An electronically enhanced chiral sum frequency generation vibrational spectroscopy study of lipid-bound cytochrome c†
Khoi Tan
Nguyen
ab
aSchool of Chemical Engineering, The University of Queensland, Brisbane, QLD 4072, Australia. E-mail: k.nguyen9@uq.edu.au
bSchool of Biotechnology, International University, Vietnam National University, Ho Chi Minh City, Vietnam
First published on 3rd November 2014
Abstract
Electronically enhanced chiral SFG spectroscopy was employed to study the lipid bound cyt c in situ. It was directly observed that upon interacting with anionic phospholipids, the amino acid residues around the heme adopted the β-sheet conformation. In addition, the orientation of this newly formed β-sheet structure was found to be sensitive to the bulk pH.
Normally bound to cardiolipin (CL) in the inner mitochondrial membrane, cytochrome c (cyt c) is generally known to play the crucial role of electron transportation in the complex III–IV redox process within the respiration process. In addition, cyt c is also involved in the cell apoptosis signaling, the very first event that triggers the apoptosis pathway. This apoptosis triggering is believed to start with the CL peroxidation, which then facilitates the release of cyt c into the cytosol. However, the mechanism of how this protein is set free from the IMM remains unclear despite much research being carried out over the past three decades.1–4 In an effort to shed light on how the tightly bound cyt c can be released from CL, many studies have focused on correlating the structural conformation of CL bound cyt c with the peroxidase function of the cyt c–CL complex. For instance, it has been suggested that cyt c is bound to CL at one or two distinct sites (A and L) and this depends on the pH of the surrounding medium.5,6
Alternatively, using resonance Raman spectroscopy, Balakrishnan et al. simulated the effect of CL on cyt c under destabilizing conditions (pH 3, heating up to 80 °C) and suggested that the cyt c partly unfolds and converts into a β-sheet structure upon binding with CL.7 However, the destabilizing conditions applied in their study might have caused complete denaturation of the protein as observed by electronic sum frequency generation at pH 2.8 Also, the authors were unable to locate the β-sheet conformational switch. More recently, it was suggested by fluorescence anisotropy measurements that cyt c adopts either the compact or extended conformers upon binding to CL containing liposomes, while its secondary structure content is almost conserved.2,9
In our study, we employed electronically enhanced sum frequency generation spectroscopy to study the structure of CL-bound cyt c under physiological conditions. Our SFG data not only confirm the β-sheet structure conversion upon cyt c–CL interaction, but also indicate its location within the molecule. Importantly, the chiral structure conversion of cyt c was also observed when the protein binds to the saturated phospholipid 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG).
The experimental setup of the SFG system has been described in our previous study and hence will not be detailed here.10 Equine cyt c was purchased from Sigma Aldrich (BioUltra, ≥99%) and used without further purifications. Bovine CL, and hydrogenated and deuterated DPPG were purchased from Avanti Polar Lipids Inc (Alabaster, AL). Deposition of the lipid bilayers onto CaF2 prisms was prepared by sequentially depositing the distal (dDPPG) and proximal (bovine CL or DPPG) layers using a 622 Nima LB trough. The protein reservoir with a volume of 4 ml was placed below the lipid bilayer deposited CaF2 prisms during the SFG measurement. The 500 nM cyt c solution was stirred using a magnetic micro-stirrer at a rate of 40 rpm during the protein–lipid interaction. Freshly purified water (by an Ultrapure Milli-Q unit from Millipore, USA) with a resistivity of 18.2 MΩ cm was used to prepare all solutions used in the experiments. The pH of the bulk was controlled by 20 mM phosphate buffer saline (PBS) of pH 5.5 and 7.2. The pH of the bulk was adjusted by multiple flushing with the corresponding PBS buffers. All experiments were performed at room temperature (∼23 °C). The possibility of having optical leakage caused by the polarizers was eliminated by indiscernible SFG signals in the amide I band of a helical peptide alamethicin collected in both spp and psp polarization combinations.
