Rapid discovery and global characterization of multiple constituents from Kai-Xin-San using an integrated MSE data acquisition mode strategy based on ultra-performance liquid chromatography coupled to electrospray ionization/quadrupole-time-of-flight mass spectrometry†
Hui
Sun
,
Chang
Liu
,
Ai-hua
Zhang
,
Ying
Han
,
Guang-li
Yan
,
Ping
Wang
and
Xi-jun
Wang
*
National TCM Key Laboratory of Serum Pharmacochemistry, Laboratory of Metabolomics and Chinmedomics, Department of Pharmaceutical Analysis, Heilongjiang University of Chinese Medicine, Heping Road 24, Harbin 150040, China. E-mail: aihuatcm@163.com; xijunwangls@126.com; Fax: +86-451-82110818; Tel: +86-451-82110818
First published on 6th October 2014
Abstract
Kai-Xin-San (KXS) is a traditional Chinese medicine (TCM) formula consisting of four herbs, Ginseng Radix, Polygalae Radix, Poria and Acori Tatarinowii Rhizoma. KXS has been used to treat Alzheimer's disease and Parkinson's disease. In this study, a UPLC/QTOF MS system with automated MSE (E represents collision energy) data analysis software (MetaboLynx™) was used to analyze and identify the chemical components of KXS. Mass spectrometry was performed in a full-spectrum (m/z 100–2000) mode using an MSE acquisition of both molecular and fragment ion data at low (10–30 eV) and ramped (30–50 eV) collision energies. With the use of this approach, a total of 107 compounds were identified from KXS; 32 compounds were from Ginseng Radix; 19 compounds were from Poria; 33 compounds were from Polygalae Radix; and 21 compounds were from Acori Tatarinowii Rhizoma. The developed method is fast, accurate and reliable due to its high resolution and high efficiency, a result of an integrated MSE data acquisition mode strategy based on UPLC/QTOF MS that is a powerful tool for global detection and identification of complex components in herbal prescriptions. The results provided helpful chemical information for further pharmacology and active mechanism research on KXS and other TCMs.
1. Introduction
Traditional Chinese medicine (TCM) preparations have been widely used and have become increasingly popular worldwide. The comprehensive identification of the chemical components in TCMs is important for revealing their therapeutic effects.1 However, the methods for detection and structural characterization of each chemical constituent contained in herbal prescriptions are often unrealistic and impractical. The newly developed, sensitive, reliable analytical methods and technology have been proven to be efficient tools for the rapid on-line analysis of the known compounds and elucidation of unknown compounds in complex TCMs. Recently, high-resolution, ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC/QTOF MS) has improved the analytical ability for the study of complex herbal medicines.2–5 The well-developed UPLC/QTOF MS system renders it possible to launch a more comprehensive and thorough chemical profiling by utilizing versatile ionization techniques and/or different ion modes. In particular, it provides accurate precursor and/or product ion information with a mass error of less than the permitted ppm, which substantially enhances the characterization reliability of constituents.6 Substantial progress has been made in using the LC/MS-based fingerprint technique in conjunction with automated MSE (E represents collision energy) data analysis software (MetaboLynx™). MSE technique can be used to predict the structures of unknown compounds in TCM based on the accurate molecular weight and the relationship between precursor and product ions.7–9Kai-Xin-San (KXS), first recorded in the Chinese ancient medical prescription book Qian-Jin-Yao-Fang around 1300 years ago, is a TCM formula consisting of four herbs, Ginseng Radix, Polygalae Radix, Poria, and Acori Tatarinowii Rhizoma (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2).10 It has been used for thousands of years in China and other Asian countries to treat Alzheimer's disease, Parkinson's disease, etc.11–13 Pharmacological studies showed that KXS has anti-oxidative, anti-depressive, and learning and memory improvement activities.14,15 The chemical analysis of KXS has rarely been reported. Lv determined polygalaxanthone III, ginsenoside Rb1, ginsenoside Rd, ginsenoside Re, and ginsenoside Rg1 in the plasma of rat and beagle dog after oral administration of KXS by ultra-fast liquid chromatography with tandem mass spectrometry.16 However, studies on the chemical components in KXS have been mainly based on identification of chemical constituents in the individual herbs; further investigation has been hindered due to the limited knowledge of their chemical components. An integrated MSE data acquisition mode strategy based on UPLC/QTOF MS is powerful and reliable for the global detection and identification of complex components in herbal prescriptions. Therefore, in this study, UPLC/QTOF MS method with automated MSE data analysis was firstly developed to systematically characterize the chemical constituents of KXS.
