DNA adducts and the total sum of at-risk DNA repair alleles in the nasal epithelium, a target tissue of tobacco smoking-associated carcinogenesis
Marco E. M.
Peluso
* and
Armelle
Munnia
Cancer Risk Factor Branch, Cancer Prevention and Research Institute, Florence, Italy. E-mail: m.peluso@ispo.toscana.it; Fax: +39 055 326 97 879
First published on 11th September 2013
Abstract
Tobacco smoking is a leading cause of death and disability. Interindividual variability in DNA adducts has been shown in subjects exposed to similar amounts of environmental carcinogens, including tobacco smoke. We have investigated the effects of smoking on DNA adducts in the nasal epithelium instead of peripheral blood in 42 volunteers, considering a panel of at-risk alleles by 32P-postlabeling and PCR. In detail, we have studied the association of DNA damage with tobacco smoke habits considering genes involved in DNA repair, including X-ray repair cross complementing protein 1 (XRCC1) Arg194Trp, XRCC protein 3 Thr241Met, and excision repair cross complementation group 2/xeroderma pigmentosum D (ERCC2/XPD) Lys751Gln polymorphisms. Then, we have analysed the combinations of the variant alleles of XRCC1 and ERCC2/XPD together with the wild type allele of XRCC3 by calculating the sum of at-risk alleles for lung cancer. DNA adducts were significantly higher in smokers with respect to nonsmokers (P < 0.001). An overall significant increase in adducts in heavy and long term smokers was found (P-values for trend <0.001, respectively). Multivariate regression analysis showed that adducts were linearly correlated to the number of cig per day (P < 0.001). Individuals with XRCC1 194Trp variant have a significant increment of DNA damage (P < 0.05), whereas XRCC3 241Met variant was inversely associated with adducts (P < 0.05). A null association was found with ERCC2/XPD. The levels of DNA adducts in participants with ≥4 at-risk alleles were two-fold increased with respect to those with one or fewer alleles (P < 0.01). A significant trend was observed (P-value for trend <0.05). Particularly, these results indicate a functional role for the XRCC1 and XRCC3 polymorphisms in genotoxic susceptibility related to the sensitivity to mutagens contained in tobacco smoke. Interindividual differences in at-risk alleles influence significantly the repair of damage in a tissue target of tobacco smoking-associated carcinogenesis. Our results support a functional role for the studied polymorphisms related to differences in cellular response to tobacco smoke mutagens. Phenotypes characterized by high levels of damage in susceptible individuals may be due to disorders in mechanisms designed to maintain cell homeostasis and DNA integrity.
Introduction
Worldwide, tobacco smoking is a leading cause of death and disability. Furthermore, smoking attributable deaths are projected to increase over the next two decades.1 Inhalation of tobacco smoke is the major source of exposure for smokers. Indeed, cigarette smoke contains 73 tumor-initiating carcinogens either in humans and/or animal models, such as benzo(a)pyrene [B(a)P] and other polycyclic aromatic hydrocarbons (PAHs).2 In particular, PAHs are capable of binding DNA covalently, forming aromatic DNA adducts and causing nucleotide alterations. Unless repaired, DNA damage leads to mutations in oncogenes and tumour suppressor genes.3 DNA adducts are a biomarker of air pollution and tobacco smoke exposure.3–9 Elevated levels of bulky DNA adducts have been also associated with increased cancer risk, including lung, larynx, bladder and gastrointestinal cancers.10–14 A significant relationship between adducts and increased cancer risk has been shown in smokers.12 In that study, smokers with high levels of DNA adducts have increased risk of lung cancer after a follow-up of several years.Most pathways involved in carcinogenesis of tobacco smoke are governed by strong interindividual variability with a resulting variation in the level of DNA adducts in subjects exposed to similar amounts of carcinogens.3 Also, our group has examined the association between polymorphisms and DNA adducts in peripheral blood.15–20 As an example, we suggested a novel function of X-ray repair cross complementing protein 1 (XRCC1) and XRCC protein 3, representing the base excision repair (BER) and the double strand break (DBS) repair pathways, in the repair of aromatic DNA adducts.19,20 Recently, polymorphisms in the 15q25 locus have been associated with DNA adducts as well as TP53 mutations in the adjacent histologically normal lung tissue and in the lung tumors of lung cancer patients.21 Therefore, we decided to investigate the effects of smoking together with a panel of at-risk alleles of DNA repair genes on the generation of DNA adducts in the nasal epithelium rather than in peripheral blood cells. The nose is a target tissue of tobacco smoking-associated carcinogenesis, where higher amounts of DNA adduct accumulation are expected.4 Additionally, increased amounts of volatile particles are generally found on the head of the inferior turbinate of the nose due to air flow turbulence behind the nasal valve and to limited mucociliary transport. The levels of DNA adducts and polymorphisms were determined in a group of healthy individuals living in North Italy using 32P-postlabeling and PCR techniques.13,22