Typically, under physiological conditions, chiral-specific SFG signals arising from the adsorbed non-aggregated protein molecules at interfaces are rather weak and difficult to probe directly due to low protein population and the absence of the additional enhancement arising from the electronic resonances.11–13 Recently, Yan's research group has reported direct detection of the chiral-specific SFG signal of an amyloid peptide (h-IAPP), which is well-known to aggregate fairly easily into large fibrous/sheet structures.14,15 Furthermore, the approximately three fold smaller Fresnel factors of the spp and psp as compared to the ssp polarization combination can be considered as some of the major experimental obstacles in detecting the chiral-specific SFG signals directly. Such chiral-specific SFG signals have been detected indirectly via an interference technique proposed by Belkin et al.11,12,16 However, cyt c is a heme protein which exhibits a characteristic adsorption Q band at 530 nm.17 The protein is thus electronically excited by the incident visible green beam at 532 nm, which is capable of enhancing the possible chiral signals in all spp (s-polarized SF output, p-polarized visible and p polarized tunable IR beams, respectively), psp and pps polarization combinations.18 This electronic enhancement was experimentally confirmed by the strong achiral ssp yet negligible psp/spp chiral SFG signals obtained when the visible beam was tuned to 355 nm (Fig. S3, ESI†). The chiral SFG signals were obtained in spp and psp polarization combinations, which are mathematically described by the following relationships
χ(2)psp = Lzz(ωSFG)Lyy(ωvis)Lxx(ωIR)sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) αSFG cos αSFG cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) αIRχ(2)zyx − Lxx(ωSFG)Lyy(ωvis)Lzz(ωIR)cos αIRχ(2)zyx − Lxx(ωSFG)Lyy(ωvis)Lzz(ωIR)cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) αSFG sin αSFG sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) αIRχ(2)xyz αIRχ(2)xyz | (1) |
χ(2)spp = Lyy(ωSFG)Lzz(ωvis)Lxx(ωIR)sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) αvis cos αvis cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) αIRχ(2)yzx − Lyy(ωSFG)Lxx(ωvis)Lzz(ωIR)cos αIRχ(2)yzx − Lyy(ωSFG)Lxx(ωvis)Lzz(ωIR)cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) αSFG sin αSFG sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) αvisχ(2)yxz αvisχ(2)yxz | (2) |
The interaction between cyt c and CL is generally believed to be electrostatically driven because CL is negatively charged while cyt c has a net charge of +8 distributed in two main clusters. Since the two pKa values of CL are 2.8 and 6, CL should be mostly deprotonated at neutral pH, which strengthens its electrostatic interaction with cyt c through site A. Under more acidic conditions, cyt c has been reported to interact with CL via an additional binding site L.5,6 Being a globular peripheral membrane protein, cyt c was observed in this study to disrupt the proximal CL leaflet, but leave the distal dDPPG leaflet almost intact, as evidenced by the dramatic SFG C–H signals of the proximal CL leaflet and the persistent C–D signal (Fig. S2, ESI†).
Native cyt c consists of a α-helical, random coil and the porphyrin structures (Fig. 1), and thus in principle is unable to give rise to any chiral SFG signal. Surprisingly, strong chiral SFG signals of CL-bound cyt c were directly probed in both spp and psp polarization combinations. Generally, the SFG signals detected in psp, pps and spp polarization combinations are attributed to the interfacial chirality.14,18 While the achiral SFG signals contributed by monomeric/oligomeric helical and disordered structures can be detected in the ssp, sps, pss and ppp polarization combinations.12
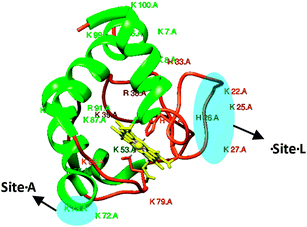 | ||
| Fig. 1 Ribbon structure of soluble cyt c. The two binding sites A and L are highlighted in blue. α-Helical motifs are presented in green, random coils are in orange and the porphyrin is in yellow. | ||
Although the chiral SFG signals can directly contribute to both the spp and psp spectra, the behaviors of the spp and psp chiral SFG signals were observed to be similar in response to the pH variations; hence, we report only the chiral SFG signals of the lipid bound cyt c collected in the psp polarization combination. We attribute the SFG chiral signal to the β-sheet structure resulting from the CL–cyt c interaction. Importantly, this newly formed β-sheet structure must be associated with either His18 or Met80 residues, which are coordinated with the heme, which aligns well with a recent resonance Raman study on cyt c in which the Met80–Fe bond rupture was related as a proposed mechanism on how the structure of this protein changes upon interacting with CL.7 Otherwise, the chiral SFG signal should not have been enhanced electronically and readily detectable without interference enhancing techniques.20 It should be noted that the chiral SFG amide I bands of CL-bound cyt c exhibited substantial differences in their response to pH changes. At pH 5.5, the chiral SFG band is dominated by the 1635 cm−1 peak, featuring the B2 mode; whilst at pH 7.2, the B1 mode at 1685 cm−1 exclusively dominates the amide I band (Fig. 2a and Fig. S1, ESI†). Interestingly, this spectral shift was reversible, changing in accordance with the pH (7.2 and 5.5) and this was regardless of the type of lipid the cyt c protein was bound to (Fig. 2).