2).10 It has been used for thousands of years in China and other Asian countries to treat Alzheimer's disease, Parkinson's disease, etc.11–13 Pharmacological studies showed that KXS has anti-oxidative, anti-depressive, and learning and memory improvement activities.14,15 The chemical analysis of KXS has rarely been reported. Lv determined polygalaxanthone III, ginsenoside Rb1, ginsenoside Rd, ginsenoside Re, and ginsenoside Rg1 in the plasma of rat and beagle dog after oral administration of KXS by ultra-fast liquid chromatography with tandem mass spectrometry.16 However, studies on the chemical components in KXS have been mainly based on identification of chemical constituents in the individual herbs; further investigation has been hindered due to the limited knowledge of their chemical components. An integrated MSE data acquisition mode strategy based on UPLC/QTOF MS is powerful and reliable for the global detection and identification of complex components in herbal prescriptions. Therefore, in this study, UPLC/QTOF MS method with automated MSE data analysis was firstly developed to systematically characterize the chemical constituents of KXS.
2. Experimental
2.1 Materials
Acetonitrile and methanol (HPLC-MS grade) were purchased from Merck (Darmstadt, Germany). Deionized water (18.2 MΩ) was further purified using a Milli-Q system (Millipore, Billerica, USA). Leucine-enkephalin was obtained from Sigma-Aldrich. Poria, Ginseng Radix, Polygalae Radix, and Acori Tatarinowii Rhizoma were purchased from Harbin Tongrentang Drug Store (Harbin, China) and authenticated by Prof. Xijun Wang, Department of Pharmacognosy of Heilongjiang University of Chinese Medicine. Voucher specimens were deposited at the authors' laboratory.2.2 Preparation of KXS samples
KXS was prepared using the following procedure: Ginseng Radix, Poria, Polygalae Radix, and Acori Tatarinowii Rhizoma in the proportion 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 were ground into crude powders, mixed, and then reflux-extracted twice in a rotary evaporator with 6 times volume of 70% ethanol for 2 h, with the temperature set at 24 °C. Then, the filtrate was freeze-dried. A 0.1 g aliquot of each powder was dispersed in 10 mL 20% aqueous methanol (v/v) and ultrasonicated in a water bath for 10 min at room temperature to prepare solutions, after which the supernatant was filtered through a 0.22 μm filter membrane. 5 μL of the filtrate was injected into the UPLC/QTOF MS system for analysis.
2 were ground into crude powders, mixed, and then reflux-extracted twice in a rotary evaporator with 6 times volume of 70% ethanol for 2 h, with the temperature set at 24 °C. Then, the filtrate was freeze-dried. A 0.1 g aliquot of each powder was dispersed in 10 mL 20% aqueous methanol (v/v) and ultrasonicated in a water bath for 10 min at room temperature to prepare solutions, after which the supernatant was filtered through a 0.22 μm filter membrane. 5 μL of the filtrate was injected into the UPLC/QTOF MS system for analysis.
2.3 Instrumentation and chromatography conditions
Chromatographic separation was performed using an UPLC system (Waters, Milford, MA, USA) equipped with a binary solvent delivery system and a photodiode-array detector. An ACQUITY™ UPLC HSS T3 Column (2.1 × 100 mm, 1.8 μm, Waters Corporation, Milford, USA) was used for the analysis. The column temperature was set at 40 °C. The flow rate was 0.3 mL min−1. The optimal mobile phase consisted of A (HCOOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 0.1
H2O = 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, v/v) and B (HCOOH
100, v/v) and B (HCOOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH3CN = 0.1
CH3CN = 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, v/v). The elution conditions were optimized as follows: 0–1.0 min, 2–20% A; 1.0–4.0 min, 20–21% A; 4.0–6.5 min, 21–30% A; 6.5–9.5 min, 30–35% A; 9.5–14.0 min, 35–50% A; 14.0–16.0 min, 50–52% A; 16.0–16.5 min, 52–82% A; 16.5–19.0 min; 82–83% A; 19.0–19.5 min, 83–100% A; 19.5–20.0 min, 100–2% A; 20.0–21.0 min, 2% A.
100, v/v). The elution conditions were optimized as follows: 0–1.0 min, 2–20% A; 1.0–4.0 min, 20–21% A; 4.0–6.5 min, 21–30% A; 6.5–9.5 min, 30–35% A; 9.5–14.0 min, 35–50% A; 14.0–16.0 min, 50–52% A; 16.0–16.5 min, 52–82% A; 16.5–19.0 min; 82–83% A; 19.0–19.5 min, 83–100% A; 19.5–20.0 min, 100–2% A; 20.0–21.0 min, 2% A.
2.4 Mass spectrometry
The MS instrument used was a Waters Synapt™ High Definition TOF Mass system (Waters Corp., USA) equipped with an ESI source in both positive and negative ion modes. The MS source temperature was set at 110 °C, and the desolvation temperature was set at 300 °C with desolvation gas flow at 600 L h−1. The capillary voltage was 3 kV. The mass spectra were recorded across the range of m/z 100 to 2700 Da, with accurate mass measurement of all mass peaks. The collision energy was set as 10–30 eV for low-energy scans, and 30–50 eV for high-energy scans. The instrument was controlled by Masslynx 4.1 (Waters Corp.).2.5 Data processing
LC-MS data of all determined samples were further processed by Markerlynx V4.1 software (Waters Corporation, Milford, USA) for peak detection and peak alignment. The method parameters for data processing were set as follows: retention time range 0.1–16.0 min, mass range 100–2000 Da, retention time tolerance 0.2 min, mass tolerance 0.05 Da, noise elimination level 6, and peak intensity threshold 50. The peak width at 5% height and the peak-to-peak baseline noise were automatically determined. Pareto scaling of data was performed prior to analysis, which generated less noise level than other scaling methods.3. Results and discussion
To improve detection, the following MS conditions were optimized: formic acid concentration in mobile phase, ion mode, desolvation temperature, capillary voltage, and desolvation gas flow, etc. The reference compounds were used as test compounds to optimize the MS conditions. A positive and negative ESI source was chosen based on preliminary experiments. These optimal MS conditions were used in all further experiments. We developed a broad-spectrum screening method on an UPLC/QTOF MS with collision energy function (MSE mode and accurate mass), which provides a unique way to incorporate fragment ion information without the need for precursor ion selection. Identification was based on the presence of one characteristic m/z ion obtained with the low collision energy function and at least one fragment ion obtained with the high collision energy function, both with mass errors of less than 5 ppm permitted. The molecular formulas with relative mass error between the measured and theoretical mass within 5 ppm were chosen and then applied to MassFragment analysis for further confirmation of their structures. The high- and low-energy MSE tool generates product ion spectra that were successfully applied to structural elucidation of detected KXS.Detailed clarification of the chemical constituents in KXS is essential for holistic quality control. In order to complete the comprehensive characterization, multiple approaches, including database searching, reference standard comparison, and QTOF-MS and MS/MS data analysis, were employed for structural characterization of the KXS constituents. The chemical formula of an unknown structure was deduced based on the high-accuracy [M − H]−/[M + HCOO]− (in the negative ion mode) or [M + H]+/[M + Na]+ (in positive ion mode) precursor ion. The compound(s) matching the determined chemical formula and considered to undergo the observed MS/MS fragmentation was selected as the final identity. As a result, a total of 107 compounds were identified or tentatively characterized from the constituents of KXS. Information regarding the 107 constituents, such as the tR (min), identity, observed m/z values, mass error (in ppm), molecular formula, and botanical source, is offered in ESI Table 1.