Nonlinear optical dye TSQ1 as an efficiently selective fluorescent probe for G-quadruplex DNA†
Yuqi
Chen
a,
Shengyong
Yan
a,
Libo
Yuan
a,
Yimin
Zhou
a,
Yanyan
Song
a,
Heng
Xiao
a,
Xiaocheng
Weng
*a and
Xiang
Zhou
*ab
aCollege of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers of Ministry of Education, Wuhan University, Wuhan, Hubei 430072, P. R. China. E-mail: xzhou@whu.edu.cn; xcweng@whu.edu.cn; Fax: (+)86-27-68756663; Tel: (+)86-27-68756663
bState Key Laboratory of Natural and Biomimetic Drugs, Peking University, Beijing, 100191, P. R. China
First published on 6th February 2014
Abstract
A squarylium dye TSQ1 shows a remarkable fluorescence enhancement selectivity for the G-quadruplex DNA structure against duplex DNA, it can be distinguished even by the naked eye. What is more important, TSQ1 did not induce the G-rich sequence folding into G-quadruplex structure which means TSQ1 can be used to detect G-quadruplex structures in cells.
G-quadruplexes are formed by G-rich DNA sequences and G-rich DNA sequences exist in many important regions such as telomere, the promoter region of some proto-oncogenes, ribosomal DNA and in the untranslated regions of mRNA. Recent reports have demonstrated that quadruplex structures play important roles in gene expression1–3 and these provide us with more opportunities for searching for new cancer therapies. Therefore, G-quadruplex detection seems to be shown an important task. Recently, some organic molecules have been discovered and synthesized, and they have fluorescence enhancement with G-quadruplexes.4–8
So far, the fluorescence induced by the G-quadruplex fluorescence probes are mostly produced by AIE or conformation change mechanisms,9 like suppressing twisting motion. However, little effect has focused on nonlinear optical (NLO) G-quadruplex fluorescence probes. NLO materials are attracting much attention because of their potential applications in clinical applications. In addition, the quantification of G-quadruplex DNA in vitro and in vivo using fluorescence probe remains a challenge, one reason is that most reported G-quadruplex probes are able to induce G-rich DNA forming G-quadruplex structures, which decreases the quantitative accuracy.7,8
Herein, we report a squarylium dye TSQ1 which is highly sensitive to G-quadruplex structures and can be used to detect G-quadruplexes with the naked eye. They can also discriminate G-quadruplexes against dsDNA and ssDNA without inducing the formation of G-quadruplex (Scheme 1). Most of the G-quadruplex fluorescent probes can induce the formation of the G-quadruplex in vivo, so, we cannot confirm that G-quadruplex exists in cells before being induced by probes.9 So, a G-quadruplex probe without inductivity is crucial for cell research. STQ1 is a nonlinear optical (NLO) squaraine consisting of two electron donating endogroups (D) and a central electron withdrawing unit (A) forming a donor–acceptor–donor (D–A–D) alignment (Scheme 1).11 It was synthesised as reported.12 The maximum absorption peak is 610 nm in aqueous solution (50 mM Tris-HCl, pH 7.0). The nonlinear responses of TSQ1 are faster than 50 fs.11 STQ1 exhibits sharp and intense absorption bands in the visible and near-IR region, so the exciting and emission lights are harmless to health.
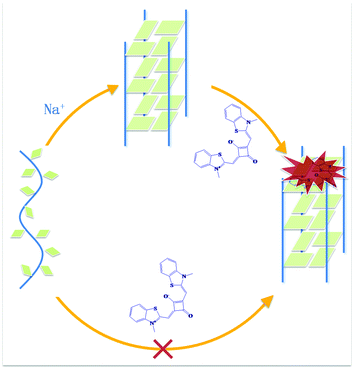 | ||
| Scheme 1 Schematic representation of the fluorescence light-up of STQ1 combining with G-quadruplex structure in the presence of Tris-HCl buffer (pH 7.0) containing 50 mM NaCl. | ||
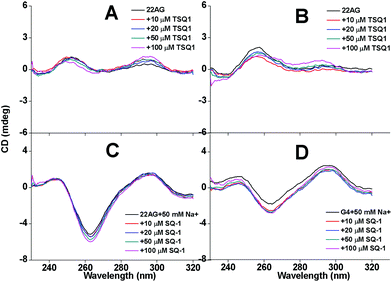
Firstly, we investigated whether TSQ1 has the G-quadruplex induced ability with telomere DNA 22AG (Fig. 1A and C) and G4TTA (Fig. 1B and D) (sequences shown in ESI†), which can not spontaneously fold into G-quadruplex in the absence of metal ion K+ or Na+, which can intensively induce the formation of G-quadruplex. Circular dichroism (CD) spectrum is a reliable method for determining the conversion of the unfolded conformation into quadruplex. Unfolded 22AG (Fig. 1A, line 1) and G4TTA (Fig. 1B, line 1) appears as a characteristic positive CD peak at 257 nm in buffer solution without the metal ions K+ or Na+ (50 mM Tris-HCl, pH 7.0). In the presence of TSQ1, 22AG (Fig. 1A, line 2–5) and G4TTA (Fig. 1B, line 2–5) there is still a characteristic CD band at 257 nm confirming its unfolded state, indicating that TSQ1 can not induce the structure exchange from unfolded state to quadruplex state. However, when 50 mM NaCl was added to the solution, two dramatic positive peaks at 245 nm and 295 nm, and one dramatic negative peak at 265 nm appear for both of them (Fig. 1C and D), indicating 22AG and G4TTA transform into antiparallel quadruplex states from single-strand states. Thus, TSQ1 does not have G-quadruplex inductivity.
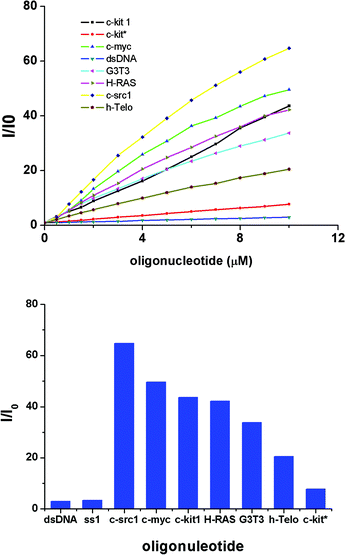
Then the fluorescence behavior of TSQ1 with G-quadruplex DNA structures were investigated, as well as the ssDNA and dsDNA. We choose several different G-quadruplex sequences, including human telomeric DNA h-Telo, G4TTA, 22AG; the promoter of oncogene c-myc, c-kit, c-src1, H-RAS and one sequence T3G3, they can form different kinds of G-quadruplex structures in the presence of sodium chloride. Meanwhile, two complementary oligonucleotides ss1, ss2 were selected as ssDNA and dsDNA. All experiments were carried out in Tris-HCl (pH = 7.0) buffer with 50 mM NaCl. The CD spectra for these G-quadruplex DNA are shown in Fig. S1.† We observe that c-src is shown to form parallel G-quadruplex, 22AG, G3T3, G4TTA and h-Telo are shown to form antiparallel G-quadruplex and c-kit*, H-RAS are shown to form mixed-type DNA G-quadruplex structures. The fluorescence spectra for all DNA are shown in Fig. S2.† Significant fluorescence enhancements were observed when STQ1 was treated with all the G-quadruplex DNAs, the maximum emission peak was 668 nm, while little fluorescence enhancements were observed with ss1 and dsDNA (ss1 + ss2) (Fig. 2, ESI Fig. S2†). When the DNA was titrated to 10 μM, the fluorescence intensity of STQ1 enhanced about 5 to 70 folds upon binding to G-quadruplex DNA, in particular, the promoter c-src1 promoted the most. The FRET melting experience increased only 2.5 folds with ss1 and dsDNA. It is necessary to highlight that the actual self-fluorescence response of STQ1 was very low, the background almost undetected. The undetected background and the light-on fluorescence make STQ1 have the potential to detect the G-quadruplex structure in vitro and in vivo. However, STQ1 has different responses to different DNA, the linear relationship between the DNA concentration and the fluorescence intensity was not very ideal for all DNA (r = 0.964–0.995) (ESI Fig. S3†). FRET melting experiments demonstrated that the STQ1 has a moderate ability to stabilise G-quadruplex structures (Tm = 12.9 °C for c-myc, ESI Fig. S4†) compared with little stability for ssDNA and dsDNA.
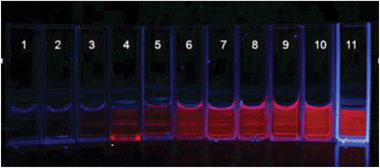
The distinction between quadruplex DNA and ssDNA, dsDNA can also easily be distinguished by the naked eye under UV light. Fig. 3 shows an image of STQ1 with different DNA structures under UV light, we can easily distinguish those system that contain G-quadruplex structures. The c-src1 DNA solution is the brightest, it is consistent with the result of the fluorescence experiment.
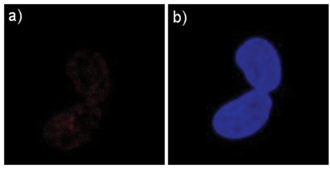
The ability of STQ1 to detect G-quadruplex in living cells was also investigated. STQ1 is virtually nonfluorescent in aqueous solution without G-quadruplex, thus leading to very weak background fluorescence. STQ1 also has little toxicity and good water solubility, these advantages allow the cell to be stained with STQ1 and imaged directly without post-processing. Confocal Laser Scanning Microscopic experiment was taken on the STQ1 treated human lung carcinoma cell A549. The fluorescence image shows that A549 cells give an obvious red fluorescence response to certain regions of the nucleus (Fig. 4a). It was reported that these regions correspond to the nucleoli where the rDNA (ribosomal DNA) undergoes transcription.10 G-rich rDNA can also form a G-quadruplex structure temporarily.13,14 We have also investigated this method in CHO and HeLa cells, similar results were observed (ESI Fig, S5†).
Conclusions
In conclusion, a turn-on fluorescence dye TSQ1 was investigated as G-quadruplex probe which shows no G-quadruplex inductivity and high selectivity between G-quadruplex and dsDNA, ssDNA. It can be distinguished by the naked eye. The TSQ1 fluorescence increases by about 70 folds at most and is about 28 times more intense than dsDNA when interacting with G-quadruplex DNA. Direct fluorescence imaging of the STQ1 in cells indicates that STQ1 can be used for the detection of G-quadruplex in vivo.Acknowledgements
The authors thank the National Basic Research Program of China (973 Program) (2012CB720600, 2012CB720603), the National Science Foundation of China (no. 91213302, 21072115, 21272181) the National Grand Program on Key Infectious Disease (2012ZX10003002-014) and the Program for Changjiang Scholars and Innovative Research Team in University (IRT1030).Notes and references
- X. D. Wang, T. M. Ou, Y. J. Lu, Z. Li, Z. Xu, C. Xi, J. H. Tan, S. L. Huang, L. K. An, D. Li, L. Q. Gu and Z. S. Huang, J. Med. Chem., 2010, 53, 4390–4398 CrossRef CAS PubMed.
- J. E. Johnson, K. Cao, P. Ryvkin, L. S. Wang and F. B. Johnson, Nucleic Acids Res., 2010, 38, 1114–1122 CrossRef CAS PubMed.
- A. Bugaut and S. Balasubramanian, Nucleic Acids Res., 2012, 40, 4727–4741 CrossRef CAS PubMed.
- J. W. Yan, W. J. Ye, S. B. Chen, W. B. Wu, J. Q. Hou, T. M. Ou, J. H. Tan, D. Li, L. Q. Gu and Z. S. Huang, Anal. Chem., 2012, 84, 6288–6292 CrossRef CAS PubMed.
- M. Nikan, M. D. Antonic, K. Abecassis, K. McLuckie and S. Balasubramanian, Angew. Chem. Int. Ed., 2013, 52, 1428–1431 CrossRef CAS PubMed.
- L. Xu, D. Zhang, J. Huang, M. G. Deng, M. Zhang and X. Zhou, Chem. Commun., 2010, 46, 743–745 CAS.
- J. Mohanty, N. Barooah, V. Dhamodharan, S. Harikrishna, P. I. Pradeepkumar and A. C. Bhasikuttan, J. Am. Chem. Soc., 2013, 135, 367–376 CAS.
- H. Ihmels and L. Thomas, Org. Biomol. Chem., 2013, 11, 480–487 CAS.
- J. Alzeer, B. R. Vummidi, P. J. C. Roth and N. W. Luedtke, Angew. Chem., Int. Ed., 2009, 48, 9362–9365 CAS.
- Y. J. Lu, S. C. Yan, F. Y. Chan, L. Zou, W. H. Chung, W. L. Wong, B. Qiu, N. Sun, P. H. Chan, Z. S. Huang, L. Q. Gu and K. Y. Wong, Chem. Commun., 2011, 47, 4971–4973 RSC.
- Z. Y. Li, S. Xu, L. Huang, X. Huang, L. H. Niu, Z. H. Chen, Z. Zhang, F. S. Zhang and K. Z. Kasatani, Chem. Phys. Lett., 2007, 441, 123–126 CrossRef CAS PubMed.
- Y. Xu, M. J. Panzner, X. Li, W. J. Youngs and Y. Pang, Chem. Commun., 2010, 46, 4073–4075 RSC.
- D. Drygin, A. Siddiqui-Jain, S. O'Brien, M. Schwaebe, A. Lin, J. Bliesath, C. B. Ho, C. Proffitt, K. Trent, J. P. Whitten, J. K. C. Lim, D. Von Hoff, K. Anderes and W. G. Rice, Cancer Res., 2009, 69, 7653–7661 CrossRef CAS PubMed.
- L. A. Hanakahi, H. Sun and N. Maizels, J. Biol. Chem., 1999, 274, 15908–15912 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3qo00048f |
| This journal is © the Partner Organisations 2014 |