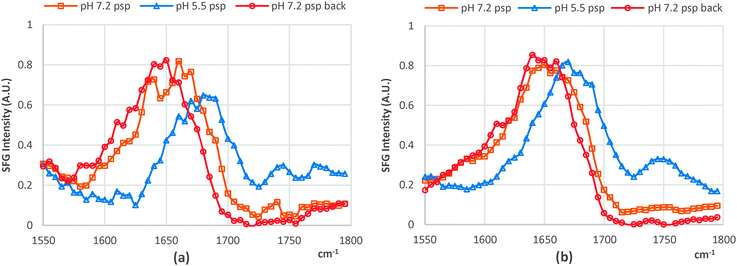 | ||
| Fig. 2 psp chiral SFG spectra when the pH was changed from 7.2 to 5.5 and back to 7.2 of (a) dDPPG/bovine CL-bound cyt c and (b) dDPPG/DPPG-bound cyt c. | ||
Since high resolution molecular details about this newly converted β-sheet structure of CL-bound cyt c are not yet available, SFG analysis is unable to provide its orientation in the macroscopic frame. However, our SFG data have provided evidence that this newly formed β-sheet structure responds to the surrounding pH by changing its orientation, enabling the B1 and B2 modes to specifically dominate the chiral SFG amide I band accordingly. The effect of the pH on the orientation of this β-sheet is likely the mechanism of how the ligands around the 5-coordinated ferrous heme facilitate the activities of the ferrous heme, which is spectroscopically observable via the strong field ligands (His18 and Met80).8 It is, however, uncertain that the orientation change of the β-sheet is self-induced or caused by a molecular orientation shift stemming from the additional binding site at acidic pH.5
In conclusion, we have successfully demonstrated that electronically enhanced chiral SFG can be employed to directly probe the structural changes in the close vicinity around the heme. In particular, it was observed that the amino acid residues coordinated with the heme group of cyt c adopt the β-sheet conformation upon the cyt c–anionic lipid interaction, even though we are uncertain if this β-sheet conversion occurred at the expense of the α-helical structures, which in fact remains controversial among recent studies on cyt c.2,7,9 Furthermore, the orientation of this β-sheet structure was found to be sensitive to the pH of the bulk. Our findings not only confirm the β-sheet structure formation caused by the protein–lipid interaction, but they also reveal its location within the molecule. We believe that this study provides an important piece of information towards accurately defining the mechanism of cyt c's peroxidase activity and the apoptosis triggering.
The author declares no competing financial interest.
This research is funded by the Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 106.16-2012.67. I sincerely thank Prof Zhan Chen at the University of Michigan for the initial inspirations and Dr Gay Marsden for her generous assistance in the manuscript preparation.
Notes and references
- V. E. Kagan, H. A. Bayir, N. A. Belikova, O. Kapralov, Y. Y. Tyurina, V. A. Tyurin, J. F. Jiang, D. A. Stoyanovsky, P. Wipf, P. M. Kochanek, J. S. Greenberger, B. Pitt, A. A. Shvedova and G. Borisenko, Free Radical Biol. Med., 2009, 46, 1439 CrossRef CAS PubMed.
- J. Muenzner, J. R. Toffey, Y. N. Hong and E. V. Pletneva, J. Phys. Chem. B, 2013, 117, 12878 CrossRef CAS PubMed.
- S. Orrenius and B. Zhivotovsky, Nat. Chem. Biol., 2005, 1, 188 CrossRef CAS PubMed.
- Y. L. P. Ow, D. R. Green, Z. Hao and T. W. Mak, Nat. Rev. Mol. Cell Biol., 2008, 9, 532 CrossRef CAS PubMed.
- C. Kawai, F. M. Prado, G. L. C. Nunes, P. Di Mascio, A. M. Carmona-Ribeiro and I. L. Nantes, J. Biol. Chem., 2005, 280, 34709 CrossRef CAS PubMed.
- M. Rytomaa and P. K. J. Kinnunen, J. Biol. Chem., 1994, 269, 1770 CAS.
- G. Balakrishnan, Y. Hu, O. F. Oyerinde, J. Su, J. T. Groves and T. G. Spiro, J. Am. Chem. Soc., 2007, 129, 504 CrossRef CAS PubMed.
- P. Sen, S. Yamaguchi and T. Tahara, J. Phys. Chem. B, 2008, 112, 13473 CrossRef CAS PubMed.
- J. Hanske, J. R. Toffey, A. M. Morenz, A. J. Bonilla, K. H. Schiavoni and E. V. Pletneva, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 125 CrossRef CAS PubMed.
- K. T. Nguyen, T. D. Nguyen and A. V. Nguyen, Langmuir, 2014, 30, 7047 CrossRef CAS PubMed.
- J. Wang, X. Y. Chen, M. L. Clarke and Z. Chen, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 4978 CrossRef CAS PubMed.
- M. A. Belkin, T. A. Kulakov, K. H. Ernst, L. Yan and Y. R. Shen, Phys. