Antimicrobial susceptibility assays in paper-based portable culture devices†
Frédérique
Deiss
,
Maribel E.
Funes-Huacca
,
Jasmin
Bal
,
Katrina F.
Tjhung
and
Ratmir
Derda
*
Department of Chemistry and Alberta Glycomics Centre, University of Alberta, Edmonton, AB T6G 2G2, Canada. E-mail: ratmir@ualberta.ca; Tel: +1 780 492 8370
First published on 8th October 2013
Abstract
To detect antibiotic-resistant bacteria in areas remote from microbiology laboratories, we designed portable culture devices performing an analogue of the Kirby–Bauer disk diffusion test inside patterned papers embedded in tape. We quantified the antibiotic susceptibility of several strains of Escherichia coli and Salmonella typhimurium by measuring blue-colored zones of inhibited growth.
Simple, disposable, and portable devices for culture assays can be used in low-resource environments without the need for an aseptic laboratory infrastructure. Portable devices that allow for the quantification of the resistance of bacteria to antibiotics could be useful for the characterization of infectious diseases in developing countries and areas remote from specialized laboratories.1 Similar devices could also allow for the assessment of food and water quality in these regions.2 Detection of bacteria resistant to antibiotics is a long-standing problem, and there are many elegant prototypes of point-of-care (POC) devices that allow for the assessment of the susceptibility of bacteria to antibiotics.3–8 To facilitate access to these tests worldwide, we aimed to design devices that could be readily produced by minimally educated personnel in any low-equipped location using readily available materials. In this report, we describe portable, paper-based devices for culture of bacteria and detection of their resistance to antibiotics using a portable version of the Kirby–Bauer antibiotic susceptibility test (AST). The classical Kirby–Bauer test is performed with antibiotic-permeated paper disks atop a slab of agar;9 we replaced the disks and agar with a sheet of paper that contained antibiotics and bacteria in pre-patterned locations. Diffusion of antibiotics into the lawn of bacteria inside the paper yielded radial ‘zones of inhibition of bacterial growth.’
We designed our devices to satisfy several criteria. (i) Materials for the production of these devices should be low-cost and readily available. (ii) The steps for the assembly and use of the devices should be easily replicated by low-educated personnel (here, high school students). (iii) The devices should have a significant shelf-life at room temperature. (iv) Devices should contain the pathogens, and it should be possible to interpret the results of the test without opening the device and exposing the user to the pathogens. Although the Kirby–Bauer AST is widely used for detecting antibiotic resistance, the assay is limited to clinical laboratories following standardized protocols. Furthermore, the assay format is fixed—it must utilize a non-portable, agar-filled Petri dish and specialized equipment such as spring-loaded cartridges of sterile antibiotic-impregnated disks. Although the disks can be deposited manually with tweezers, an automatic disk dispenser is frequently used to improve the reproducibility of the assay. Any changes to the format are difficult to implement because the components of the device, such as sterile plastic Petri dishes and disk dispensers, are produced by specialized industries that follow fixed standards. We envisioned that switching from an agar plate to a paper-based support would provide benefits such as simplicity in fabrication and flexibility in modifying the composition and the dimensions of the device. Using wax-patterned paper as a support also allows for flexibility in the deposition of cells, nutrients or chemicals in specific locations.10–12
The core of the portable paper-based AST is a previously reported portable culture device made from tape, PDMS, and patterned paper.13 The performance of our portable device was similar, in many aspects, to that of a regular Petri dish: inside the paper, bacteria grew as compact colonies, and the transport of molecules, such as resazurin, was limited solely to diffusion.13 We envisioned these paper-based culture devices to be suitable as analogues of the Kirby–Bauer AST.14,15 Reproducible diffusion of antibiotics in agar made it possible to build susceptibility-resistance tables for different microorganisms and antibiotics and to standardize AST worldwide.16–18 Similarly, paper-based AST could be standardized if they yield a reproducible diffusion of small molecules (Fig. S1†) and reproducible zones of inhibition of bacterial growth that correlate with the results observed with the Kirby–Bauer AST. This report describes improvements to previously reported paper-based culture devices that allow for a uniform diffusion profile of antibiotics and yield reproducible AST results. We believe that AST in portable culture devices could be easily adopted by clinical microbiologists given the similarities in procedure between paper devices and the common Kirby–Bauer test.
To validate the performance of these improved devices, we used several strains of bacteria (details available in ESI†): (i) laboratory strains of Escherichia coli K12 ER2738 that contained tetracycline or ampicillin resistance plasmids, (ii) laboratory-evolved strains of E. coli K12 ER2738 that had varying susceptibilities to ampicillin, and (iii) pathogenic E. coli O157:H7 and Salmonella typhimurium LT2. For all strains, we compared AST results obtained from paper-based devices and agar-filled dishes.
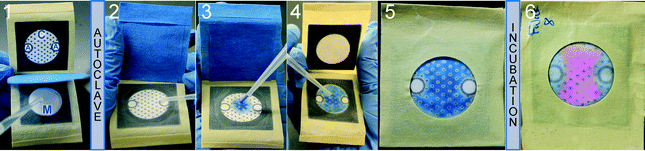
The steps for the preparation and application of the AST device are described in Fig. 1. The device has a zone for deposition of media (M-zone), a culture zone (C-zone) and two zones for deposition of antibiotics (A-zones). The preparation of the device consists of five steps: (i) assemble the device (see movie S1 in ESI†);13 (ii) add culture media to the M-zone and cover it with a protective layer (blue painter tape in Fig. 1);19 (iii) close the device and sterilize it; (iv) open the device after autoclaving and spot antibiotics into the A-zones; and (v) pack the device into moisture-impermeable bags for storage if needed. To complete the AST, a user needs to perform three simple steps: (i) open the device and add PrestoBlue™ and a bacterial inoculum to the C-zone; (ii) remove the protective layer and seal the device; and (iii) incubate it at a suitable temperature (here, 37 °C).
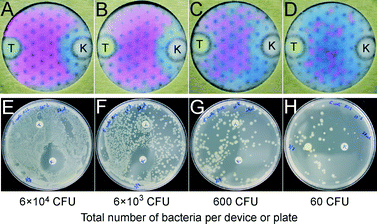
As uniform seeding of bacteria is very important in the Kirby–Bauer AST, we had to optimize the design of the culture zone. To this end, we patterned the culture zone paper with hydrophobic spots (Fig. 2B and D) to facilitate uniform capillary spreading of the bacterial inoculum and to avoid ‘coffee ring’ effects.20 We performed AST for tetracycline (T) and ampicillin (A) by depositing 106 CFU E. coli K12 ER2738 in each device (Fig. 2). In a representative result described in Fig. 2C, the majority of the bacteria were concentrated by the capillary effect at the periphery of the device, forming a pink halo of turned-over PrestoBlue™. The zone of inhibition was not visible because the central region did not have a sufficient number of bacteria. The assays were poorly reproducible, and identification of the zone of inhibition was often not possible. In a similar device with an array of hydrophobic dots, the inoculum spread uniformly (Fig. 2D). The susceptibility of E. coli K12 ER2738 to ampicillin and its resistance to tetracycline were clearly visible.
The grid of hydrophobic spots in our devices served another purpose: it allowed simple digitization of the bacterial resistance after culture. Due to the uniform shape, high contrast of the dots, and color difference between the blue and pink areas, an image of a device could be analyzed by software to count the number of dots in both areas. We tested devices with different arrangements of hydrophobic spots in the culture zone (some examples are presented in Fig. S2†), and we selected the hexagonal grid described in Fig. 2D as the arrangement that yielded the most optimal spreading, contrast and spacing for quantification. The deposition of the visualization reagent (PrestoBlue™) also influenced the quality and the contrast of the colorimetric readouts. Deposition of the dye before the culture step, as opposed to spraying this reagent after the culture step as we reported for previous culture devices,13 significantly increased the reproducibility of the assay. It yielded high-contrast images even with low quality cameras under poor illumination conditions (Fig. S2†). Such images should be amenable to analysis by smart phones and smart phone software, such as the image acquisition software for Android platforms that we described in our previous report.13
To make the portable AST devices practical for use in remote settings, they must be amenable to storage under ambient conditions. We tested the storage of ready-to-use devices that contained sterilized LB culture media (400 μL) and antibiotics in the A-zones (2 μL per 5 mm circle) at room temperature in sealed, moisture-impermeable and puncture-resistant bags for up to 70 days. Devices either contained PrestoBlue™ (resazurin) during storage or were supplemented with this dye after storage; in both cases, we added 50 μL of the dye to the culture zone. Devices stored without PrestoBlue™ for 40 days yielded results similar to those obtained from devices prepared on the same day of inoculation (Fig. S3†). The devices stored with PrestoBlue™ had a shorter shelf-life: after 14 days of storage, the dye became inactive as it was no longer turned over by bacteria (Fig. S4†, the culture zone remained blue in the presence of viable bacteria). Although resazurin remains active for up to one year of storage at room temperature in liquid form, the dye must be freely diffusible to be turned over by cells. We hypothesized that the apparent loss of activity of this dye is caused by the partial drying on the culture zone side over time and the irreversible adsorption of dye to the cellulose; similar irreversible binding of resazurin to bovine serum albumin has been shown to decrease the efficiency of this dye.21 Following these observations, we stored all devices without PrestoBlue™ and added it at the inoculation step as described in Fig. 1.
