Naphthyridine based fluorescent receptors for the recognition of uric acid†
Swapan
Dey
*a,
Dibyendu
Sain
a and
Shyamaprosad
Goswami
*b
aDepartment of Applied Chemistry, Indian School of Mines, Dhanbad, Jharkhand 826 004, India. E-mail: deyswapan77@yahoo.com; Fax: +91 326 2296563; Tel: +91 326 2235607
bDepartment of Chemistry, Bengal Engineering and Science University, Shibpur, Howrah 711 103, West Bengal, India. E-mail: spgoswamical@yahoo.com; Fax: +91 3326 682916; Tel: +91 3326 684561
First published on 21st November 2013
Abstract
Naphthyridine based fluorogenic receptors (R1, R2, R3 and R4) have been synthesised for the recognition of uric acid (UA). The receptors are very useful for potential applications arising from complexation reactions as demonstrated by 1H NMR, UV-vis and fluorescence studies. The association constants (Ka) between the receptors and UA have been reported using UV and fluorescence techniques. The minimisation energy calculations and molecular modelling studies for the host–guest assemblies have been discussed in this context.
Introduction
Hydrogen bonds are particularly useful for the involuntary self-assembly of small molecular components to generate supramolecular structures with predefined structural features because of their directionality and specificity.1 To further enhance the strength, directionality, and specificity of hydrogen bonding interactions, there is intense research interest in designing molecular assemblies which have arrays of hydrogen bond donor and acceptor sites.2 In addition to the shape and size compatibilities, effective molecular recognition requires a precise alignment of the binding groups on the receptor with complementary binding regions on the guest substrate. Heterocycles like naphthyridines, which belong to an important class of N donor molecules, are of considerable pharmacological interest due to their use as antihypertensives, antiarrhythmics, herbicide safeners and also as immuno-stimulant compounds.3 Functionalized naphthyridines like 2-amino-1,8-naphthyridine and 2,7-diamino-1,8-naphthyridine are unambiguously important in the field of molecular recognition.4,5
UA is the end product of purine metabolism and the role of UA in the human body is very significant. Depending on the level of UA in the blood, people may suffer from diseases like kidney stones, arthritis, gout and other physiological disorders. Kelly et al. first synthesised and described the binding pattern of UA with synthetic rigid receptors.6 A non-enzymatic method was developed for the selective detection and quantification of serum UA using 2-thiouracil adapted Au nanoparticles, which showed an instantaneous visible colour change.7 Measuring the level of UA in a clinical laboratory is difficult because of the enzyme and reagents concerned.8 It is therefore important to design and synthesise some water-soluble fluorescent receptors to utilise in the molecular recognition of UA. Using the chemistry of naphthyridine, we have successively synthesised some fluorescent receptors which recognise UA in water and also in polar organic solvents, namely DMSO.9 To design the receptors, we concentrated on the hydrogen-bonding pattern that would fulfil the criteria of hydrogen bonding complementarity as well as ensuring that they exhibited fluorescence activity. Using our method, one can identify the presence of UA with a ppm level precision.
Synthesis of the receptors
Receptor 1 (R1) was synthesised by the condensation of 2,6-diaminopyridine and DL-malic acid in the presence of conc. H2SO4.5R1 was then treated with POCl3, followed by NaN3 and Zn–acetic acid to afford R2 [Scheme 1]. | ||
| Scheme 1 Reaction conditions: (i) DL-malic acid, conc. H2SO4, 80–90 °C, 3 h; (ii) POCl3, reflux, 1 h. (iii) NaN3, CH3CN, reflux, 4 h. (iv) Zn dust, acetic acid, reflux, 3 h. | ||
The yellow coloured solid, R2, was treated with salicylaldehyde and the acid chloride of salicylic acid separately to get R3 and R4, accordingly [Scheme 2].