In particular, we assessed the association of DNA adducts with polymorphisms in (1) XRCC1 Arg194Trp (rs1799782, C/T), a gene that stimulates the DNA kinase at damaged DNA termini and accelerates the overall repair reaction;23 (2) XRCC3 Thr241Met (rs861539, C/T), a gene that participates in DBS repair and maintaining chromosome stability;24 (3) excision repair cross complementation group 2/xeroderma pigmentosum D (ERCC2/XPD) Lys751Gln (rs13181, G/T), a gene involved in nucleotide excision repair (NER) that recognizes and repairs also aromatic DNA adducts.25 DNA polymorphisms were selected because of their well known association with lung cancer risk. The XRCC1-194Trp and the ERCC2/XPD-751Gln variants were linked to increased lung cancer risk; on the other hand, the XRCC3-241Met variant was associated with a modest protective effect.26–28
Materials and methods
Study population
The study was conducted in nasal epithelium of 42 healthy individuals, 33 males and 9 females living in North Italy who underwent an otolaryngological examination for respiratory nasal occlusion after traumatic deviation of the nasal septum. 24 volunteers reported to be current smokers, 8 ex-smokers and 10 never smokers. The mean age was 45.2 ± 16 years. Smokers have smoked 15.4 ± 9.5 cig per day for at least one year. Former smokers smoked 31.7 ± 21 cig per day. Ex-smokers reported to have quitted smoking cigarettes since at least seven years ago. A questionnaire aimed to collect standard demographic and lifestyle data, including age, gender, and smoking habit, was administered to the participants before sampling. Biological samples were collected at recruitment. The questionnaire was administered to the participants before biological sampling and after signed informed consent was obtained. The study was conducted according to the guidelines of the Declaration of Helsinki. Ethical clearance for this study was obtained by the relevant ethical committee.6Nasal sampling and DNA extraction
Nasal epithelium was obtained after contact anesthesia of the nasal cavities, 2% mepivacaine with epinephrine, by punch biopsy of the inferior or middle turbinate head. This technique has proven to be well tolerated and easily performed. Nasal epithelium was frozen shortly after removal, and kept at 80 °C until DNA extraction. DNA was extracted and purified using a method that requires digestion with ribonuclease A, ribonuclease T1 and proteinase K treatment and extraction with saturated phenol, phenol–chloroform–isoamyl alcohol (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), chloroform–isoamyl alcohol (24
1), chloroform–isoamyl alcohol (24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and ethanol precipitation.29 DNA concentration and purity were determined using a spectrophotometer. Coded DNA samples were subsequently stored at −80 °C until laboratory analyses.
1) and ethanol precipitation.29 DNA concentration and purity were determined using a spectrophotometer. Coded DNA samples were subsequently stored at −80 °C until laboratory analyses.
32P-DNA postlabeling assay
The levels of DNA adducts were analysed using the 32P-DNA postlabeling technique.22,30DNA polymorphism
The choice of XRCC1 Arg194Trp, XRCC3 Thr241Met, and ERCC2/XPD Lys751Gln polymorphisms was based on the known effects on enzyme activity,23–25 and their association with lung cancer risk.27–29 PCR followed by enzymatic digestion was used for genotyping.13Statistical analysis
DNA adduct levels were expressed as adducted nucleotides per 108 nt. Given the right-skewed distribution of the DNA adduct levels, the data were log transformed to stabilize the variance and normalize the distribution. The association between DNA adducts and smoking habit was evaluated by the analysis of covariance, including age (continuous) and gender as predictive variables. Subsequently, a multiple regression model adjusted for age and sex was used to evaluate the correlation between the levels of DNA adducts with the number of cig per day. The different genotypes were coded as wild-type (major allele homozygote), and variant genotypes (heterozygote and minor allele homozygote). The individuals were then grouped according to the number of alleles relevant for lung cancer risk. The effects of genotypes that were involved in DNA repair and the combinations of lung cancer at-risk alleles on the generation of DNA adducts were examined by the analysis of covariance, including age, gender and tobacco smoking as independent variables. Post hoc LSD and Dunnett tests were performed for multiple comparisons among variable levels. All statistical tests were two-sided and P less than 0.05 was considered to be statistically significant. The data were analyzed using SPSS 13.0 (IBM SPSS Statistics, New York, NY).Results
DNA adducts and tobacco smoking
ImageQuant analysis of the chromatograms of 32P-postlabeled digests of DNA from current and former smokers revealed a so-called diagonal radioactive zones (DRZ). Generally, the DRZ pattern is considered to be typical of DNA damage induced by complex mixtures of carcinogens such as cigarette smoke.22 No clear qualitative differences in DNA patterns between smokers and former-smokers could be observed, but only differences in intensities. Conversely, fainter DRZ or spot-patterns were found in the plates of never smokers.Table 1 reports the average levels of DNA adducts measured in the nasal epithelium of the study participants. After adjusting for age and sex, there was a statistically significant increase in the levels of DNA adducts in current smokers with respect to never smokers with intermediate amounts in ex-smokers. The association with smoking habit was strengthened by a statistically significant increment of DNA damage in heavy and long-term smokers. After correction for age and gender, the multivariate regression analysis showed that the generation of DNA damage was linearly correlated to the number of cig per day.