We compared AST in portable culture devices to disk diffusion on agar plates for different concentrations of E. coli K12 ER2738 with tet-plasmid (6 × 104, 6 × 103, 600, and 60 CFU) using kanamycin (10 μg) and tetracycline (10 μg) as model antibiotics (Fig. 3). For the paper-based culture devices, we determined the susceptibility from the size of the area without viable cells, indicated by the blue-colored zones around the antibiotic reservoirs (A-zones), after overnight incubation in the presence of PrestoBlue™ (Fig. 3A–D). The sizes of these blue areas correlated with the diameters of the zones of inhibited growth observed around the antibiotic-impregnated disks on agar dishes after overnight incubation (Fig. 3E–H). Both platforms confirmed that E. coli K12 ER2738 is resistant to tetracycline and is more susceptible to kanamycin. Fig. 3C, however, displays the limit of AST in portable culture devices; as observed with a low number (600 CFU per device or 1.2 × 104 CFU mL−1) of E. coli K12 ER2738, the visualization of antibiotic diffusion was unreliable. A similar limitation was observed on agar Petri dishes for 6000 CFU (i.e., 1.2 × 105 CFU mL−1, Fig. 3F). The portable culture devices developed in this work allow for a lower standard concentration of bacterial inoculum of 105 CFU mL−1. Inoculum concentration is an important parameter for standardization of Kirby–Bauer AST. At high inoculum concentration, the sizes of the zones of inhibition decrease (Fig. S4†) because the assay is a reaction–diffusion process (the ‘reaction’ component being the binding of antibiotic to bacteria).9 Standard clinical protocols typically require a titer of 1.5 × 108 CFU mL−1 (McFarland 0.5) and we envision a similar requirement for AST in paper devices. For example, to test a patient sample such as stool (in the case of E. coli or Salmonella infections), the extracted bacteria should be brought to a titer of ~105–106 CFU mL−1 before inoculation of the device to obtain results gaugeable with reference tables.
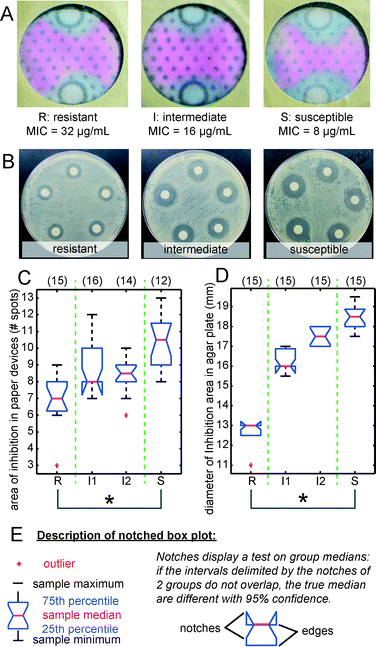
To demonstrate that AST in portable culture devices could distinguish ‘resistant’ from ‘susceptible’ behaviors, as defined by the CLSI,17,18 we evolved strains of E. coli K12 ER2738 that exhibit these behaviors. Evolution of these bacteria was performed as reported by Cira et al.3 Briefly, we plated >109 CFU non-resistant E. coli K12 strain ER2738 on agar containing 20 μg mL−1 of ampicillin, which is twice the minimal inhibitory concentration of this strain for ampicillin. We then isolated the emerging resistant clones and characterized them by micro-broth dilution to categorize each clone as ‘resistant’ or ‘susceptible’ and, alternatively, as ‘intermediate’ following the typical CLSI protocol.22 We compared images resulting from AST in paper devices (Fig. 4A) and on agar plates (Fig. 4B). For paper devices, we counted the number of spots in the blue zones (Fig. 4C), and for agar plates, we measured the diameters of the zones of inhibited growth (Fig. 4D). We observed that AST both on agar and in portable culture devices could distinguish ‘resistant’ from ‘susceptible’ strains according to ANOVA and a multiple comparison procedure (MATLAB™). Both AST exhibited a class of ‘intermediate’ strains that could be categorized neither as ‘resistant’ nor as ‘susceptible’ (Fig. 4). AST in paper devices was reproducible even when using different types of paper (Fig. S5†).