 | ||
| Scheme 2 Reaction conditions: (i) salicylaldehyde, dry methanol, reflux, 24 h (ii) (a) salicylic acid, SOCl2, benzene, reflux, 4 h; (b) Et3N, CH2Cl2, 12 h, r.t. | ||
Results and discussion
In recent years, a new generation of pre-organized host–guest systems have emerged in which unfavourable intermolecular repulsive interactions between D–D and A–A sites have been minimized.10UA contains hydrogen bond donor (D)–acceptor (A) patterns like DADA and ADDA in a cyclic array. Therefore we used ADAD and DAAD type receptors, which were designed for favourable complexation. Molecules of ADAD type are self-complementary with DADA molecules, i.e. they may easily undergo dimerisation to form hetero-complexes. The complexation is possible only when the receptors have a high affinity for the guests. Some novel work by Zimmerman et al. motivated us to extensively study multiple hydrogen bonding.11 We envisaged that the receptors (1–4) might be capable of solubilising the guest in solvents where the guest is relatively less soluble by complexation. Amino-naphthyridines and naphthyridone moieties have the ability to be soluble in aqueous solvents as well as in organic solvents like chloroform, dichloromethane, etc. In all cases, model studies suggested that the guest molecule would be well arranged in the receptor’s cavity.For the theoretical calculations and molecular modelling studies, the complex structures of all four receptors including the dimeric structure of R1 were optimized using the semi-empirical AM1 method with the RHF wave function. All calculations were executed using the Gaussian03 program package with the aid of the Gauss-View and Chemcraft visualization programs.
Complexation studies using NMR spectroscopy, theoretical calculations and molecular modelling
Receptor 1 contains an ADAD arrangement, which is self-complementary. However, it also has the binding sites required to bind UA with its DADA arrangement, involving the CO and NH groups as the main binding points [Fig. 1]. Moreover, this type of arrangement prefers self-association over complexation with guests like UA. In the NMR spectrum, no changes in the peak positions were found for the complex R1C. The lactam proton and the amine protons of R1 appeared at δ 11.55 and δ 6.75 ppm respectively and these positions did not change on complexation with UA in d6-DMSO (see ESI†). When R1 was converted to a derivative of pivaloylamide using pivaloyl chloride, the resulting compound was found to possess an ADAD–DADA dimeric structure via quadruple hydrogen bonding in the solid-state.12 This result also suggests that R1 prefers the stable self-assembled dimeric structure over the hetero-assembled R1C complex structure (Fig. 1).Theoretical calculations also supported the greater stability of the dimeric structure (R1D) compared to the complex structure (R1C). Molecular modelling studies showed that the minimised energy of R1D was 0.0376 a.u., which is lower than the minimised energy of R1C (0.1089 a.u.). Also, there is a quadruple H-bonding interaction in R1D whereas R1C has only three H-bonding interactions (Fig. 1).
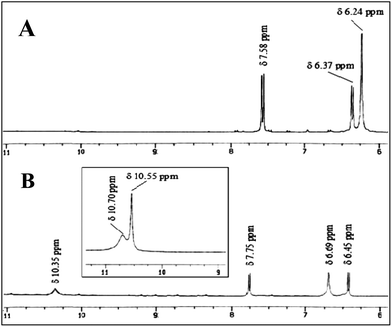
On the other hand, R2 has a DAAD binding array to associate with UA, which has a complementary ADDA hydrogen bonding array (Fig. 2). The energy minimised complex structure of R2 with UA (R2C) clearly shows the DAAD–ADDA arrangements through the formation of quadruple H-bonding. R2C is highly stable with a low energy (0.0083 a.u.). From the NMR data of R2 in d6-DMSO, the peak for the amine protons (NH) appeared at δ 6.24 ppm and was shifted to δ 6.69 ppm (Δδ 0.45 ppm) upon 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexation with UA. The aromatic protons observed as doublets at δ 7.58 and δ 6.37 ppm were shifted downfield to δ 7.75 and δ 6.45 ppm (Δδ 0.17 and 0.08 ppm) respectively. The four UA protons which exhibited two broad singlets at δ 10.70 and δ 10.55 ppm, appeared as a wide broad singlet at δ 10.35 ppm when UA binds with R2 (Fig. 3).
1 complexation with UA. The aromatic protons observed as doublets at δ 7.58 and δ 6.37 ppm were shifted downfield to δ 7.75 and δ 6.45 ppm (Δδ 0.17 and 0.08 ppm) respectively. The four UA protons which exhibited two broad singlets at δ 10.70 and δ 10.55 ppm, appeared as a wide broad singlet at δ 10.35 ppm when UA binds with R2 (Fig. 3).
The chemistry of imine compounds are well known in inorganic coordination chemistry.13 A salicylaldehyde condensed naphthyridine based receptor, R3, was also good for molecular recognition. R3 exists not only in the imine form (R3) in solution, but may also exist as tautomeric structures like R3A and R3B (Fig. 4).
Molecular modelling studies concluded that among the three tautomeric structures, the R3 form was the most stable, but after complexation with UA, the tautomeric structure R3B provided the energetically most stable complex (R3B-C). The energy minimised model structures of the complexes with the three different tautomeric modes are given in Fig. 5. The comparative minimised energy values are given in Table 1.
| Tautomeric structure of R3 | Minimised energy (a.u.) | |
|---|---|---|
| Receptor | Complex | |
| 3 | 0.1433 | 0.1095 |
| 3A | 0.1546 | 0.1130 |
| 3B | 0.2196 | 0.0950 |
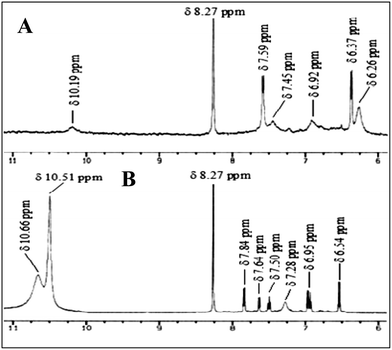
1H NMR analysis showed one broad singlet peak at δ 10.19 ppm for the phenolic –OH groups (R3). A sharp peak appeared at δ 8.27 ppm, indicating the imine (C–H) protons, but all aromatic peaks for the receptor did not clearly appear in the 1H NMR spectrum (d6-DMSO).14 When we added the guest UA to R3, the 1H NMR spectrum of the receptor part of the complex was clearly resolved and there was no peak for the phenolic –OH group, but a broad peak at δ 7.28 ppm was observed. The protons in UA experienced a slight upfield shift (Δδ 0.04 ppm) upon complexation (Fig. 6). This observation strongly supports the formation of the complex structure R3B-C from recognition induced tautomerisation.
Receptor R4 has four H-bond donor points, i.e. two –OH and two –NH–CO groups, as well as two H-bond acceptor points, i.e. the pyridine ring N atoms. Therefore it can mutually bind UA using the donor–acceptor points. Due to conformational flexibility, R4 exist in the R4A conformer, where intra-molecular hydrogen bonding is present in solution. This phenomenon was very significant as it could be identified by the NMR chemical shifts of the complex (R4C) peaks upon binding with UA. UA breaks the intra-molecular hydrogen bonds in R4A and forms a new complex (R4C) (Fig. 7).
 | ||
| Fig. 7 Structure of R4 along with its intra-molecular hydrogen bonded form (R4A) and the energy minimised complex structure (R4C). | ||
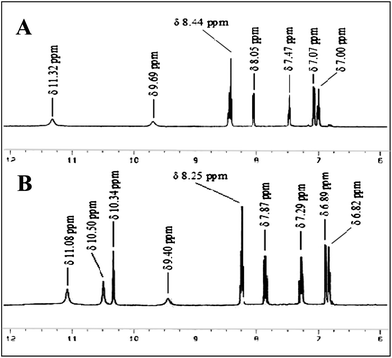
This can easily be explained through simple 1H-NMR spectral data. The phenolic –OH protons in R4 appeared at δ 11.32 ppm, which is abnormally more downfield from their normal position. This is due to the obvious formation of a strong six-membered intra-molecular hydrogen bond in the structure R4A. When the receptor formed a complex with UA (R4C), the amide protons shifted upfield from δ 9.69 ppm to δ 9.40 ppm (Δδ 0.29 ppm) as they hydrogen bonded with the nitrogen atoms in the naphthyridine ring. A remarkable upfield shift was also found for the phenolic –OH protons (δ 11.32 ppm to δ 11.08 ppm, Δδ 0.24 ppm). The aromatic protons also moved upfield (Fig. 8). It is also clear that the phenolic hydroxyl group is a good H-bond donor, but a poor H-bond acceptor.
Complexation studies by UV-vis and fluorescence methods
To study the complexation reactions and calculate the binding constants using UV-vis and fluorescence techniques, separate stock solutions of the receptors and the guest (UA) were prepared in the range of 10−5 M to 10−4 M in H2O.A representative set of UV-vis titration spectra is shown in Fig. 9A, where R2 shows an absorption maxima at λmax = 351 nm. As the UA solution was gradually added (upto 3 equiv.), the spectra showed a red shift in the absorbance from λmax 351 nm to λmax 360 nm (Δλ = 9.0 nm), indicating the complexation. A new peak also appeared around 290 nm, which signifies the presence of excess UA in solution. Upon the gradual addition of UA, the other receptors (R1, R3 and R4) possessed the same responses in the titrations. The UV-vis titration spectra for R1, R3 and R4 with UA are reported in the ESI.†
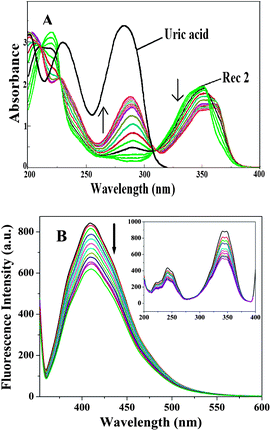 | ||
| Fig. 9 A representative set of titration spectra for R2 with UA in water: (A) UV-vis spectra; (B) fluorescence emission spectra [inset: fluorescence excitation spectra]. | ||
Fluorescence techniques have been widely used to investigate host–guest interactions due to their high sensitivity.15 Complexation studies of the receptors with UA were also carried out using fluorescence spectroscopic analysis in the same H2O solvent, maintaining the same concentrations as mentioned for the UV-vis experiments. When R2 was excited at 351 nm, the emission spectrum had a peak maxima at 409 nm. Significant quenching of the fluorescence intensity was observed upon the gradual addition of UA (upto 3 equiv.) (Fig. 9B). The fluorescence quenching, which is due to a considerable modification of the excited state of the receptor, obviously confirms the complex formation between R2 and UA. The most probable mechanism for the fluorescence quenching effect involves an inversion between the strongly emissive π–π* and the poorly emissive n–π* states of the fluorophore. Such a fluorescence quenching occurs due to hydrogen bonding interactions between the receptor and UA, which leads to a greater stabilization of the n–π* state with respect to the π–π* state and results in a significant decrease in the fluorescence emission intensity.16 Significant fluorescence quenching effects were observed for the other receptors too and the fluorescence spectra of these receptors are in the ESI.†
Fig. 10 shows the actual stoichiometries of the complexes formed during the UV-vis titrations of the receptors (R2, R3 and R4) with UA in water. When we plotted the concentration ratio of guest to host ([G]/[H]) against the intensity change in the UV-vis absorbance (ΔI), the curve, after [G]/[H] = 1, was parallel to the x-axis for R2, implying that the complex formed in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (R2C). Initially the intensity of the absorption changed sharply, then very slowly (Fig. 10). No such clear break in the titration curves was observed for the other receptors R3 and R4 with UA as there were weaker interactions compared to R2 with UA.
1 ratio (R2C). Initially the intensity of the absorption changed sharply, then very slowly (Fig. 10). No such clear break in the titration curves was observed for the other receptors R3 and R4 with UA as there were weaker interactions compared to R2 with UA.