| Bulky DNA adducts | |||
|---|---|---|---|
| N a | Mean ± SE | P-valueb | |
| a Some figures do not add up to the total because of some missing values. b From the analysis for covariance model including terms for age, sex, and smoking habits. | |||
| Age | 42 | 2.8 ± 0.4 | 0.311 |
| Gender | |||
| Female | 9 | 2.6 ± 0.9 | Reference |
| Male | 33 | 4.3 ± 0.4 | 0.091 |
| Smoking status | |||
| Never smokers | 10 | 1.4 ± 0.2 | Reference |
| Ex-smokers | 8 | 2.5 ± 0.4 | 0.002 |
| Current smokers | 24 | 5.5 ± 0.5 | <0.001 |
| P-value for trend | <0.001 | ||
| Number of cig per day | |||
| <15 cig per day | 11 | 4.7 ± 0.8 | Reference |
| ≥15 cig per day | 13 | 6.2 ± 0.6 | 0.033 |
| P-value for trend | <0.001 | ||
| Smoking years | |||
| <15 smoking years | 6 | 3.9 ± 0.9 | Reference |
| ≥15 smoking years | 17 | 6.0 ± 0.6 | 0.077 |
| P-value for trend | <0.001 | ||
In detail, the levels of DNA adducts were higher in former smokers and current smokers with respect to nonsmokers. A statistically significant trend was present (P-value for trend <0.001 vs. nonsmokers). The levels of DNA damage were significantly increased in smokers of ≥15 cig per day as compared to the individuals who smoke less than 15 cig per day (P < 0.05). A statistically significant trend was found (P-value for trend <0.001 vs. nonsmokers). A borderline statistically significant increment was found in subjects who reported to smoke for 15 or more years with respect to those who smoked less than 15 years (P = 0.077). A statistically significant trend was found (P-value for trend <0.001 vs. nonsmokers). No significant effect of modification was observed for age and sex.
When we examined the association between the levels of DNA adducts and the self-reported number of cigarettes smoked per day, the multivariate regression analysis showed that the levels of DNA adducts were statistically significantly correlated with the amounts of cig per day, regression coefficient (β) = 0.575 ± 0.004 (SE), P < 0.001, after correction for age and gender. Former smokers were not included in the analysis. A scatter plot of these data is reported in Fig. 1.
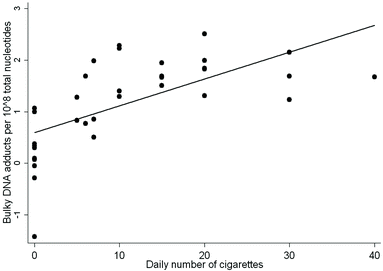 | ||
| Fig. 1 Relationship between individual DNA adduct levels and the number of cig per day, R2 = 0.723, P < 0.001. The former smokers were not included in the analysis. | ||
DNA adducts and polymorphisms
Table 2 shows the mean levels of DNA adducts measured in the nasal epithelium of the study participants and the sum of at-risk alleles. Table 2 reports that higher levels of DNA adducts were found in individuals with the homozygous XRCC1 194Trp genotype and the Arg/Trp genotype with respect to those with the wild type alleles (P < 0.05 and P < 0.05 vs. wild type, respectively, P-value for trend = 0.212 vs. wild type).| Bulky DNA adducts | |||
|---|---|---|---|
| Polymorphisms | N a | Mean ± SE | P-valueb |
| a Some figures do not add up to the total because of some missing values. b From the analysis for covariance model including terms for age, sex, and smoking habits. | |||
| XRCC1 Arg194Trp (rs1799782) | |||
| Arg/Arg | 21 | 3.5 ± 0.6 | Reference |
| Arg/Trp | 14 | 4.6 ± 0.9 | 0.011 |
| Trp/Trp | 6 | 4.2 ± 0.8 | 0.033 |
| Trp/Trp + Arg/Trp | 20 | 4.4 ± 0.6 | 0.958 |
| P-value for trend | 0.212 | ||
| XRCC3 Thr241Met (rs861539) | |||
| Thr/Thr | 12 | 5.3 ± 1.0 | Reference |
| Thr/Met | 19 | 3.2 ± 0.5 | 0.003 |
| Met/Met | 9 | 3.7 ± 0.6 | 0.145 |
| Met/Met + Thr/Met | 28 | 3.4 ± 0.4 | 0.699 |
| P-value for trend | 0.482 | ||
| ERCC2/XPD Lys751Gln (rs13181) | |||
| Lys/Lys | 13 | 3.6 ± 0.6 | Reference |
| Lys/Gln | 20 | 4.1 ± 0.7 | 0.447 |
| Gln/Gln | 8 | 4.3 ± 0.9 | 0.167 |
| Gln/Gln + Lys/Gln | 28 | 4.2 ± 0.5 | 0.730 |
| P-value for trend | 0.507 | ||
| Sum of at-risk alleles | |||
| ≤1 at-risk allele | 19 | 3.1 ± 0.4 | Reference |
| 3 at-risk alleles | 14 | 4.1 ± 0.8 | 0.340 |
| ≥4 at-risk alleles | 8 | 5.9 ± 1.1 | 0.003 |
| P-value for trend | 0.045 | ||
In analyzing the XRCC3 Thr241Met polymorphism, subjects carrying the Met/Met genotype and the Thr/Met genotype had levels of DNA damage decreased with respect to those with the wild type alleles (Table 2). The difference reached statistical significance in the subjects with the Thr/Met genotype but not in those with the Met/Met genotype, mainly due to low statistical power for detecting a moderate effect (P < 0.01 and P = 0.145 vs. wild type, respectively, P-value for trend = 0.482 vs. wild type).
When the ERCC2/XPD Lys751Gln polymorphism was considered, individuals carrying the Gln/Gln genotype and the Lys/Gln genotype had levels of DNA damage slightly higher than those with the wild type genotype (P = 0.167 and P = 0.447, respectively, P-value for trend = 0.507 vs. wild type).
DNA adducts and sum of at-risk alleles
Next, we analysed the association of combinations of the XRCC1-194Trp and ERCC2/XPD-751Gln allelic variants with the XRCC3-241Thr wild type allele on the levels of DNA adducts to explore the effects of the sum of at-risk alleles by the analysis of covariance, including age, sex and smoking habit as predictive variables. Table 2 shows that the levels of DNA adduct were about two-fold increased in subjects with unfavorable genetic traits. In detail, the individuals with 3 and ≥4 at-risk alleles had levels of DNA damage higher than the reference category. After correction for confounding factors, the levels of DNA adducts in subjects with ≥4 at-risk alleles were significantly different from those without these genetic characteristics (P < 0.01 vs. reference level). A trend was also found with the highest levels in the subjects carrying ≥4 lung cancer at-risk alleles (P-value for trend <0.05 vs. baseline).Discussion
Tobacco smoke is a complex mixture of more than 5000 identified chemical compounds.2 In the present study, the levels of bulky DNA damage were significantly higher in the upper respiratory tract of current smokers with respect to former smokers and nonsmokers. This is in line with previous reports from our group that showed elevated levels of DNA adducts in the laryngeal, nasal and bronchial mucosa of current smokers.4,6,31 Also, a number of studies have explored the influence of tobacco smoking on the generation of DNA adducts in humans to study the mechanisms of smoking-related diseases. For example, the levels of DNA adducts were consistently higher in lung tissue of smokers as compared with nonsmokers.3 The detection of DNA adducts chromatographically similar to those derived from hydroquinone and benzenetriol metabolites of benzene was also reported in smokers.3 Conversely, a significant correlation with smoking was not apparent in other studies.3,32 Indeed, these studies suffered from the fact that peripheral blood was used as a source of DNA. In the current study, the levels of aromatic DNA adducts in the nasal epithelium of former smokers show intermediate values with respect to current and never smokers. Previous reports have reported that smoking-induced DNA adducts persist for a long time and could still be detectable after quitting.33,34Our findings show that there was an overall significant increase in the levels of bulky DNA adducts in heavier and long term smokers. Furthermore, the relationship of DNA adducts with tobacco smoking tended to reach some kind of saturation point at higher number of self-reported cig per day. Commonly, reactive metabolites tend to be intercepted and detoxified by cytoplasmic nucleophiles, such as reduced glutathione, and by conjugating and detoxifying enzyme activities, such as GSH S-transferases. However, cytoplasmic nucleophiles, detoxifying and DNA repair enzymes may reach saturation at increasing carcinogen concentrations. We previously observed a flattening of the relationship between carcinogen exposures and DNA damage.35 In that study, we demonstrated, by a meta-analytic approach, that the association between DNA adducts and external concentrations of B(a)P was approximately linear at low doses and sublinear at high doses.
The levels of DNA adducts primarily reflect exposures to carcinogens but their levels are also influenced by genetic susceptibilities. Our next results show that subjects carrying the XRCC1 194Trp have a significant increment of genetic damage, whereas XRCC3 241Met variant was inversely associated with adducts. A null relationship was found with the ERCC2/XPD 751Gln variant. Particularly, these results indicate a functional role for the XRCC1 and XRCC3 polymorphisms in genotoxic susceptibility related to the sensitivity to mutagens contained in tobacco smoke. The main pathway for removal of DNA adducts is NER, but BER and DSB repair mechanisms may participate in repair of bulky DNA adducts.10,13,20,36 Our findings are in line with previous studies, which showed that individuals with variant alleles of XRCC1 Arg399Gln polymorphism have elevated levels of DNA damage.13,20 In keeping with our findings, Lu et al. have found a null association of B(a)P–DNA adducts with ERCC2/XPD Lys751Gln polymorphism.37 Mateuca et al. have previously analyzed the influence of occupational exposure, including to styrene, ionizing radiation, cobalt/hard metal, welding fumes and inorganic arsenite compounds, considering polymorphisms in NER and DSB repair genes.38,39 In these studies, workers carrying the XRCC3 241Met allele have been shown to have increased levels of micronuclei frequencies compared to their referent counterparts. Interestingly, carriers of low-activity alleles in NER genes seemed to benefit from a four week dietary supplementation of antioxidants.40
Polymorphisms in NER genes have been associated with reduced lung cancer risk and a modulation of repair capacity.41,42 A lung cancer case–control study has suggested a protective effect conferred by XRCC3 241Met allele carriers against the generation of DNA adducts.13 However, nonsmokers with XRCC3 241Met alleles and N-acetyltransferase type 2 slow genotype showed an opposite trend on the generation of blood DNA adducts.10 Further studies are needed to confirm the association between DNA damage and XRCC3 polymorphism in target tissues. Interaction between DNA repair polymorphisms may also contribute to the genetic susceptibility of cancer. Indeed, Vogel et al. have shown that XPD Lys751Gln polymorphism may positively interact with xeroderma pigmentosum group A (XPA) A23G and xeroderma pigmentosum complementation group C (XPC) Lys939Gln polymorphisms.43