† The ESI base peak ion (BPI) chromatogram of the KXS by UPLC/QTOF MS is shown in Fig. 1. By UPLC/QTOF MS analysis, a total of 107 compounds, including 61 in the negative ion mode and 59 in the positive ion mode, were identified from KXS; 32 compounds, including 15 ginsenosides (44, 49, 51, 54–61, 63, 64, 77, and 80) were from Ginseng Radix; 19 compounds, including 7 triterpenoids (78, 79, 89, 91, 92, 98, and 106) were from Poria; 33 compounds were from Polygalae Radix; 21 compounds were from Acori Tatarinowii Rhizoma. Generally, MSE provides sufficient full scan MS and MS/MS spectral information. The characteristic MSn fragments of the 107 compounds are shown in ESI Table 2.†
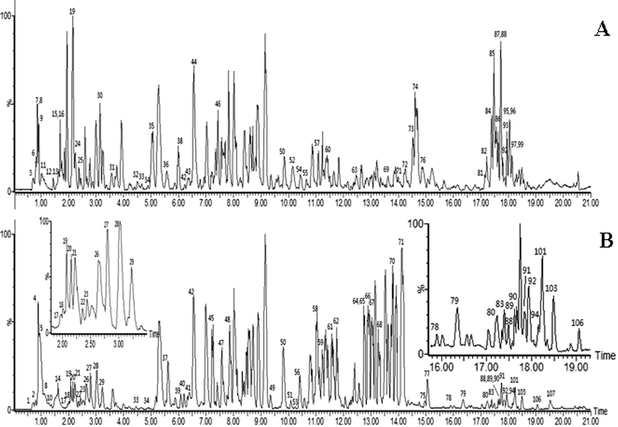 | ||
| Fig. 1 ESI base peak ion (BPI) chromatogram of the KXS analyzed by UPLC/QTOF MS. Positive ion mode (A), negative ion mode (B). (The peak numbers are displayed in Table 1†). | ||
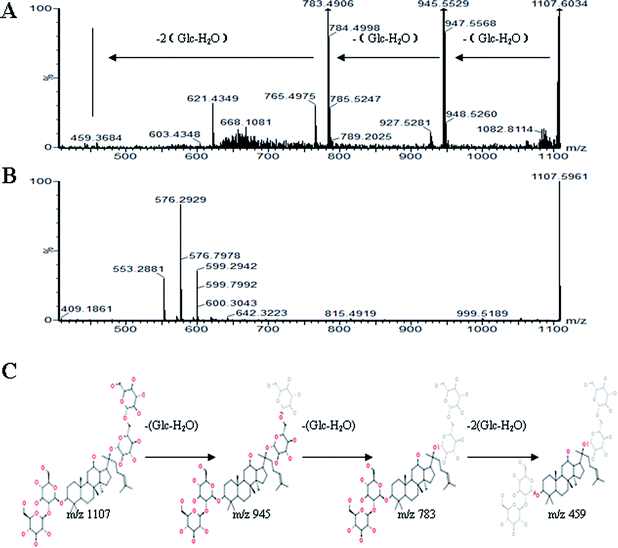
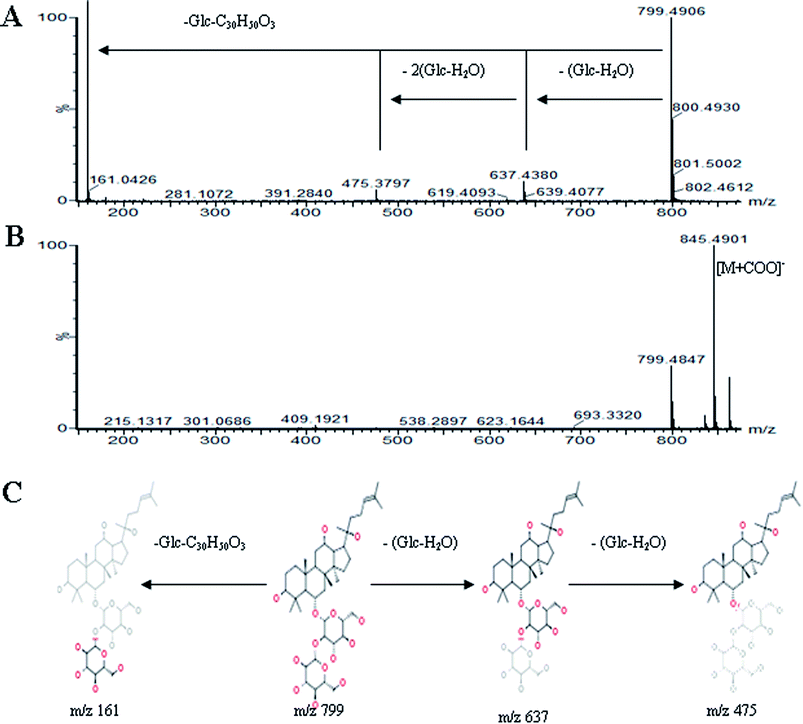
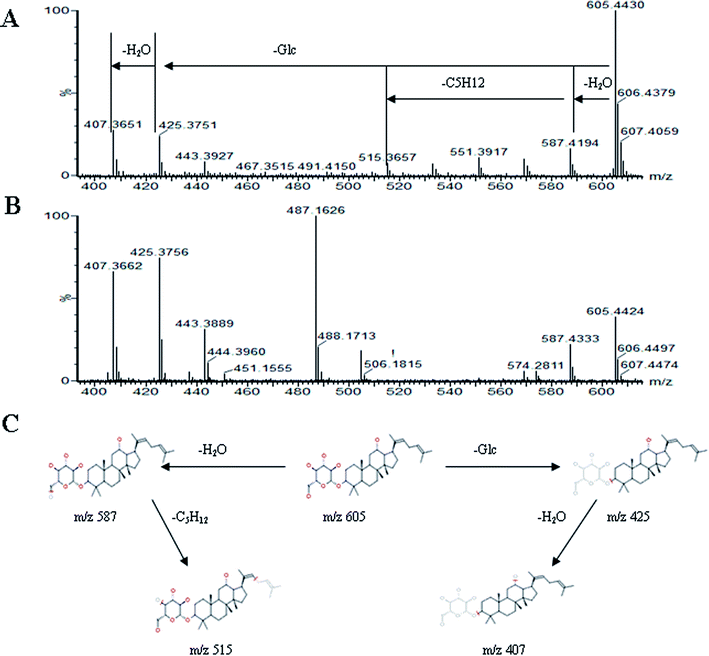
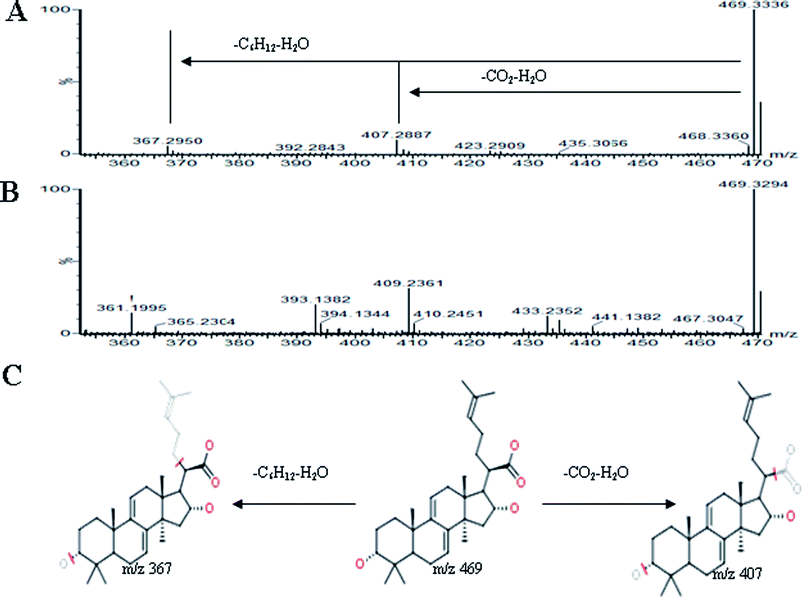
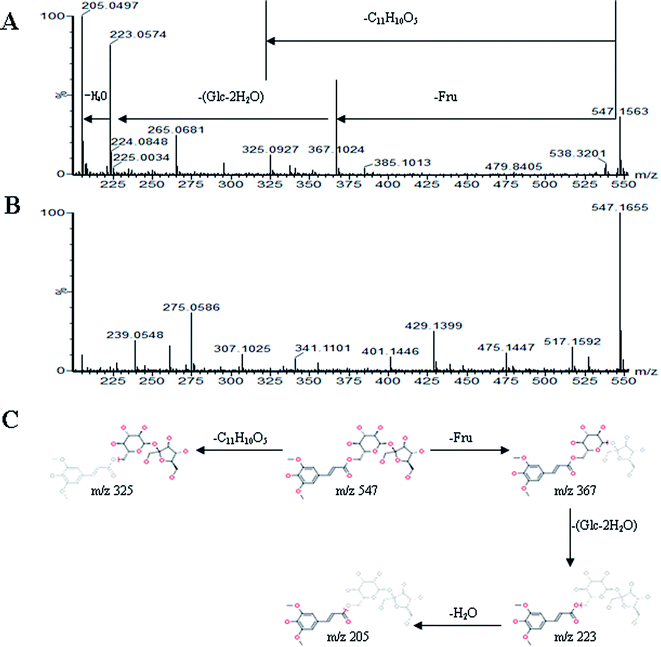
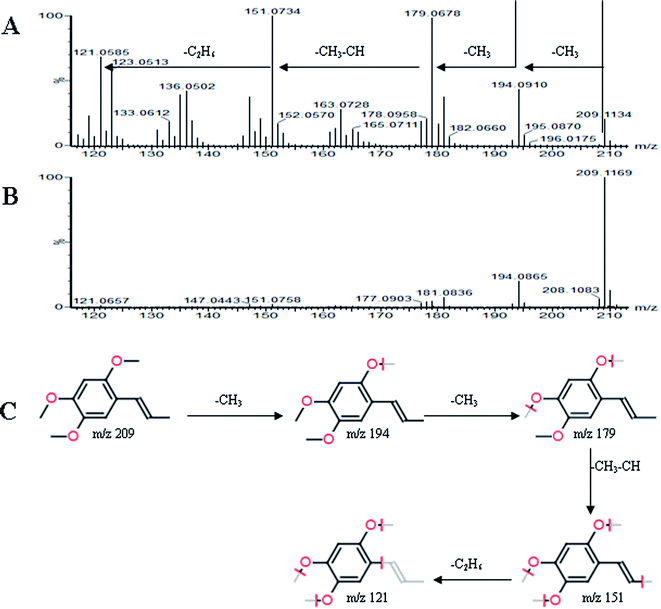
All MS data obtained from the robust UPLC-ESI-Q-TOF-MS analysis that was performed using the aforementioned protocol indicated the retention time and precise molecular mass and provided the MS/MS data, which was necessary for structural identification. Taking an example, in ESI negative ion mode, at low CE voltage, peak 56 produced a prominent [M − H]− ion at m/z 1107.5961. At high CE voltage, it produced fragment ions at m/z 945 [M − H − (Glc–H2O)]−, 783 [M − H − 2(Glc–H2O)]− and 459 [M − H − 4(Glc–H2O)]− (Fig. 2), respectively. Thus, compound 56 was consistent with the MS/MS information of, and tentatively identified as, ginsenoside Rb1 by comparing with a reference standard. Peak 49 gave an [M − H]− ion at m/z 799.4906, as shown in ESI Table 1.† In the high CE scan, 637 [M − H − (Glc–H2O)]−, 475 [M − H − 2(Glc–H2O)]−, and 161 [M − H − Glc − C30H50O3]− were observed as the main fragmentation pathways (Fig. 3), which was identical to that of ginsenoside Rf. Peak 55 at 10.79 min gave an [M + H]+ ion at m/z 605.4430. In the high CE scan, 587 [M + H − H2O]+, 515 [M + H − C5H12 − H2O]+, 425 [M + H − Glc]+, and 407 [M + H − Glc − H2O]+ were observed as the main mass fragments (Fig. 4). The results suggest compound 55 might be ginsenoside Rh3. Peak 88 gave an [M − H]− ion at m/z 469.3318. It produced a similar MS/MS spectrum to that of 3β,16α-dihydroxylanosta-7,9(11),24-trien-21-oic acid (Fig. 5). Peak 23 at 2.41 min gave an [M − H]− ion at m/z 547.1663. In the high CE scan, it showed main fragment ions at m/z 385.1013, 367.1024, 325.0927, 223.0574, and 205.0497 (Fig. 6). All the ions were similar to those of compound sibiricose A1. By comparing the retention time with that of sibiricose A1, compound 23 was tentatively identified as sibiricose A1. Compound 73 was identified to be α-asarone, which was confirmed by comparison with a reference compound. In the high CE scan, it produced the same fragment ions at m/z 194.0910, 179.0678, 151.0734, and 121.0586 as those of α-asarone (Fig. 7). Similarly, other compounds could also be characterized according to the above-mentioned methods. The results demonstrate the potential of UPLC/QTOF MS coupled with the MarkerLynx™ software and MSE tool as an efficient and convenient method to screen and identify constituents in herbal medicine.
4. Conclusion
In this work, we demonstrated the use of UPLC/QTOF MS coupled with automated MSE data analysis for the first structural characterization of global compounds in KXS. Using MSE approach and UPLC/QTOF MS technique, a total of 107 compounds were tentatively characterized; 32 compounds, including 15 ginsenosides, were from Ginseng Radix; 19 compounds, including 7 triterpenoids, were from Poria; 33 compounds were from Polygalae Radix; 21 compounds were from Acori Tatarinowii Rhizoma. These findings provide informative groundwork for further pharmacological studies of the KXS prescription. This strategy could greatly increase the knowledge on the components of TCMs; we expect that this approach would be useful for the screening and characterization of compounds in other famous herbal medicines.Competing financial interests
The authors declare no competing financial interests.Acknowledgements
This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant no. 81302905, 81173500, 81373930, 81102556, 81202639, 81430093), National Key Technology Research and Development Program of the Ministry of Science and Technology of China (Grant no. 2011BAI03B03, 2011BAI03B06, 2011BAI03B08).References
- X. Wang, A. Zhang, G. Yan, Y. Han and H. Sun, UHPLC-MS for the analytical characterization of traditional Chinese medicines, Trends Anal. Chem., 2014 DOI:10.1016/j.trac.2014.05.013.
- A. Zhang, D. Zou, G. Yan, Y. Tan, H. Sun and X. Wang, Identification and characterization of the chemical constituents of Simiao Wan by ultra high performance liquid chromatography with mass spectrometry coupled to an automated multiple data processing method, J. Sep. Sci., 2014, 37(14), 1742–1747 CrossRef CAS PubMed.
- Y. Y. Zhao, L. Zhang, Y. L. Feng, D. Q. Chen, Z. H. Xi, X. Du, X. Bai and R. C. Lin, Pharmacokinetics of 2,3,5,4′-tetrahydroxystilbene-2-O-β-D-glucoside in rat using ultra-performance LC-quadrupole TOF-MS, J. Sep. Sci., 2013, 36(5), 863–871 CrossRef CAS PubMed.
- H. Wang, G. Yan, A. Zhang, Y. Li, Y. Wang, H. Sun, X. Wu and X. Wang, Rapid discovery and global characterization of chemical constituents and rats metabolites of Phellodendri amurensis cortex by ultra-performance liquid chromatography-electrospray ionization/quadrupole-time-of-flight mass spectrometry coupled with pattern recognition approach, Analyst, 2013, 138(11), 3303–3312 RSC.
- D. Zou, A. Zhang, G. Yan, Y. Tan, H. Sun and X. Wang, UPLC/MS coupled with a dynamic multiple data processing method for comprehensive detection of chemical constituents of herbal San-Miao-Wan formula, Anal. Methods, 2014, 6, 2848–2854 RSC.
- W. Dong, P. Wang, X. Meng, H. Sun, A. Zhang, W. Wang, H. Dong and X. Wang, Ultra-performance liquid chromatography-high-definition mass spectrometry analysis of constituents in the root of Radix Stemonae and those absorbed in blood after oral administration of the extract of the crude drug, Phytochem. Anal., 2012, 23(6), 657–667 CrossRef CAS PubMed.
- D. Moran, T. Cross, L. M. Brown, R. M. Colligan and D. Dunbar, Data-independent acquisition (MSE) with ion mobility provides a systematic method for analysis of a bacteriophage structural proteome, J. Virol. Methods, 2014, 195, 9–17 CrossRef CAS PubMed.
- L. X. Fang, A. Z. Xiong, R. Wang, S. Ji, L. Yang and Z. T. Wang, A strategy for screening and identifying mycotoxins in herbal medicine using ultra-performance liquid chromatography with tandem quadrupole time-of-flight mass spectrometry, J. Sep. Sci., 2013, 36(18), 3115–3122 CAS.
- Y. Y. Zhao, P. Lei, D. Q. Chen, Y. L. Feng and X. Bai, Renal metabolic profiling of early renal injury and renoprotective effects of Poria cocos epidermis using UPLC Q-TOF/HSMS/MSE, J. Pharm. Biomed. Anal., 2013, 81–82, 202–209 CAS.
- Y. Cao, Y. Hu, P. Liu, H. X. Zhao, X. J. Zhou and Y. M. Wei, Effects of a Chinese traditional formula Kai Xin San (KXS) on chronic fatigue syndrome mice induced by forced wheel running, J. Ethnopharmacol., 2012, 139(1), 19–25 CrossRef PubMed.
- K. Y. Zhu, Q. Q. Mao, S. P. Ip, R. C. Choi, T. T. Dong, D. T. Lau and K. W. Tsim, A standardized chinese herbal decoction, kai-xin-san, restores decreased levels of neurotransmitters and neurotrophic factors in the brain of chronic stress-induced depressive rats, J. Evidence-Based Complementary Altern. Med., 2012, 2012, 149256 Search PubMed.
- Y. Hu, Y. Cao, M. Liu, P. Liu, H. Cui and G. Dai-Hong, Behavioral and biochemical effects of a formulation of the traditional Chinese medicine, Kai-Xin-San, in fatigued rats, Exp. Ther. Med., 2013, 6(4), 973–976 Search PubMed.
- Y. Hu, M. Liu, P. Liu, J. J. Yan, M. Y. Liu, G. Q. Zhang, X. J. Zhou and B. Y. Yu, Effect of kai xin san on learning and memory in a rat model of paradoxical sleep deprivation, J. Med. Food, 2013, 16(4), 280–287 CrossRef PubMed.
- K. Y. Zhu, S. L. Xu, R. C. Choi, A. L. Yan, T. T. Dong and K. W. Tsim, Kai-xin-san, a chinese herbal decoction containing ginseng radix et rhizoma, polygalae radix, acori tatarinowii rhizoma, and poria, stimulates the expression and secretion of neurotrophic factors in cultured astrocytes, J. Evidence-Based Complementary Altern. Med., 2013, 2013, 731385 Search PubMed.
- X. J. Zhou, M. Liu, J. J. Yan, Y. Cao and P. Liu, Antidepressant-like effect of the extracted of Kai Xin San, a traditional Chinese herbal prescription, is explained by modulation of the central monoaminergic neurotransmitter system in mouse, J. Ethnopharmacol., 2012, 139(2), 422–428 CrossRef PubMed.
- C. Lv, Q. Li, X. Zhang, B. He, H. Xu, Y. Yin, R. Liu, J. Liu, X. Chen and K. Bi, Simultaneous quantitation of polygalaxanthone III and four ginsenosides by ultra-fast liquid chromatography with tandem mass spectrometry in rat and beagle dog plasma after oral administration of Kai-Xin-San: application to a comparative pharmacokinetic study, J. Sep. Sci., 2014, 37(9–10), 1103–1110 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ay01954g |
| This journal is © The Royal Society of Chemistry 2015 |