We also demonstrated that portable culture devices could be used with pathogenic bacteria: E. coli O157:H7 (Fig. 5) and S. typhimurium LT2 (Fig. S6†). We tested two antibiotics, ampicillin (A) and tetracycline (T), in the devices (Fig. 5A, S6A†) and agar plates (Fig. 5B, S6B†) inoculated with 104 CFU of bacteria. Both strains of pathogenic Enterobacteriaceae displayed greater susceptibility to ampicillin than to tetracycline in both platforms. The size of the inhibition area around ‘A’ was significantly different (p < 0.05) from the area adjacent to ‘T’ as determined by ANOVA (Fig. 5C–D and S6C–D†).
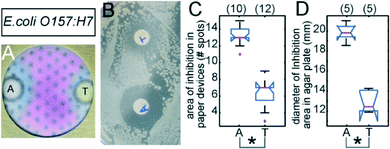 | ||
| Fig. 5 AST with E. coli O157:H7. Representative images of culture devices (A) and Petri dishes (B) inoculated with 104 CFU E. coli O157:H7 and exposed to ampicillin ‘A’ or tetracycline ‘T’ (10 μg per zone or disk) after 18 hours of culture at 37 °C. Notched box plots display the number of spots in blue-colored areas of inhibited growth in paper culture devices (C) and the diameters of the zones of inhibited growth on agar plates (D). See Fig. 4 for the description of the notched box plots. | ||
Conclusions
This report demonstrated that AST in paper-based portable culture devices displays performance, reproducibility, and predictive capacity similar to the Kirby–Bauer AST on agar-filled Petri dishes. A complete characterization of portable devices with different classes of bacteria and antibiotics is beyond the scope of this communication because standards such as the CLSI M100S series used for the interpretation of the Kirby–Bauer test are the results of international collaborations carried out for over a decade.9 Upon further characterization, these devices could find use in the estimation of the resistance of bacteria in biological, environmental, or food-borne samples and could be tailored for specific applications. For example, we observed that assays of highly susceptible strains yield high variability in the radius of inhibition; however, decreasing the concentration of the antibiotics yields smaller inhibition areas and mitigates the problem. We believe that the devices reported in this communication are not only functional prototypes of portable AST for remote locations but also one of the first examples of devices suitable for POC production. The flexibility of the paper-based format will facilitate the integration of the described culture platform with other paper-based tests that rely on an external pre-culture of bacteria.23–25Acknowledgements
We thank Christine Szymanski for the samples of E. coli O157:H7 and S. typhimurium LT2. We thank Alyson Wu, Pavithra Rajendran, Falak Shah and Jessica Wickware from Harry Ainlay High School (Edmonton, AB) for testing the AST prototypes. This research was supported by the Alberta Glycomics Centre, SENTINEL Bioactive paper network and Grand Challenges Canada Rising Star Award in Global Health (to RD).Notes and references
- R. W. Peeling, P. G. Smith and P. M. M. Bossuyt, Nat. Rev. Microbiol., 2008, 6, S2–6 CrossRef CAS PubMed.
- C. Bouki, D. Venieri and E. Diamadopoulos, Ecotoxicol. Environ. Saf., 2013, 91, 1–9 CrossRef CAS PubMed.
- N. J. Cira, J. Y. Ho, M. E. Dueck and D. B. Weibel, Lab Chip, 2012, 12, 1052–1059 RSC.
- R. Takagi, J. Fukuda, K. Nagata, Y. Yawata, N. Nomura and H. Suzuki, Analyst, 2013, 138, 1000–1003 RSC.
- K. P. Kim, Y. G. Kim, C. H. Choi, H. E. Kim, S. H. Lee, W. S. Chang and C. S. Lee, Lab Chip, 2010, 10, 3296–3299 RSC.
- M. Kalashnikov, J. C. Lee, J. Campbell, A. Sharon and A. F. Sauer-Budge, Lab Chip, 2012, 12, 4523–4532 RSC.
- J. Q. Boedicker, L. Li, T. R. Kline and R. F. Ismagilov, Lab Chip, 2008, 8, 1265–1272 RSC.
- C. D. Chin, T. Laksanasopin and Y. K. Cheung, et al. , Nat. Med., 2011, 17, 1015–U1138 CrossRef CAS PubMed.
- A. Wanger, in Antimicrobial Susceptibility Testing Protocols, ed. R. Schwalbe, L. Steele-Moore and A. C. Goodwin, CRC press, Boca Raton, FL, 2007, pp. 53–73 Search PubMed.
- R. Derda, S. K. Y. Tang, A. Laromaine, B. Mosadegh, E. Hong, M. Mwangi, A. Mammoto, D. E. Ingber and G. M. Whitesides, PLoS One, 2011, 6, e18940 CAS.
- F. Deiss, A. Mazzeo, E. Hong, D. E. Ingber, R. Derda and G. M. Whitesides, Anal. Chem., 2013, 85, 8085–8094 CrossRef CAS PubMed.
- E. Carrilho, A. W. Martinez and G. M. Whitesides, Anal. Chem., 2009, 81, 7091–7095 CrossRef CAS PubMed.
- M. Funes-Huacca, A. Wu, E. Szepesvari, P. Rajendran, N. Kwan-Wong, A. Razgulin, Y. Shen, J. Kagira, R. Campbell and R. Derda, Lab Chip, 2012, 12, 4269–4278 RSC.
- A. W. Bauer, W. M. M. Kirby, J. C. Sherris and M. Turck, Am. J. Clin. Pathol., 1966, 45, 493–496 CAS.
- A. W. Bauer, D. M. Perry and W. M. M. Kirby, Arch. Intern. Med., 1959, 104, 208–216 CrossRef CAS.
- K. E. Cooper and D. Woodman, J. Pathol. Bacteriol., 1946, 58, 75–84 CrossRef CAS.
- Clinical and Laboratory Standards Institute, Performance standards for antimicrobial susceptibility testing; Seventeenth informational supplement, Wayne, PA, 2007 Search PubMed.
- These categories are defined as the likelihood of an infection caused by a specific organism to respond (‘susceptible’) or not (‘resistant’) to treatment with a standard dosage of antibiotic. The ‘intermediate’ category is used to indicate no clear therapeutic options and to avoid major categorical errors due to slight changes in zones of inhibition. The diameter of the zone corresponding to each breakpoint will vary depending on the antibiotic, the standard dosage, and the class of bacteria..
- The protective layer allows for separation of the wet M-zone from both the dry C-zone and A-zones and thus prevents unwanted diffusion of antibiotics. It can be made from any non-toxic, autoclavable, and flexible layer..
- R. D. Deegan, O. Bakajin, T. F. Dupont, G. Huber, S. R. Nagel and T. A. Witten, Nature, 1997, 389, 827–829 CrossRef CAS PubMed.
- P. Goegan, G. Johnson and R. Vincent, Toxicol. In Vitro, 1995, 9, 257–266 CrossRef CAS.
- S. Qaiyumi, in Antimicrobial Susceptibility Testing Protocols, ed. R. Schwalbe, L. Steele-Moore and A. C. Goodwin, CRC press, Boca Raton, FL, 2007, pp. 75–79 Search PubMed.
- J. C. Jokerst, J. A. Adkins, B. Bisha, M. M. Mentele, L. D. Goodridge and C. S. Henry, Anal. Chem., 2012, 84, 2900–2907 CrossRef CAS PubMed.
- J. Hoorfar, APMIS, Suppl., 2011, 119, 1–24 CrossRef PubMed.
- S. M. Z. Hossain, C. Ozimok, C. Sicard, S. D. Aguirre, M. M. Ali, Y. F. Li and J. D. Brennan, Anal. Bioanal. Chem., 2012, 403, 1567–1576 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary figures S1–S6, protocols, MATLAB™ scripts and a movie describing the fabrication of the devices. See DOI: 10.1039/c3lc50887k |
| This journal is © The Royal Society of Chemistry 2014 |