through orthogonal click chemistry. Functionalization of polystyrene-b-polyethylene oxide block copolymers (PS-PEO) with azide (PS-PEO-N3) and aminooxy (PS-PEO-ONH2) moieties permitted the use of orthogonal click chemistry protocols to sequentially add desired bioactive moieties. Thereafter, we show that co-self-assembly of non-functionalised PS-PEO with different amounts of these functionalized PS-PEOs produces polymer films having well-defined, hexagonally arrayed PEO nanocylinder domains, of near constant diameter (∼17 nm diameter) and lateral spacing (∼35 nm). The invariant diameters and lateral spacing of the nanodomains with changes in the amounts of PS-PEO-N3 and PS-PEO-ONH2 confirmed our ability to tune the number density of these functional groups locally within each PEO nanodomain. Stepwise conjugation of alkyne-terminated IKVAV or aldehyde-terminated RGD to the azide and aminooxy decorated nanodomains produced a series of substrates with increasing local number density of grafted adhesion peptides in each nanodomain. We then systematically investigated the impacts of ligand affinity and availability (leading to differing levels of redundancy) on cell integrin binding and adhesion behaviours. We show that with increasing numbers of single peptides (IKVAV or RGD) or with changes in the ratio of IKVAV and RGD peptides within each of the ∼17 nm nanodomains of these films, there was significant changes in the number of hMSCs adhered and substantial modulation of cell morphology, cytoskeletal actin stress fibres and focal adhesion maturation. We observed that increases in the ratio of RGD to IKVAV peptides within the constrained surface nanodomains greatly enhanced hMSC adhesion, and effectively modulated hMSC morphology, cytoskeletal actin structures and focal adhesion number and maturity between the two extremes noted for the single peptides. The results presented suggest that these self assembled block copolymer substrates regulate hMSC adhesion and morphology through modulation of ligand affinity and ligand redundancy, and hence the effectiveness of integrin binding and mechanotransduction signalling. These novel 2D polymer substrates offer encoded and defined cues for cell adhesion at length scales previously unrealised and the results of this investigation expose a new parameter set by which the surfaces of biomaterials may be tailored for stem cell culture, selection and fate.
1. Introduction
Human mesenchymal stem cells (hMSCs) have emerged as a clinically relevant cell source for regenerative medicine owing to their potential to differentiate into several mesenchymal cell types, such as cartilage, bone, muscle and fat.1,2 The status and fate of a hMSC in culture is modulated by the culture microenvironment, which can be tailored to mimic interactions normally provided through cell–cell and cell–extracellular matrix (cell–ECM) interactions in the tissue microenvironment, including both biochemical and biophysical cues.3,4 To elucidate the permissive cues required to drive hMSCs differentiation, a wide range of synthetic biomaterials including two-dimensional (2D) substrates and three-dimensional (3D) scaffolds have been constructed to mimic various components of the ECM in the targeted tissue.5 In particular, nanostructured 2D substrates have received significant attention most recently as they provide flexible platforms to probe the relative impacts of surface nano-presentation of cell adhesive molecules,6–9 surface functionalities,10 surface topographies,11,12 and the mechanical modulus of the polymer substrate13,14 on a range of stem cell behaviours or fate choices, such as adhesion, spreading, differentiation and apoptosis.Polymeric films offer a great deal of flexibility in terms of encouraging cell adhesion, as they permit the conjugation of ECM-derived peptides on their surfaces through a range of versatile chemical schemes. A variety of covalent mechanisms have been used to tether peptides onto 2D polymer films, including carbodiimide condensation and Michael-type addition, depending on the functionalities presented on the film surfaces.15,16 These mechanisms, however, are subject to a pragmatic limitation: due to cross-reactivity it is difficult to covalently tether multiple, distinct peptides in a precisely controllable manner on a single substrate. To address this limitation, we need to develop chemically orthogonal substrates that enable one to conjugate ECM-derived peptides through highly selective and efficient reactions. To this end, recently developed “click” chemistries, such as azide–alkyne Huisgen cycloaddition, oxime ligation, the Diels–Alder reaction, and the thiol–yne coupling reaction, provide us with effective solutions, by taking advantages of their features of extreme selectivity, orthogonality, high efficiency, and the requirement for mild reaction conditions.17–19 Hence, through the use of distinct, orthogonal click reactions, a substrate with controllable numbers of different peptides can be produced by simply immobilizing multiple, distinct peptides on a chemically-orthogonal substrate bearing the corresponding “clickable” moieties. These resulting substrates, presenting controlled ratios of different cell adhesion molecules may prove to be a promising culture surface for hMSCs, with a built-in capability to modulate their fate choices (adhesion, migration, differentiation etc.).
Previously, through the rapid kinetically constrained self-assembly of polystyrene-b-poly(ethylene oxide) (PS-PEO) block copolymers on a hydrophobic substrate, it has been demonstrated that it is possible to produce a thin film consisting of separated, vertically oriented cylindrical PEO nanodomains in a matrix of PS.20,21 Furthermore, when the terminal end of the PEO chain was functionalised with various chemical moieties, including a maleimide group, the PS-PEO was shown to exhibit similar phase-separation behaviour and self-organized into well-defined, spatially separated nanocylinders presenting the desired functional group within these cylinders, permitting further covalent binding of cell adhesive peptides to encourage cell adhesion on a ‘blank slate’, low fouling background.22 Moreover, the lateral spacing of these cylindrical PEO domains can be effectively controlled by simply blending functionalised PS-PEO with homopolymer PS.8,22 Recently, by blending an adamantane-functionalised PS-PEO with a non-functionalised PS-PEO block copolymer, it was shown that one could not only modulate the number of adamantane-functionalised PEO chains within each nanocylinder, but through subsequent defined host–guest interactions with peptide-functionalised cyclodextrins, the impact of ligand availability within each nanocylinder on cell adhesion could be explored.23 It is commonly thought that integrins are stably adhered to ligands within focal adhesions, however Rossier et al. recently showed that this is not the case, and that they are constantly switching between active and inactive states, undergoing repeated anchoring cycles.24 Based on this recent insight, Li et al.23 suggested that ligand ‘redundancy’ (a result of the relative availability of ligands within a defined spatial location for integrin binding), may play a role in cell adhesion and spreading. Together, these previous investigations have provided new insight into the versatility of these PS-PEO substrates, with the intrinsic capacity for controlled nano-presentation of different surface functional motifs for tethering different ECM-derived peptides through covalent coupling or host–guest interactions.
To further extend the capability of this self-assembling cell culture system, and to thereafter to explicitly investigate the impact of ligand availability/redundancy and type within local nanodomains of ∼10–20 nm (approximately the size of a pair of cell integrins, the transmembrane proteins used by cells to bind to ECM molecules in vitro and in vivo)25 on stem cell adhesion and spreading, we have developed a new self-assembling block copolymer system that permits explicit control over the number of functional groups within each nanodomain and further, the step-wise coupling of different adhesion peptides through orthogonal chemistries.