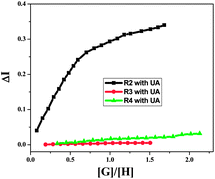 | ||
| Fig. 10 Stoichiometric UV-vis titration curves of R2, R3 and R4 with UA for their wavelength maxima at around 350 nm. | ||
The association constants (Ka) were calculated from linear regression analysis17 and are summarised in Table 2. Comparing the Ka values with the semi-empirical energy values calculated theoretically, it is clear that R2 and R3 bind to UA more strongly than the other two receptors R1 and R4. The self-complementary nature of R1 and the strong intra-molecular H-bond formation in R4 result in lower Ka values compared to R2 and R3. The limits of detection (LOD) estimated by the fluorescence method also indicated that among all four receptors, R2 can sense the lowest concentration of UA (Table 2).
| Complex | UV-vis method (Ka M−1) | Fluorescence method (Ka M−1) | Limit of Detection (M) | Semi-empirical energy (a.u.) |
|---|---|---|---|---|
| Complex R1C | 0.996 × 104 | 0.188 × 104 | 19.5121 × 10−8 | 0.1089 |
| Complex R2C | 6.114 × 104 | 1.767 × 104 | 9.0609 × 10−8 | 0.0083 |
| Complex R3B-C | 3.542 × 104 | 1.279 × 104 | 9.7194 × 10−8 | 0.0950 |
| Complex R4A-C | 0.158 × 104 | 0.319 × 104 | 9.9682 × 10−8 | 0.1369 |
Conclusions
This work was predominately focused upon the development of host–guest systems that are able to form an array of donor and acceptor atoms. Naphthyridine based water soluble receptors (R1–R4) are in fact, proficient in binding to UA in water. These receptors are significant for their use as fluorogenic sensors. The detection and estimation of UA in water using synthesised fluorosensors are very important issues from medicinal and clinical points of view.Acknowledgements
SD acknowledges DST (Govt. of India) for awarding him the DST-FAST Track young scientist project (SR/S1/CS-108/2009). DS is sincerely thankful to Indian School of Mines for the junior research fellowship.Notes and references
- (a) M. M. Conn and J. Rebek, Jr, Chem. Rev., 1997, 97, 1647 CrossRef CAS PubMed; (b) A. Kumar, S.-S. Sun and A. J. Lees, Coord. Chem. Rev., 2008, 252, 922 CrossRef CAS PubMed.
- (a) C. Schmuck and W. Wienand, Angew. Chem., Int. Ed., 2001, 40, 4363 CrossRef CAS; (b) L. Brunsveld, B. J. B. Folmer, E. W. Meijer and R. P. Sijbesma, Chem. Rev., 2001, 101, 4071 CrossRef CAS PubMed.
- (a) A. S. Tomcufcik, W. E. Meyer and J. W. Marsico, Eur. Pat. Appl., EP 446604, 1991; (b) US Appl. 494387, 1990; (c) Chem. Abstr., 1992, 116, 235628p; (d) Chem. Abstr., 1991, 114, 81808s; (e) C. Cotrel, C. Guyon, G. Roussel and G. Taurand, Eur. Pat. Appl., EP 208621 1987; (f) FR Appl. 85/10619, 1985; (g) Chem. Abstr., 1987, 107, 39780g.
- (a) P. S. Corbin, S. C. Zimmerman, P. A. Thiessen, N. A. Hawryluk and T. J. Murray, J. Am. Chem. Soc., 2001, 123, 10475 CrossRef CAS PubMed; (b) T. R. Kelly, G. J. Bridger and C. Zhao, J. Am. Chem. Soc., 1990, 112, 8024 CrossRef CAS.
- (a) C. J. Chandler, L. W. Deady, J. A. Reiss and V. Tzimos, J. Heterocycl. Chem., 1982, 19, 1017 CrossRef CAS; (b) C. J. Chandler, L. W. Deady and J. A. Reiss, J. Heterocycl. Chem., 1981, 18, 599 CrossRef CAS; (c) S. P. Goswami, R. Mukherjee, R. Mukherjee, S. Jana, A. C. Maity and A. K. Adak, Molecules, 2005, 10, 929 CrossRef CAS PubMed.
- T. R. Kelly and M. P. Maguire, J. Am. Chem. Soc., 1987, 109, 6549 CrossRef CAS.
- R. K. Bera and C. R. Raj, Chem. Commun., 2011, 47, 11498 RSC.
- (a) V. J. Pileggi, J. D. Giorgio and D. K. Wybenga, Clin. Chim. Acta, 1972, 37, 141 CrossRef CAS; (b) G. F. Domagk and H. H. Schlicke, Anal. Biochem., 1968, 22, 219 CrossRef CAS.
- (a) R. A. Poole, F. Kielar, S. L. Richardson, P. A. Stenson and D. Parker, Chem. Commun., 2006, 4084 RSC; (b) M. D. Cowart, I. Sucholeiki, R. R. Bukownik and C. S. Wilcox, J. Am. Chem. Soc., 1988, 110, 6204 CrossRef CAS PubMed; (c) E. Klein, M. P. Crump and A. P. Davis, Angew. Chem., Int. Ed., 2005, 44, 298 CrossRef CAS PubMed; (d) P. Rzepecki and T. Schrader, J. Am. Chem. Soc., 2005, 127, 3016 CrossRef CAS PubMed; (e) T. Grawe, G. Schafer and T. Schrader, Org. Lett., 2003, 5, 1641 CrossRef CAS PubMed.
- R. P. Sijbesma and E. W. Meijer, Chem. Commun., 2003, 5 RSC.
- J. R. Quinn and S. C. Zimmerman, Org. Lett., 2004, 6, 1649 CrossRef CAS PubMed.
- S. P. Goswami, S. Dey, J. F. Gallagher, A. J. Lough, S. G. Granda, L. T. Fernandez, I. Alkorta and J. Elguero, J. Mol. Struct., 2007, 846, 97 CrossRef CAS PubMed.
- (a) T. G. Larocque, S. Dastgir and G. G. Lavoie, Organometallics, 2013, 32, 4314 CrossRef CAS; (b) P. N. Basa, A. Bhowmick, L. M. Horn and A. G. Sykes, Org. Lett., 2012, 14, 2698 CrossRef CAS PubMed; (c) G. R. Owen, A. J. P. White and R. Vilar, Organometallics, 2009, 28, 5783 CrossRef CAS.
- S. P. Goswami, A. K. Mahapatra and R. Mukherjee, J. Chem. Soc., Perkin Trans. 1, 2001, 2717 RSC.
- (a) H. Tong, G. Zhou, L. X. Wang, X.-B. Jing, F.-S. Wang and J.-P. Zhang, Tetrahedron Lett., 2003, 44, 131 CrossRef CAS; (b) L. Fabbrizzi, M. Licchelli, G. Rabaioli and A. Taglietti, Coord. Chem. Rev., 2000, 205, 85 CrossRef CAS.
- C. Bazzicalupi, A. Bencini, A. Bianchi, L. Borsari, A. Danesi, C. Giorgi, C. Lodeiro, P. Mariani, F. Pina, S. Santarelli, A. Tamayo and B. Valtancoli, Dalton Trans., 2006, 4000 RSC.
- (a) P.-T. Chou, G.-R. We, C.-Y. Wei, C.-C. Cheng, C.-P. Chang and F.-T. Hung, J. Phys. Chem. B, 2000, 104, 7818 CrossRef CAS; (b) J.-H. Liao, C.-T. Chen, H.-C. Chou, C.-C. Cheng, P.-T. Chou, J.-M. Fang, Z. Slanina and T. J. Chow, Org. Lett., 2002, 4, 3107 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1H-NMR spectra of all receptors and their complexes, titration spectra, association constant calculation curves, LOD calculations and experimental procedures. See DOI: 10.1039/c3ra45197f |
| This journal is © The Royal Society of Chemistry 2014 |