Our study indicates that the combinations of multiple lung cancer at-risk alleles predispose to a significant increment of aromatic DNA adducts in a target tissue of smoking-associated carcinogenesis. The individuals carrying ≥4 at-risk alleles in genes involved in DNA repair showed increased levels of DNA damage as compared with those not possessing these genetic characteristics. Thus, our findings show that interindividual differences in at-risk alleles may influence significantly the repair of DNA damage in the nasal epithelium, a tissue target of tobacco smoking-associated carcinogenesis. Ketelslegers et al. have recently studied the association between multiple genetic polymorphisms in genes involved in carcinogen metabolism, DNA repair, and oxidant metabolism and the levels of DNA adducts in smokers.7 In that study, the total number of alleles that were considered as high-risk alleles were associated with high levels of DNA adducts, indicating that the interindividual variation in DNA damage upon exposure to tobacco smoke may in part be explained by the genetic polymorphisms in genes involved in processes that affect the generation of DNA adducts. Therefore, one could also argue that genetic variations were in part modulated by tobacco addiction of the subjects, since the levels of DNA adducts were linearly associated with the self-reported number of cig per day. The augmented levels of DNA adducts in subjects carrying multiple at-risk alleles may be due to interconnected alterations in critical DNA repair pathways, which are essential for maintaining genome integrity. In addition, the association between the levels of DNA adducts and the total number of at risk alleles was stronger than that observed with each single DNA repair polymorphism suggesting a functional link. A similar picture emerged from previous studies of DNA damage and at-risk alleles in subjects exposed to environmental risk factors, including tobacco smoke.18 In this review, elevated levels of DNA adducts were found in occupationally-exposed and smoking subjects carrying multiple at-risk alleles with respect to the reference categories. Our results support a functional role for the studied polymorphisms related to differences in cellular responses to tobacco smoke mutagens. Phenotypes characterized by high levels of damage in susceptible individuals may be due to disorders in mechanisms designed to maintain cell homeostasis and DNA integrity. In keeping with our hypothesis, the combination of minor alleles of excision repair cross complementation group 1 with ERCC2/XPD has been recently associated with a reduced DNA repair efficiency in the generation of bulky DNA adducts induced from B(a)P in vitro experiments.37
A strength of our study is that the levels of DNA damage were analyzed in the epithelium of the upper respiratory tract, rather than in blood cells. Indeed, the enzymatic patterns of Phase I enzymes can be qualitatively different in the respiratory tract and peripheral blood.35 Although statistical power was sufficient in the overall analysis, a clear limitation of the present study is the restricted sample size. DNA damage was analyzed by the 32P-postlabeling assay known to be sensitive to the detection of a wide range of aromatic adducts in addition to those induced by PAHs, such as nitro-aromatic compounds and heterocyclic amines.44 A good repeatability of bulky DNA adduct measurements has been also reported for this assay.45 In particular, Reddy and Randerath have greatly enhanced the sensitivity of the 32P-postlabeling assay by introducing the postincubation of DNA digest with Penicillium citrinum nuclease P1 before 32P-labeling.46 This modification enhanced the sensitivity of the 32P-postlabeling technique to 1 adduct in approximately 1010 nt for a 10 μg DNA samples. The new procedure was found to be applicable to the detection of aromatic or bulky non-aromatic DNA adducts formed with structurally different carcinogens, including B(a)P, 7,12-dimethyl-ben(a)anthracene, dibenzo(c,g)carbazole, 4-aminobiphenyl, safrole, and mitomycin C.
However, 32P-postlabeling is an assay that is unable to determine the structure of the adducts under study; higher specificity may be obtained if the technique is combined with appropriate internal standards,44 or coupled with MALDI-TOF mass spectrometry techniques, such as in the case of malondialdehyde–deoxyguanosine.9 Knowledge of the nature of DNA adducts under study gives relevant data regarding the mutational effects that may result from particular environmental exposures. As part of this process, 32P-postlabeling interlaboratory trials using DNA standards modified by B(a)P, 4-aminobiphenyl, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, and N-methyl-N-nitrosourea have been carried out from twenty-five laboratories in Europe and USA to lead to a standardized 32P-postlabeling assay to be used in biomonitoring studies.30 A method using HPLC combined with electrospray tandem mass spectrometry (ES-MS/MS) was recently developed to detect and quantify trans-7,8-dihydroxy-anti-9.10-epoxy-7,8,9,10-tetrahydrobenzo(a)pyrene (BPDE), the major DNA adducts resulting from exposure to the carcinogenic B(a)P metabolite.47 The HPLC-ES-MS/MS method was successfully applied to analyze the presence of dG-BPDE in human lung DNA samples at levels comparable to those determined by 32P-postlabeling and immunoassay techniques. New radioisotope-postlabeling techniques incorporating [(14)C] isotope into specific DNA adducts after formation are also being developed to study environmental carcinogen exposures.48
Our study suggests that the levels of aromatic DNA adducts measured in the nasal epithelium are a good marker of exposure to cigarette smoking and uphold its use in gene–environment interaction studies. Similarly, Godschalk et al. have shown that the use of monocytes plus lymphocytes (MNC) as the source of DNA for DNA adduct measurements represents a good indicator for exposure to the carcinogens contained in tobacco smoke as compared to granulocytes and to the cells obtained by broncho-alveolar lavages (BAL).49 In this study, the levels of aromatic DNA adducts were significantly increased in MNC as compared with granulocytes after in vitro incubation with B(a)P. Furthermore, in MNC a significant linear relationship was observed between exposure to cigarette tar and the levels of aromatic DNA adducts, whereas no correlation was found in BAL cells.
Conclusion
Our results corroborate data of epidemiological studies showing that the cancer risk and mortality of smokers increase with the intensity and duration of smoking and decline slowly after cessation of smoking. Additionally, when we asked whether unfavorable genetic traits may influence the levels of DNA damage, we found that the total number of lung cancer at-risk alleles promotes significantly the generation of DNA adducts despite null or borderline effects in single polymorphism associations. Interindividual differences in at-risk DNA repair alleles influenced significantly the repair of DNA damage in the nasal epithelium, a tissue target of tobacco smoking-associated carcinogenesis, indicating that genetic variations were in part modulated from smoking habits of the subjects. Phenotypes characterized by high levels of DNA damage in susceptible individuals may be due to disorders in mechanisms designed to maintain cell homeostasis and DNA integrity. Multiple unfavorable low penetrant alleles in susceptible individuals may contribute to conferring increased cancer risk.Acknowledgements
This work was in part supported by the Tumor Institute of Tuscany and by the “Associazione Italiana per la Ricerca sul Cancro”.References
- C. D. Mathers and D. Loncar, Projections of global mortality and burden of disease from 2002 to 2030, PLoS Med., 2006, 3, e442 Search PubMed.
- International Agency for Research on Cancer. A review of human carcinogens: personal habits and indoor combustions, Monogr. Eval. Carcinog. Risks Hum, 2012, 100E Search PubMed.
- D. H. Phillips and S. Venitt, DNA and protein adducts in human tissues resulting from exposure to tobacco smoke, Int. J. Cancer, 2012, 131, 2733–2753 CrossRef CAS PubMed.
- M. Peluso, E. Amasio, S. Bonassi, A. Munnia, F. Altrupa and S. Parodi, Detection of DNA adducts in human nasal mucosa tissue by 32P-postlabeling analysis, Carcinogenesis, 1997, 18, 339–344 CrossRef CAS PubMed.
- F. Merlo, C. Bolognesi, M. Peluso, F. Valerio, A. Abbondandolo and R. Puntoni, Airborne levels of polycyclic aromatic hydrocarbons: 32P-postlabeling DNA adducts and micronuclei in white blood cells from traffic police workers and urban residents, J. Environ. Pathol., Toxicol. Oncol., 1997, 16, 157–162 CAS.
- M. Peluso, M. Neri, G. Margarino, C. Mereu, A. Munnia and M. Ceppi, et al. Comparison of DNA adduct levels in nasal mucosa, lymphocytes and bronchial mucosa of cigarette smokers and interaction with metabolic gene polymorphisms, Carcinogenesis, 2004, 25, 2459–2465 CrossRef CAS PubMed.
- H. B. Ketelslegers, R. W. Gottschalk, A. M. Knaapen, F. J. van Schooten, R.-F. Vlietinckm and J. C. Kleinjans, et al. Interindividual variations in DNA adduct levels assessed by analysis of multiple genetic polymorphisms in smokers, Cancer Epidemiol. Biomarkers Prev., 2006, 15, 624–629 CAS.
- M. Peluso, P. Srivatanakul, A. Munnia, A. Jedpiyawongse, M. Ceppi and S. Sangrajrang, et al. DNA adduct formation among workers in a Thai industrial estate and nearby residents, Sci. Total Environ., 2008, 389, 283–288 CrossRef CAS PubMed.
- M. Peluso, A. Munnia, M. Ceppi, R. W. Giese, R. W. L. Godschalk and D. Catelan, et al. Malondialdehyde-deoxyguanosine and aromatic DNA adducts among schoolchildren living near the Sarroch industrial estate, Mutagenesis, 2013, 28, 315–321 CrossRef CAS PubMed.
- G. Matullo, S. Guarrera, S. Carturan, M. Peluso, C. Malaveille and L. Davico, et al. DNA repair gene polymorphisms, bulky DNA adducts in white blood cells and bladder cancer in a case-control study, Int. J. Cancer, 2001, 92, 562–567 CrossRef CAS PubMed.
- L. Airoldi, P. Vineis, A. Colombi, L. Olgiati, C. Dell'Osta and R. Fanelli, et al. 4-Aminobiphenyl-hemoglobin adducts and risk of smoking-related disease in never smokers and former smokers in the European Prospective Investigation into Cancer and Nutrition prospective study, Cancer Epidemiol. Biomarkers Prev., 2005, 14, 2118–2124 CAS.
- F. Veglia, S. Loft, G. Matullo, M. Peluso, A. Munnia and F. Perera, et al. DNA adducts and cancer risk in prospective studies: a pooled analysis and a meta-analysis, Carcinogenesis, 2008, 29, 932–936 CrossRef CAS PubMed.
- M. Peluso, A. Munnia, S. Piro, A. Armillis, M. Ceppi and G. Matullo, et al. Smoking, DNA adducts and number of risk DNA repair alleles in lung cancer cases, in subjects with benign lung diseases and in controls, J. Nucleic Acids, 2010, 386798 Search PubMed.
- A. Agudo, M. Peluso, A. Munnia, L. Luján-Barroso, M. J. Sánchez and E. Molina-Montes, et al. Aromatic DNA adducts and risk of gastrointestinal cancers: a case-cohort study within the EPIC-Spain, Cancer Epidemiol. Biomarkers Prev., 2012, 21, 685–692 CrossRef CAS PubMed.
- A. Agudo, M. Peluso, N. Sala, G. Capellá, A. Munnia and S. Piro, et al. Aromatic DNA adducts and polymorphisms in metabolic genes in healthy adults: findings from the EPIC-Spain cohort, Carcinogenesis, 2009, 30, 968–976 CrossRef CAS PubMed.
- D. Palli, G. Masala, M. Peluso, L. Gaspari, V. Krogh and A. Munnia, et al. The effects of diet on DNA bulky adduct levels are strongly modified by GSTM1 genotype: a study on 634 subjects, Carcinogenesis, 2004, 25, 577–584 CrossRef CAS PubMed.
- M. Peluso, P. Srivatanakul, A. Jedpiyawongse, S. Sangrajrang, A. Munnia and S. Piro, et al. Aromatic DNA adducts and number of lung cancer risk alleles in Map-Ta-Phut Industrial Estate workers and nearby residents, Mutagenesis, 2013, 28, 57–63 CrossRef CAS PubMed.
- M. Peluso, A. Munnia, P. Srivatanakul, A. Jedpiyawongse, S. Sangrajrang and M. Ceppi, et al. DNA adducts and number of lung cancer risk alleles in environmentally-exposed and smoking subject, Environ. Mol. Mutagen., 2013, 54, 375–383 CrossRef CAS PubMed.
- G. Matullo, D. Palli, M. Peluso, S. Guarrera, S. Carturan and E. Celentano, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and 32P-DNA adducts in a sample of healthy subjects, Carcinogenesis, 2001, 22, 1437–1445 CrossRef CAS PubMed.
- D. Palli, A. Russo, G. Masala, C. Saieva, S. Guarrera and S. Caturan, et al. DNA adducts levels and DNA repair polymorphisms in traffic-exposed workers and a general population, Int. J. Cancer, 2001, 94, 121–127 CrossRef CAS PubMed.
- X. Tekpli, N. E. Landvik, V. Skaug, A. Gulsvik, A. Haugen and S. Zienolddiny, Functional effect of polymorphisms in 15q25 locus on CHRNA5 mRNA, bulky DNA adducts and TP53 mutations, Int. J. Cancer, 2013, 132, 1811–1820 CrossRef CAS PubMed.
- A. Munnia, F. Saletta, A. Allione, S. Piro, M. Confortini and G. Matullo, et al. 32P-Post-labelling method improvements for aromatic compound-related molecular epidemiology studies, Mutagenesis, 2007, 22, 381–385 CrossRef CAS PubMed.
- C. J. Whitehouse, R. M. Taylor, A. Thistlethwaite, H. Zhang and F. Karimi-Busheri, et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair, Cell, 2001, 104, 107–117 CrossRef CAS.
- N. Liu, J. E. Lamerdin, R. S. Tebbs, D. Schild, J. D. Tucker and M. R. Shen, et al. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages, Mol. Cell, 1998, 1, 783–793 CrossRef CAS.
- E. Braithwaite, X. Wu and Z. Wang, Repair of DNA lesion mechanisms and relative repair efficiencies, Mutat. Res., 1999, 424, 207–219 CrossRef CAS.
- R. J. Hung, D. C. Christiani, A. Risch, O. Popanda, A. Haugen and S. Zienolddiny, et al. International lung cancer consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways., Cancer Epidemiol. Biomarkers Prev., 2008, 17, 3081–3089 CrossRef CAS PubMed.
- Z. Feng, Y. Ni, W. Dong, H. Shen and J. Du, Association of ERCC2/XPD polymorphisms and interaction with tobacco smoking in lung cancer susceptibility: a systemic review and meta-analysis, Mol. Biol. Rep., 2012, 39, 57–69 CrossRef CAS PubMed.
- S. Zienolddiny, D. Campa, H. Lind, D. Ryberg, V. Skaug and L. Stangeland, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer, Carcinogenesis, 2006, 27, 560–567 CrossRef CAS PubMed.
- A. Munnia, S. Bonassi, A. Verna, R. Quaglia, D. Pilucco and M. Ceppi, et al. Bronchial malondialdehyde DNA adducts, tobacco smoking, and lung cancer, Free Radicals Biol. Med., 2006, 41, 1499–14505 CrossRef CAS PubMed.
- D. H. Phillips and M. Castegnaro, Standardization and validation of DNA adduct postlabelling methods: report of interlaboratory trials and production of recommended protocols, Mutagenesis, 1999, 14, 301–315 CrossRef CAS.
- A. Munnia, M. E. Amasio and M. Peluso, Exocyclic malondialdehyde and aromatic DNA adducts in larynx tissues, Free Radicals Biol. Med., 2004, 37, 850–858 CrossRef CAS PubMed.
- F. Ricceri, R. Godschalk, M. Peluso, D. H. Phillips, A. Agudo and P. Georgiadis, et al. Bulky DNA adducts in white blood cells: a pooled analysis of 3600 subjects, Cancer Epidemiol. Biomarkers Prev., 2010, 19, 3174–3181 CrossRef CAS PubMed.