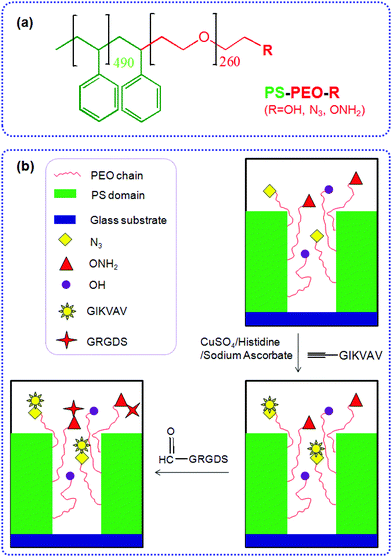
Herein we detail the method by which we can precisely control the numbers of azide and aminooxy groups (on the terminal end of the PEO chains) within well-defined PEO nanodomains, by co-self-assembly of azide terminated PS-PEO (PS-PEO-N3), aminooxy terminated PS-PEO (PS-PEO-ONH2), and pristine PS-PEO in various ratios. Mediated by the surface-presented N3 and ONH2 motifs within these nanodomains, alkyne terminated IKVAV and aldehyde terminated RGD peptides (two commonly utilised peptide sequences found within ECM proteins, including collagens and fibronectin (RGD) and laminin (IKVAV)) can be immobilized onto the polymer substrates through an azide–alkyne Huisgen cycloaddition reaction and an aldehyde-aminooxy oxime ligation reaction, respectively, forming substrates capable of presenting distinct stoichiometry of ECM-derived peptides within well defined, spatially separated, geometrically coordinated, nanocylindrical domains (Fig. 1). These novel 2D polymer substrates offer encoded and defined cues for cell adhesion at length scales previously untested. As an exemplar, we thereafter assessed the ability of these substrates to effect differences in stem cell fate choice, evidenced by differences in hMSC adhesion, morphology, and cytoskeletal organisation when exposed to these PS-PEO-based substrates presenting these two peptides over differing levels of availability/redundancy and a range of stoichiometries.
2. Experimental section
2.1 Materials
PS-PEO block copolymer, with a PS block molecular weight of 51 kDa and a PEO block of 11.5 kDa was purchased from Polymer Source Pty. Ltd (Montreal, PQ, Canada). CHO-CO-NH-GRGDS and (L-propargylglycine)-GGIKVAV were purchased from ChinaPeptides Co., Ltd (Shanghai, China) and used as received. Tosyl chloride (TsCl), sodium azide, N-hydroxyphthalimide, diisopropyl azodicarboxylate (DIAD), triphenylphosphine (TPP), hydrazine monohydrate, anhydrous N,N-dimethylformamide (DMF), and dichloromethane were purchased from Sigma-Aldrich and used as supplied. All materials for hMSCs culture, staining and imaging were purchased from Sigma unless otherwise stated.2.2 Synthesis of PS-PEO-N3
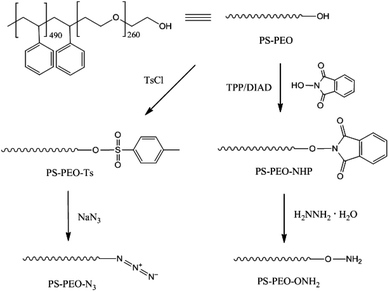
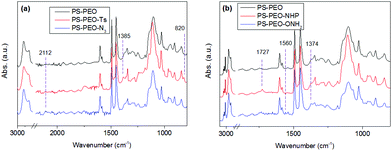
The synthesis of azide terminated PS-PEO (PS-PEO-N3) is shown in Fig. 2. In a typical reaction, PS-PEO (1.0 g) was dissolved in 40 mL of anhydrous DMF. Followed by the addition of 60 mg of tosyl chloride, the reaction was performed at room temperature for 24 h. The solvent was removed by distillation under vacuum. The crude product was redissolved in 15 mL of chloroform and extracted with MilliQ water twice. After the removal of solvent by rotor-evaporation, solid tosylated PS-PEO (PS-PEO-Ts) was collected and further dried under vacuum overnight. To synthesize PS-PEO-N3, the obtained PS-PEO-Ts was firstly dissolved in 30 mL of anhydrous DMF. Upon the addition of 42 mg of sodium azide, the reaction was allowed to proceed at 70 °C for 3 days. After distilling off DMF under vacuum, the crude product was obtained and then redissolved in chloroform. The resultant solution was further dialyzed against chloroform for 3 days. PS-PEO-N3 was collected by first evaporating off the chloroform, followed by further drying under vacuum overnight.The successful synthesis of chain-end functionalized PS-PEO was evidenced by FTIR spectra using the diamond ATR-unit of a FT-IR spectrometer (spectrum One, Perkin-Elmer, Germany). Compared with pristine PS-PEO, PS-PEO-Ts exhibits two new sharp bands at 1385 and 820 cm−1, corresponding to anti-symmetrical vibration of SO2, and bending vibration of benzene ring,26,27 respectively (Fig. 3a). Upon substitution with azide, both peaks at 1385 and 820 cm−1 vanished. The appearance of a relatively weak peak situated at 2112 cm−1 is characteristic of the formation of azide. These changes in the FTIR spectra evidenced the successful synthesis of PS-PEO-N3.
2.3 Synthesis of PS-PEO-ONH2
In a typical reaction for synthesizing the oxyamine terminated PS-PEO (PS-PEO-ONH2, Fig. 2), 25 mg of N-hydroxyphthalimide, 84 mg of TPP, and 65 mg of DIAD were sequentially added to a PS-PEO solution in anhydrous DMF (25 mL, 0.64 mM). The reaction was performed at room temperature for 24 h. After the completion of reaction, the reaction solution was subjected to dialysis against dichloromethane for 3 days. The resultant polymer solution was then transferred into a flask containing 40 μL of hydrazine monohydrate. After stirring the mixture at room temperature for 20 h, the resultant reaction solution was dialyzed against dichloromethane for 3 days. PS-PEO-ONH2 was collected after evaporating off the solvent and subsequently drying under vacuum overnight.As shown in Fig. 3b, after immobilization of the NHP group at the PEO chain end of PS-PEO through a Mitsunobu reaction, the functionalized polymer exhibits two new peaks at 1727 and 1374 cm−1, corresponding to the benzene conjugated C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching, and N–O symmetrical stretching.28 However, after being reacted with hydrazine monohydrate, the peak centred at 1727 cm−1 disappeared, while a small new band, situated at 1560 cm−1 assigned to the characteristic band of a primary amine, appears. The peak at 1374 cm−1 remained. These results evidenced the successful synthesis of PS-PEO-ONH2.
O stretching, and N–O symmetrical stretching.28 However, after being reacted with hydrazine monohydrate, the peak centred at 1727 cm−1 disappeared, while a small new band, situated at 1560 cm−1 assigned to the characteristic band of a primary amine, appears. The peak at 1374 cm−1 remained. These results evidenced the successful synthesis of PS-PEO-ONH2.
2.4 Preparation of peptide-immobilized polymer surfaces using orthogonal chemistries
Glass slides (13 mm in diameter) were exposed to UV/ozone for 20 min to remove any organics. The slides were then rendered hydrophobic by boiling in benzyl alcohol for 4 h, and further rinsed thoroughly in isopropanol and dried under a stream of nitrogen. Polymer films were generated by spin casting solutions of the polymer in toluene (2% wt/vol) onto the treated glass slides at 2000 rpm. By blending the different polymers including PS-PEO, PS-PEO-N3, and PS-PEO-ONH2 in a controllable ratio, a variety of polymer substrates with precisely tunable nanospacings and orthogonal chemistries were prepared.IKVAV was immobilized onto polymer surfaces through the Huisgen azide–alkyne cycloaddition reaction. In a typical reaction, PS-PEO-N3 coated glass slides were immersed in an aqueous solution containing (L-propargylglycine)-GGIKVAV (0.13 mM), L-histidine (0.13 mM), sodium ascorbate (1.3 mM), and copper(II) sulfate pentahydrate (0.13 mM). After incubation at room temperature for 24 h, the surfaces were thoroughly rinsed with MilliQ water. To immobilize RGD onto polymer surfaces, PS-PEO-ONH2 coated glass slides were immersed in an aqueous solution containing CHO-CO-NH-GRGDS (0.13 mM). Followed by incubation at room temperature for 4 h, the surfaces were thoroughly rinsed with MilliQ water. Following similar procedures, both IKVAV and RGD can be introduced sequentially (or simultaneously) onto these orthogonal surfaces which present azide and oxyamine motifs in different ratios within each defined nanodomain.
2.5 Characterization of polymer films
Characterisation of the peptide conjugated polymer films was performed using a number of techniques. Surface Raman measurements were performed on the films in air using an Alpha 300 Raman/AFM (WITek GmbH, Ulm, Germany) equipped with a 100×, N.A. 0.90 objective (Nikon). A frequency-doubled continuous-wave Nd:YAG laser stabilized at 532 nm was used for excitation. Atomic force microscopy (AFM) imaging of the self-organized polymer films on glass slides was carried out on a Multimode Nanoscope-IIIa Scanning Probe Microscope (Digital Instrument Co., Ltd, U.S.A.) equipped with a NT-MDT silicon cantilever (NSCII, radius <10 nm, resonance frequency = 300 kHz, nominal spring constant = 42 N m−1, vertical resolution <0.03 nm, lateral resolution <2 nm) by using AC mode at room temperature. The AFM is mounted on an anti-vibrational table (Herzan) and operated within an acoustic isolation enclosure (TMC, USA).2.6 hMSC culture, staining and imaging
3. Results and discussion
3.1 hMSCs on substrates with controllable nano-presentation of IKVAV peptide
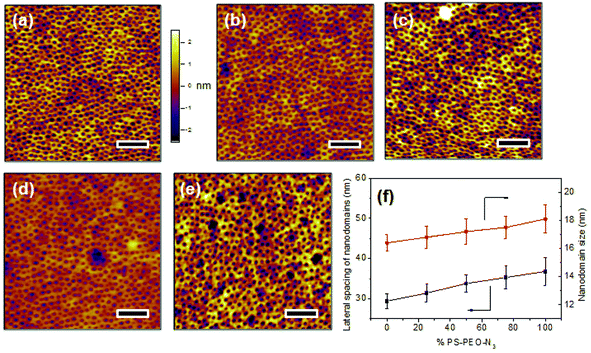
Upon rapid self-assembly, PS-PEO can self-organize into a thin film with well-defined cylindrical PEO nanodomains uniformly distributed into PS matrix (Fig. 4a). As determined by AFM observation, the average lateral spacing of the nanodomains is 29.4 ± 1.8 nm, and the average size of the nanodomains was 16.4 ± 0.6 nm. To highlight the utility of the polymer films of controlled surface nano-presentation of peptide sequences, polymer surfaces were fabricated by simply blending together different ratios of PS-PEO-N3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PS-PEO, and encouraging co-self-assembly. This changed the number of azide groups in each PEO nanodomain, and thus controlled the number of possible conjugated IKVAV peptide sequences (and hence their availability for cell integrin binding) within each nanodomain. When 25 wt% of PS-PEO-N3 was mixed with the unmodified PS-PEO, a polymer film with similarly well-organized cylindrical PEO nanodomains was achieved (Fig. 4b). The lateral spacing and size of those PEO nanodomains were 31.4 ± 2.2 and 16.8 ± 0.8 nm, respectively. Following a similar blending technique, further increases in the amount of PS-PEO-N3 up to 50 wt% and 75 wt% resulted in the formation of polymer films with similar nanodomain diameters (50 wt%: 17.2 ± 0.9 nm, and 75 wt%: 17.5 ± 0.8 nm) but with slightly increased average lateral spacing of nanodomains, being 34.3 ± 2.1 nm and 35.3 ± 2.8 nm, respectively. There was however no statistical difference between these lateral spacings. Overall, all of the resultant blended polymer films presented well-defined cylindrical PEO nanodomains of similar diameter and lateral spacing incorporating varying ratios of N3 moieties (Fig. 4c and 4d). Further AFM analysis confirmed that the self-assembly of pure PS-PEO-N3 resulted in a well-defined film with 36.8 ± 3.5 nm of lateral spacing of PEO nanodomains and 18.1 ± 1.0 nm of domain size (Fig. 4e). These insignificant changes in the lateral spacing and nanodomain size suggests that this method leads to controlled nano-presentation of varying levels of azide moieties within each nanodomain on the resulting polymer substrates.
PS-PEO, and encouraging co-self-assembly. This changed the number of azide groups in each PEO nanodomain, and thus controlled the number of possible conjugated IKVAV peptide sequences (and hence their availability for cell integrin binding) within each nanodomain. When 25 wt% of PS-PEO-N3 was mixed with the unmodified PS-PEO, a polymer film with similarly well-organized cylindrical PEO nanodomains was achieved (Fig. 4b). The lateral spacing and size of those PEO nanodomains were 31.4 ± 2.2 and 16.8 ± 0.8 nm, respectively. Following a similar blending technique, further increases in the amount of PS-PEO-N3 up to 50 wt% and 75 wt% resulted in the formation of polymer films with similar nanodomain diameters (50 wt%: 17.2 ± 0.9 nm, and 75 wt%: 17.5 ± 0.8 nm) but with slightly increased average lateral spacing of nanodomains, being 34.3 ± 2.1 nm and 35.3 ± 2.8 nm, respectively. There was however no statistical difference between these lateral spacings. Overall, all of the resultant blended polymer films presented well-defined cylindrical PEO nanodomains of similar diameter and lateral spacing incorporating varying ratios of N3 moieties (Fig. 4c and 4d). Further AFM analysis confirmed that the self-assembly of pure PS-PEO-N3 resulted in a well-defined film with 36.8 ± 3.5 nm of lateral spacing of PEO nanodomains and 18.1 ± 1.0 nm of domain size (Fig. 4e). These insignificant changes in the lateral spacing and nanodomain size suggests that this method leads to controlled nano-presentation of varying levels of azide moieties within each nanodomain on the resulting polymer substrates.
Using an azide–alkyne Huisgen click reaction, alkyne terminated IKVAV was selectively conjugated in the azide decorated PEO domains, resulting in a range of PS-PEO-IKVAV substrates with precisely controlled spatial distribution of IKVAV peptides of varying number density within each nanodomain.
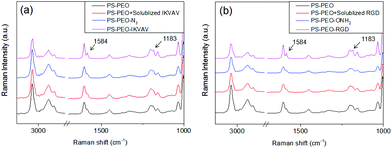
Successful immobilization of IKVAV on these polymer substrates was evidenced by Surface Raman Spectroscopy. As shown in Fig. 5a, in comparison to the bare PS-PEO-N3 films, the PS-PEO-IKVAV substrate showed two new sharp shoulder Raman shifts centred at 1584 and 1184 cm−1, corresponding to the characteristic Raman signature of Amide II and Amide III bonds derived from N–H bending, C–N stretching, and C–C stretching vibrations in the peptide sequences.29 In contrast, no feature Raman shifts were observed on the PS-PEO surface, nor the PS-PEO surface that was incubated with alkyne terminated IKVAV for 12 h followed by a thorough rinse with MilliQ water. This fact further proved that the immobilization of IKVAV on PS-PEO-N3 surface is through the selective azide–alkyne click chemistry rather than nonspecific physisorption.
Differences in MSC attachment and morphology on these polymer substrates with controllable nano-presentation of IKVAV peptide were then investigated. Attachment was quantified by an MTT assay after 4 h of culture on these surfaces. As shown in Fig. 6, rather low cell adhesion was observed on PS-PEO alone, on PS-PEO-N3 alone, or on PS-PEO surfaces (PS-PEO + IKVAV) that had been incubated with alkyne terminated IKVAV for 12 h, followed by thorough rinse with PBS. In contrast, surfaces made from 25% of PS-PEO-N3, post conjugation with IKVAV (PS-PEO-IKVAV), exhibited significant increased levels of cell attachment. This result suggests that hMSC adhesion is successfully mediated specifically by the surface conjugated IKVAV, rather than by the physisorption of the IKVAV onto the PS-PEO polymer surfaces. Further increases in the content of PS-PEO-N3 in the polymer substrates to 50% and 75%, followed by conjugation of the IKVAV peptide, resulted in further increases in hMSC attachment. The highest level of hMSC attachment was achieved on the 100% PS-PEO-N3 IKVAV conjugated substrates, which was statistically indifferent to that of our control tissue culture plastic substrate (TCP, positive control). Due to the similarity in lateral spacing of similarly sized nanodomains, the observed increase in hMSC attachment as a function of increasing PS-PEO-N3 content in these substrates and subsequent conjugation of the IKVAV peptide is believed to be a result of increases in the local density (availability) of IKVAV moieties within the PEO nanodomains.
An increase in the availability of any ligand within the spatial context provided by these ∼17 nm dia. nanocylinders will result in increased ligand redundancy for integrin-ligand binding. As shown by Rossier et al., integrins within focal adhesions undergo repeated anchoring cycles, constantly switching between active and inactive states – they are not stably adherent.24 The increased IKVAV ligand number density in each PEO nanodomain in these surfaces thus enables a higher probability of longer time localized integrin-ligand binding events. These longer time binding events should thus produce increased focal adhesion stability and maturation, greater force transmission and maturation of the actin cytoskeleton. As a result, we should observe increased cell adhesion and spreading with increased ligand redundancy in our nanodomains.
To confirm our hypothesis, the morphologies of the adhered hMSCs after 24 h on the surfaces containing different contents of PS-PEO-IKVAV were thus characterised using Confocal Fluorescence Microscopy. The cytoskeletal organisation of actin and the extent of focal adhesion formation (and maturation) within the adhered hMSCs on these substrates were also characterised, by staining for actin (cytoskeleton) and vinculin (focal adhesion protein). As shown in Fig. 7a and 7b, the very small number of hMSCs that adhered to bare PS-PEO and PS-PEO-N3 surfaces displayed rounded and rather less spread morphologies, were absent of stress fibres and had nascent focal complexes (indicated by small, punctate vinculin structures). In comparison, whilst hMSCs on the 25% of PS-PEO-IKVAV substrate showed little overall changes in morphology, a small number of larger vinculin clusters (indicative of more developed focal adhesions) were observed around the edges of the cells (Fig. 7c). With further increases in PS-PEO-IKVAV content in the polymer films, to 50% and 75%, hMSCs exhibited well spread morphologies with clear actin stress fibres and increasing numbers of well defined, large vinculin structures, indicative of more mature focal adhesions (Fig. 7d and 7e). hMSCs cultured on the 100% PS-PEO-IKVAV surface demonstrated increased spreading with well-developed actin stress fibres and numerous elongated vinculin structures at the end of microfilament bundles (Fig. 7f), evidencing a relatively strong level of adhesion and cytoskeletal stress within these cells on these particular surfaces. These results indicate, as hypothesised, that with increasing numbers of IKVAV peptides in each nanodomain (estimated to range from 7 (25 wt%) to 28 (100 wt%) PEO chains at the upper most surface of any PEO nanocylinder),22 there are systematic increases in hMSC adhesion, spreading, cytoskeletal tension and focal adhesion maturation.
Using ImageJ image analysis, the average spread area of the attached hMSCs was calculated and plotted as function of the content of PS-PEO-IKVAV in these PS-PEO polymer substrates (Fig. 7g). As expected, the cell spread area of hMSCs on the PS-PEO alone, PS-PEO-N3 alone, and polymer substrate containing 25% of PS-PEO-IKVAV are rather low, being 172 ± 17, 174 ± 23, and 163 ± 25 μm2, respectively. With an increase in the content of PS-PEO-IKVAV in the nanodomains of these substrates to 50%, the average spread area of hMSCs significantly increased to 530 ± 110 μm2. Further increases of PS-PEO-IKVAV content in the nanodomains of the substrate results in a steady increase of cell area from 635 ± 142 μm2 on the 75% of PS-PEO-IKVAV surface to 730 ± 111 μm2 on 100% of PS-PEO-IKVAV surface.
3.2 hMSCs on substrates with controllable nano-presentation of RGD peptide
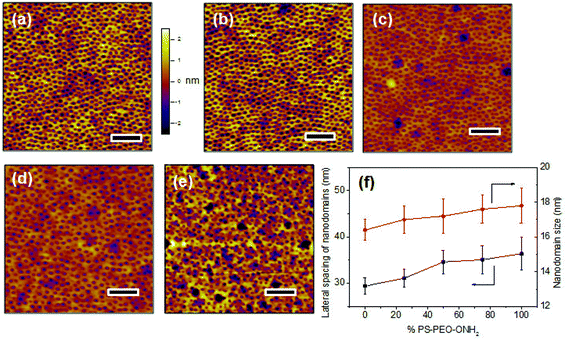
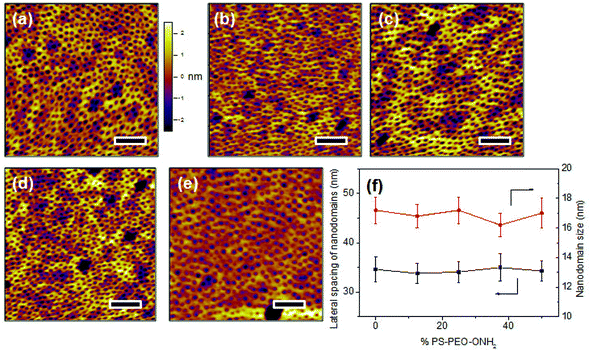
The co-self-assembly behaviours of PS-PEO with different content of PS-PEO-ONH2 were also investigated by means of AFM analysis (Fig. 8). Similar to the blended system of PS-PEO and PS-PEO-N3, all of the blended polymers self-organized into polymer films with well-defined cylindrical PEO domains embedded within the PS matrix (Fig. 8b and d). 100% PS-PEO-ONH2 also exhibited similar self-assembly behaviours, resulting in well-defined microphase-separated polymer films (Fig. 8e). Moreover, the average lateral spacing of the nanodomains is again relatively invariant (statistically), varying from 31.4 ± 2.0, 34.6 ± 2.5, 35.1 ± 3.0, to 36.4 ± 3.6 nm with the use of 25%, 50%, 75%, and 100% of PS-PEO-ONH2, respectively (Fig. 8f). Similarly, no obvious change in domain size was observed (17.0 ± 0.8–17.8 ± 1.0 nm) across the range of concentrations explored.As detailed in the Methods section, through an oxime ligation reaction, aldehyde terminated RGD peptides were immobilized onto these PS-PEO/PS-PEO-ONH2 blended polymer films, resulting in a series of substrates with well controlled, nanopresentation of immobilized RGD peptides, of varying number density within each nanodomain. The successful conjugation of RGD on the surfaces was evidenced by Surface Raman analysis (Fig. 5b). Similar to the PS-PEO-IKVAV surfaces, RGD conjugated PS-PEO-ONH2 surfaces exhibited new characteristic Raman shifts of Amide II and Amide III bonds of peptide, while bare PS-PEO, and PS-PEO incubated with solubilized aldehyde terminated RGD for 12 h followed by thorough rinsing with MilliQ water, demonstrated the same Raman shifts as that of bare PS-PEO surface (Fig. 5).
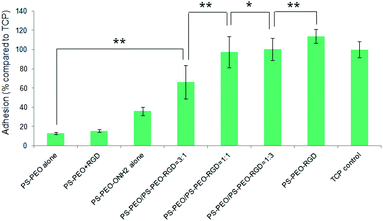
Levels of hMSC attachment on the resulting polymer substrates were next investigated. As shown in Fig. 9, PS-PEO alone, and the PS-PEO surface incubated in the presence of solubilized aldehyde terminated RGD followed by thorough rinse with MilliQ water, showed rather poor levels of cell attachment. In contrast, the 100% PS-PEO-ONH2 surface without any immobilized peptides exhibited a moderate level of cell attachment. This enhanced cell adhesion on the unfunctionalised PS-PEO-ONH2 surface can be attributed to the positively charged primary amine group being presented on these self-assembled surfaces. When cells are cultured in contact with amine-modified surfaces (such as tissue culture plate), even in the absence of serum, we would expect negatively charged pericellular protein fragments (cleaved by the use of trypsin when transferring to these surfaces) to enable adhesion.
After the immobilization of RGD peptides onto the PS-PEO substrate containing 25% PS-PEO-ONH2 through the oxime ligation reaction, cell attachment on the resulting surfaces was significantly enhanced compared with the bare PS-PEO substrate without RGD conjugation. With further increases in the content of the PS-PEO-ONH2 in the polymer film, to 50% and 75, conjugation of the RGD peptide to these substrates resulted in further increases in attachment, to a similar level to that achieved on tissue culture plastic (TCP, positive control). In particular, when the RGD peptide was immobilized onto the 100% PS-PEO-ONH2 surface, cells exhibited a slightly higher cell attachment than that of TCP positive control, although the differences were not statistically separable. The observed increases in adhesion with increases in the amount of RGD ligand within the nanodomains on these surfaces corroborate with the outcomes noted for the PS-PEO-IKVAV substrates, in terms of increases in ligand redundancy seemingly driving increased adhesion.
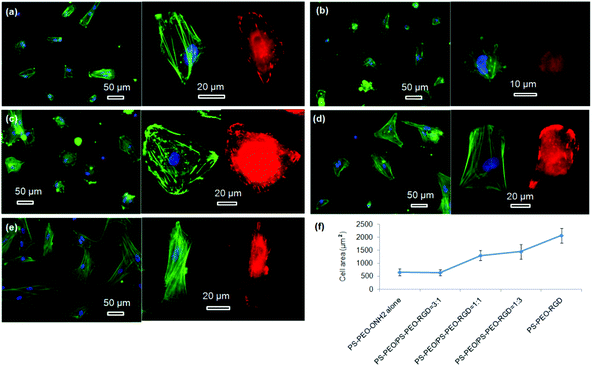
hMSC morphology on the bare PS-PEO-ONH2 surface and the RGD conjugated PS-PEO/PS-PEO-ONH2 blended polymer substrates were also investigated. hMSCs cultured on the bare PS-PEO-ONH2 substrates (without RGD conjugation) displayed a moderately spread morphology, with an average cell area of 650 ± 130 μm2 (Fig. 10a and 10f). Interestingly, even though not well spread, these cells do have a small number of longitudinally oriented stress fibres and some well-defined focal contacts. Obviously, the presence of the aminooxy groups on the PS-PEO-ONH2 surfaces rendered the surface with a greater affinity to hMSC attachment (again, likely enabled through pericellular protein association) compared with the bare PS-PEO and PS-PEO-N3 substrates (160–170 μm2, Fig. 7g).
Interestingly, once the RGD peptide was conjugated to the surfaces composed of 25 wt% of PS-PEO-ONH2 (25% PS-PEO-RGD), the cells showed a lower level of affinity for the surface (Fig. 10b), with cells exhibiting only minimal spreading. None of them displayed any substantial stress fibres nor pseudopodia, with punctate vinculin structures homogeneously displayed across the cells. The corresponding average cell spread area was calculated as 640 ± 120 μm2 (Fig. 10f). In contrast, an increase in the content of PS-PEO-RGD to 50% in the blended polymer substrate resulted in more oblate, well spread hMSCs with significantly enhanced average cell spread areas up to 1300 ± 200 μm2, with many cells displaying a number of stress fibres and substantive focal contacts around the cell periphery (Fig. 10c). Upon a further increase in the PS-PEO-RGD content in these films to 75%, the resulting substrate became substantially more favourable for cell adhesion. hMSCs now demonstrated well spread morphologies with well-developed stress fibres and a numerous, substantive vinculin structures (Fig. 10d). The average cell area was 1450 ± 290 μm2 (Fig. 10f). Interestingly, hMSCs cultured on 100% PS-PEO-RGD substrate showed highly polarised, well-spread morphologies (Fig. 10e). All of the cells showed longitudinally-oriented actin stress fibres and elongated vinculin structures, indicative of mature focal adhesions. The average cell area was further increased up to 2080 ± 290 μm2.
Based on these data, it is clear that the increase in the local number density of the RGD peptide within each constrained nanodomain of these substrates leads to enhanced adhesion, cytoskeletal tension, focal adhesion maturation and spreading in hMSCs. This data corroborates with the IKVAV-based investigation, although clear differences between IKVAV- and RGD-based surfaces in terms of cell spread area, morphology and cytoskeletal organisation were noted at the same local number density of a given ligand in nanodomains of the same size. These differences suggest that ligand affinity (as determined by the available integrins on the surface of a particular cell type), even at the same level of ligand redundancy, can also still create differential outcomes.
Overall, regardless of the ligand type being displayed, the spread area and morphologies of hMSCs can be substantially regulated by increasing not just the overall number density of adhesive ligands, as has been reported previously,8,22,23 but also by the local number density within a defined ∼17 nm nanodomain, that approximates that of the size of an individual integrin pair.25 On surfaces of constant nanodomain lateral spacing, the level of ligand redundancy within each nanodomain, in terms of availability for integrin binding and increasing the probability of longer term binding events, is thus believed to be having a significant impact on cell adhesion and morphology.
3.3 hMSCs on substrates with mixed ratios of nano-presented IKVAV and RGD peptides
The above data have confirmed that by utilising the thermodynamically-driven, co-self-assembly of PS-PEO and functionalized PS-PEO block copolymers (either PS-PEO-N3 or PS-PEO-ONH2), we can create polymer films with PEO nanodomains embedded in the PS matrix decorated with singular functional moieties (N3 and ONH2). Moreover, the fraction of functional motifs within each of these nanodomains can be precisely controlled by simply tuning the ratio of PS-PEO to functionalized PS-PEO. Based on these results, it is conceivable that a polymer film with defined ratios of distinct peptides within each of these cylindrical nanodomains may be able to be fabricated by firstly driving the co-self-assembly of mixtures of these three block copolymers, PS-PEO, PS-PEO-N3, and PS-PEO-ONH2, followed by the sequential conjugation of IKVAV and RGD peptides through orthogonal click reactions, as described above.To prove this concept, solutions of 50 wt% PS-PEO and 50 wt% of mixtures of the functional PS-PEO-N3 and PS-PEO-ONH2 (in different ratios) were blended and subjected to the same rapid co-self-assembly process previous exploited in all of this work. As expected, all of the resulting polymer films exhibited well-defined cylindrical surface nano-patterns of similar diameter (16.8 nm ± 1.3 nm) and with similar lateral spacing (34.5 nm ± 2.6 nm) of the nanodomains (Fig. 11).
Hence, by simply changing the ratio of PS-PEO-N3 to PS-PEO-ONH2, we can produce a set of polymer films of unaltered surface nanotopography and inter-domain spacing, but of tunable nanodomain content, in terms of the ratio of these two different functional moieties (N3 and ONH2) within each PEO nanodomains. Following application of the orthogonal click reactions as described above, alkyne terminated IKVAV and aldehyde terminated RGD peptides can be sequentially, selectively immobilized on these surfaces, yielding a set of substrates with completely defined, in terms of amounts and nano-presentation, distributions of IKVAV and RGD peptides. Such a novel substrate allows one to compare and contrast the impacts of ligand affinity but now at constant levels of ligand redundancy.
Further, by simply adding aldehyde or alkyne groups to a chosen ligand, this platform can be used to tether sequentially multiple other ligand pairs. More than two ligands can of course be introduced, but this would still require that a choice be made regarding which of these two endgroups would be utilised for the surface coupling reaction. However, this same concept (end group functionalization of the PS-PEO polymer) can be applied and expanded to other orthogonal chemistries, opening options for increased numbers of orthogonal sequential additions of distinct peptides or proteins.
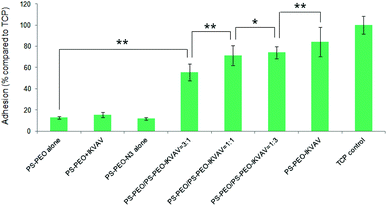
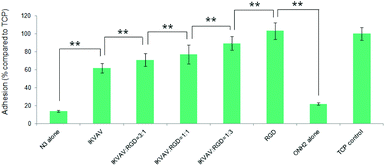
To investigate the effects of the resultant variations in the amounts of IKVAV and RGD peptides within each nanodomain on hMSC behaviours, hMSC attachment was firstly quantified. As shown in Fig. 12, the substrates consisting of 50% of PS-PEO and 50% of PS-PEO-N3 (N3 alone) showed rather poor cell attachment. After the subsequent conjugation of IKVAV peptide to this substrate, however, the level of hMSC attachment was significantly enhanced (up to 60% of the positive control (TCP plate)). In contrast, when the RGD was conjugated to surfaces containing 50% of PS-PEO and 50% of PS-PEO-ONH2, these surfaces displayed similar hMSC attachment to that achieved on TCP (positive control). Interestingly, when cells were presented with surfaces displaying both IKVAV and RGD (produced through our orthogonal click chemistries and increased fractional amounts of PS-PEO-ONH2 to PS-PEO-N3) within the 50 wt% of functionalised PS-PEO block copolymer, with increasing amounts of RGD peptide, we noted a further increase in the levels of hMSC attachment. In the absence of RGD conjugation, the corresponding substrate (ONH2 alone) demonstrated greatly reduced cell attachment compared to the control, showing similar levels to that previously reported in Fig. 9 (as expected) and slightly above that of the PS-PEO-N3 only substrate.
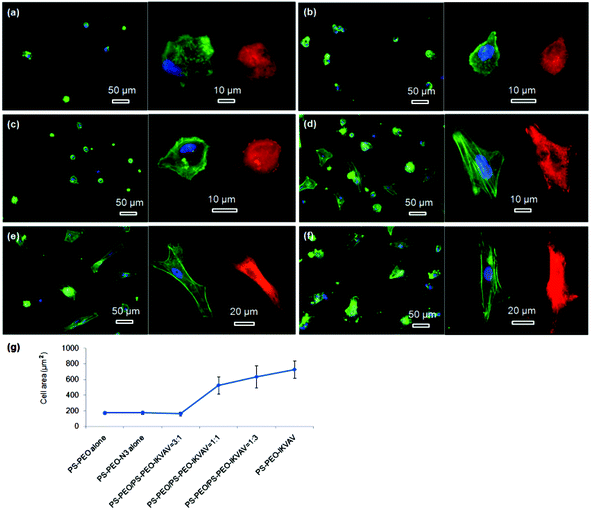
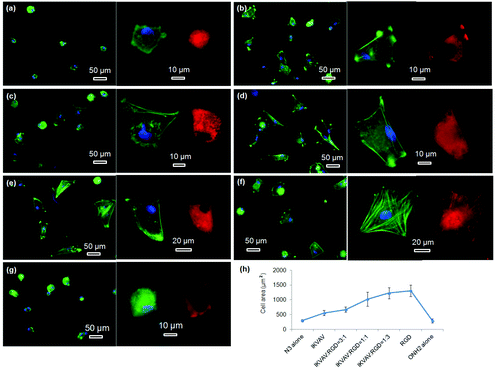
The morphologies of hMSCs on these substrates were then investigated. hMSCs on the substrates containing 50% of PS-PEO and 50% PS-PEO-N3 showed typical rounded morphologies without any actin stress fibres or substantive focal complexes being observed (Fig. 13a). The calculated average cell area is only 290 ± 40 μm2 (Fig. 13h), similar to that shown by hMSCs on 100% PS-PEO-N3 (Fig. 7). In contrast, after being conjugated with IKVAV peptide, hMSCs displayed moderate spread morphologies, however, with very few actin stress fibres and nascent focal complexes. The majority of the cells displayed rounded shapes but with slightly improved average spread area (Fig. 13b). The average cell spread area was calculated to be 550 ± 90 μm2, nearly two fold that of the spread areas of cells on the same substrate without IKVAV conjugation, and similar to that observed in Fig. 7 for this same system configuration. Whilst no obvious changes in overall cell morphology were observed when a small amount of RGD peptide (RGD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IKVAV = 1
IKVAV = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) was incorporated into the nanodomains on the surface (Fig. 13c), through the substitution of PS-PEO-ONH2 for the PS-PEO-N3 in a similar ratio and subsequent stage-wise conjugation of IKVAV and then RGD, this incorporation of 25% of RGD peptide (by mass) to the total amount of peptide in these nanodomains (the remaining 75% is IKVAV) led to a slight increase in the average cell spread area to 670 ± 90 μm2 (although not statistically significant). With a further increase in the relative amount of RGD to IKVAV peptides in each nanodomain, to 1
3) was incorporated into the nanodomains on the surface (Fig. 13c), through the substitution of PS-PEO-ONH2 for the PS-PEO-N3 in a similar ratio and subsequent stage-wise conjugation of IKVAV and then RGD, this incorporation of 25% of RGD peptide (by mass) to the total amount of peptide in these nanodomains (the remaining 75% is IKVAV) led to a slight increase in the average cell spread area to 670 ± 90 μm2 (although not statistically significant). With a further increase in the relative amount of RGD to IKVAV peptides in each nanodomain, to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 3
1 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the hMSCs displayed improved cell spreading, with further increases in cell spread area of 1010 ± 240 and 1230 ± 190 μm2, respectively. Few rounded hMSCs were observed on these substrates (Fig. 13d and e). However, on closer inspection, whilst cell spread areas increased, most of the cells adhered to these substrates have very few stress fibres (excepting along their periphery) and poorly developed focal adhesions (indicated by the punctate and small vinculin structures). In contrast, hMSCs grown on the RGD only peptide conjugated substrate, composed of 50% PS-PEO and 50% PS-PEO-ONH2, exhibited well-spread, oblate morphologies with well-developed actin stress fibres and substantive vinculin structures around the edges of the cells and across the cells (Fig. 13f). The average cell spread area is 1320 ± 200 μm2, which is the highest observed within the investigated surfaces and in agreement with that previous observed for this same system configuration (Fig. 10). Without RGD conjugation, however, there was rather poor adhesion of the hMSCs to the bare 50% PS-PEO/50% PS-PEO-N3 substrate, with an average cell spread area of 280 ± 70 μm2, rounded morphologies, without any actin fibres or substantive focal adhesions being observed (Fig. 13g).
1, the hMSCs displayed improved cell spreading, with further increases in cell spread area of 1010 ± 240 and 1230 ± 190 μm2, respectively. Few rounded hMSCs were observed on these substrates (Fig. 13d and e). However, on closer inspection, whilst cell spread areas increased, most of the cells adhered to these substrates have very few stress fibres (excepting along their periphery) and poorly developed focal adhesions (indicated by the punctate and small vinculin structures). In contrast, hMSCs grown on the RGD only peptide conjugated substrate, composed of 50% PS-PEO and 50% PS-PEO-ONH2, exhibited well-spread, oblate morphologies with well-developed actin stress fibres and substantive vinculin structures around the edges of the cells and across the cells (Fig. 13f). The average cell spread area is 1320 ± 200 μm2, which is the highest observed within the investigated surfaces and in agreement with that previous observed for this same system configuration (Fig. 10). Without RGD conjugation, however, there was rather poor adhesion of the hMSCs to the bare 50% PS-PEO/50% PS-PEO-N3 substrate, with an average cell spread area of 280 ± 70 μm2, rounded morphologies, without any actin fibres or substantive focal adhesions being observed (Fig. 13g).
At a constant level of ligand redundancy (∼14 IKVAV ligands per nanodomain), and nanodomain lateral spacing (∼35 nm), these results clearly show that systematically substituting the IKVAV ligand with linearly increasing numbers of RGD peptides (0, ∼3, 7, ∼10, 14) within the PEO nanodomains encourages increasing levels of attachment, spreading, cytoskeletal tension and focal adhesion maturation in hMSCs, in a similarly linear fashion. These substrates thus allow one to encode into a surface defined cues for cellular interaction at levels of control previously untested. Given previous insights into the significant correlations between hMSC differentiation and shape/stress indicators (such as spread area/morphology, cytoskeletal stress, and focal adhesion maturation), these novel substrates open up many new opportunities to explore a new cue, the impact of ligand redundancy on hMSC differentiation and lineage commitment.
4. Conclusions
In this work, a variety of polymer substrates with well-controlled local number density of peptides within defined cylindrical PEO nanodomains of similar size (∼17 nm) to a single integrin pair were fabricated by co-self-assembly of PS-PEO with different amount of PS-PEO-N3 or PS-PEO-ONH2, followed by IKVAV and RGD conjugation through azide–alkyne Huisgen cycloaddition reaction and aldehyde-aminooxy oxime ligation reaction, respectively. It was found that increases in the local number density, and hence the level of redundancy for integrin binding, of IKVAV and RGD peptides within each nanodomain of these substrates significantly impacted hMSC adhesion and enabled effective modulation of hMSC morphology, from rounded shapes with rather poor adhesion to well-spread cells with well-developed actin stress fibres and the number of mature focal adhesions. A set of surfaces with controllable presentation of both IKVAV and RGD peptides within each PEO nanodomain were also constructed by co-self-assembling 50% of PS-PEO and 50% of the mixture of PS-PEO-N3 and PS-PEO-ONH2 in different ratios, permitting orthogonal coupling of IKVAV and RGD peptides by sequential Huisgen click and oxime ligation reactions. At a constant level of ligand redundancy, these novel surfaces encoding defined ratios of peptides in each nanodomain enabled the impacts of ligand affinity on hMSC adhesion and morphology to be elucidated. Such differences in hMSC spreading and cytoskeletal organisation have been shown to influence the differentiation of hMSC populations. These novel self-assembling cell culture substrates thus provide extremely flexible platforms to probe the impacts of ligand redundancy and affinity on surface-mediated regulation of hMSC fate choices.Acknowledgements
The authors would like to acknowledge the financial support provided by the Australian Research Council Discovery Grant Scheme (DP1095429). This work was performed in part at the Queensland node of the Australian National Fabrication Facility, a company established under the National Collaborative Research Infrastructure Strategy to provide nano and microfabrication facilities for Australian researchers.Notes and references
- J. J. Rogers, H. E. Young, L. F. Adkison, P. A. Lucas and A. C. Black, Am. Surg., 1995, 61, 231 CAS.
- A. I. Caplan, J. Orthop. Res., 1991, 9, 641 CrossRef CAS PubMed.
- C. Cha, W. B. Liechty, A. Khademhosseini and N. A. Peppas, ACS Nano, 2012, 6, 9353 CrossRef CAS PubMed.
- B. Joddar and Y. Ito, J. Mater. Chem., 2011, 21, 13737 RSC.
- G. A. Hudalla and W. L. Murphy, Soft Matter, 2011, 7, 9561 RSC.
- M. Schvartzman, M. Palam, J. Sable, J. Abramson, X. Hu, M. Sheetz and S. J. Wind, Nano Lett., 2011, 11(3), 1306 CrossRef CAS PubMed.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, D. D. Wilkinson and R. O. C. Oreffo, Nat. Mater., 2007, 6, 997 CrossRef CAS PubMed.
- J. E. Frith, R. J. Mills and J. J. Cooper-White, J. Cell Sci., 2012, 125, 317 Search PubMed.
- V. C. Hirschfeld-Warneken, M. Arnold, A. Cavalcanti-Adam, M. López-García, H. Kessler and J. P. Spatz, Eur. J. Cell Biol., 2008, 87, 743 CrossRef CAS PubMed.
- J. E. Phillips, T. A. Petrie, F. P. Creighton and A. J. García, Acta Biomater., 2010, 6, 12 CrossRef CAS PubMed.
- M. Guvendiren and J. A. Burdick, Biomaterials, 2010, 31, 6511 CrossRef CAS PubMed.
- O. Jeon and E. Alsberg, Adv. Funct. Mater., 2013, 23, 4765 CAS.
- J. Fu, Y.-K. Wang, M. T. Yang, R. A. Desai, X. Xu, Z. Liu and C. S. Chen, Nat. Methods, 2010, 7, 733 CrossRef CAS PubMed.
- A. R. Cameron, J. E. Frith and J. J. Cooper-White, Biomaterials, 2011, 32, 5979 CAS.
- H. Yang and W. J. Kao, Int. J. Nanomed., 2007, 2, 89 CrossRef CAS.
- B. T. Houseman, E. S. Gawalt and M. Mrksich, Langmuir, 2003, 19, 1522 Search PubMed.
- Y.-X. Chen, G. Triola and H. Waldmann, Acc. Chem. Res., 2011, 44, 762 CrossRef CAS PubMed.
- J. E. Moses and A. D. Moorhouse, Chem. Soc. Rev., 2007, 36, 1249 RSC.
- C. E. Hoyle, A. B. Lowe and C. N. Bowman, Chem. Soc. Rev., 2010, 39, 1355 RSC.
- Z. Q. Lin, D. H. Kim, X. D. Wu, L. Boosahda, D. Stone and L. LaRose, Adv. Mater., 2002, 14, 1373 CrossRef CAS.
- P. A. George and J. J. Cooper-White, Eur. Polym. J., 2009, 45, 1065 Search PubMed.
- P. A. George, M. R. Doran, T. I. Croll, T. P. Munro and J. J. Cooper-White, Biomaterials, 2009, 30, 4732 CrossRef CAS PubMed.
- H. Li, J. Frith and J. J. Cooper-White, Biomacromolecules, 2014, 15, 43 CrossRef CAS PubMed.
- O. Rossier, V. Octeau, J.-B. Sibarita, C. Leduc, B. Tessier, D. Nair, V. Gatterdam, O. Destaing, C. Albigès-Rizo, R. Tampé, L. Cognet, D. Choquet, B. Lounis and G. Giannone, Nat. Cell Biol., 2012, 14, 1057 Search PubMed.
- I. D. Campbell and M. J. Humphries, Cold Spring Harbor Perspect. Biol., 2011, 3 CAS , a004994.
- E. V. Titov, V. I. Rybachenko and R. A. Makarova, Zhurnal Prikladnoi Specktroskopii (J. Appl. Spectrosc.), 1979, 31, 844 CAS.
- Y. Wu, F. Zuo, Z. Zheng, X. Ding and Y. Peng, Nanoscale Res. Lett., 2009, 4, 738 CrossRef CAS PubMed.
- S. Rajora, D. Khatri and G. L. Talesara, Asian J. Chem., 1999, 11, 59 CAS.
- I. K. Lednev, V. V. Ermolenkov, S. Higashiya, L. A. Popova, N. I. Topilina and J. T. Welch, Biophys. J., 2006, 91, 3805 CrossRef CAS PubMed.
Footnote |
| † Current address: CSIRO, Division of Process and Engineering, Clayton South, Victoria 3169, Australia. |
| This journal is © The Royal Society of Chemistry 2014 |