- J. K. Wiencke, S. W. Thurston, K. T. Kelsey, A. Varkonyi, J. C. Wain and E. J. Mark, et al. Early age at smoking initiation and tobacco carcinogen DNA damage in the lung, J. Natl. Cancer Inst., 1999, 91, 614–619 CrossRef CAS PubMed.
- L. Chen, M. Wang, P. W. Villalta, X. Luo, R. Feuer and J. Jensen, et al. Quantitation of an acetaldehyde adducts in human leukocyte DNA and the effect of smoking cessation, Chem. Res. Toxicol., 2007, 20, 108–113 CrossRef CAS PubMed.
- M. Peluso, M. Ceppi, A. Munnia, R. Puntoni and S. Parodi, Meta analysis of thirteen 32P-DNA postlabeling studies of occupational cohorts exposed to air pollution, Am. J. Epidemiol., 2001, 153, 546–558 CrossRef CAS PubMed.
- H. Yu, H. Zhao, L. E. Wang, Z. Liu, D. Li and Q. Wei, Correlation between base-excision repair gene polymorphisms and levels of in-vitro BPDE-induced DNA adducts in cultured peripheral blood lymphocytes, PLoS One, 2012, 7, e40131 CAS.
- X. Lu, Y. Liu, T. Yu, S. Xiao, X. Bao and L. Pan, et al. ERCC1 and ERCC2 Haplotype Modulates Induced BPDE-DNA Adducts in Primary Cultured Lymphocytes, PLoS One, 2013, 8, e60006 CAS.
- R. Mateuca, P. V. Aka, M. De Boeck, R. Hauspie, M. Kirsch-Volders and D. Lison, Influence of hOGG1, XRCC1 and XRCC3 genotypes on biomarkers of gentoxicity in workers exposed to cobalt or hard metal dusts, Toxicol. Lett., 2005, 156, 277–288 CrossRef CAS PubMed.
- R. A. Mateuca, M. Roelants, G. Iamarcovai, P. V. Aka, L. Godderis and A. Tremp, et al. hoGG1(326), XRCC1(399) and XRCC3(241) polymorphisms influence micronuclei frequencies in human lymphocytes in vivo, Mutagenesis, 2008, 23, 35–41 CrossRef CAS PubMed.
- S. A. Langie, L. C. Wilms, S. Hamalainen, J. C. Kleinjas, R. W. Godschalk and F. J. van Schooten, Modulation of nucleotide excision repair in human lymphocytes by genetic and dietary factors, Br. J. Nutr., 2010, 103, 490–501 CrossRef CAS PubMed.
- X. Wu, H. Zhao, Q. Wei, C. L. Amos, K. Zhang and Z. Guo, et al. XPA polymorphism associated with reduced lung cancer risk and a modulating effect on nucleotide excision repair capacity, Carcinogenesis, 2003, 24, 505–509 CrossRef CAS PubMed.
- Y. Zhu, H. Yang, Q. Chen, J. Lin, H. B. Grossman and C. P. Dinney, Modulation of DNA damage/DNA repair capacity by XPC polymorphisms, DNA Repair, 2008, 7, 141–148 CrossRef CAS PubMed.
- U. Vogel, K. Overvad, H. Wallin, A. Tionneland, B. A. Nexo and O. Raaschou-Nielsen, Combinations of polymorphisms in XPD, XPC and XPA in relation to risk of lung cancer, Cancer Lett., 2005, 222, 67–74 CrossRef CAS PubMed.
- M. Peluso, M. Castegnaro, C. Malaveille, G. Talaska, P. Vineis and F. Kadlubar, et al. 32P-postlabelling analysis of DNA adducted with urinary mutagens from smokers of black tobacco, Carcinogenesis, 1990, 11, 1307–1311 CrossRef CAS PubMed.
- M. Peluso, P. Hainaut, L. Airoldi, H. Autrup, A. Dunning and S. Garte, et al. Methodology of laboratory measurements in prospective studies on gene-environment interactions: the experience of GENAIR, Mutat. Res., 2005, 574, 92–104 CrossRef CAS PubMed.
- M. V. Reddy and K. Randerath, Nuclease P1-mediated enhancement of sensitivity of 32P-postlabeling test for structurally diverse DNA adducts, Carcinogenesis, 1986, 7, 1543–1551 CrossRef CAS PubMed.
- F. A. Beland, M. I. Churchwell, L. S. Von Tungeln, S. Chen, P. P. Fu and S. J. Culp, et al. High-performance liquid chromatography electrospray ionization tandem mass spectrometry for the detection and quantification of benzo(a)pyrene-DNA adducts, Chem. Res. Toxicol., 2005, 18, 1306–1315 CrossRef CAS PubMed.
- P. B. Farmer, K. Brown, E. Tompkins, V. L. Emms, D. J. Jones and R. Singh, et al. DNA adducts: mass spectrometry methods and future prospects, Toxicol. Appl. Pharmacol., 2005, 207, 293–301 CrossRef CAS PubMed.
- R. W. L. Godschalk, L. M. Maas, N. Van Zandwijk, L. J. van ‘t Veer, A. Breedijk and P. J. A. Born, et al. Differences in aromatic-DNA adduct levels between alveolar macrophages and subpopulations of white blood cells from smokers, Carcinogenesis, 1998, 19, 819–825 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |
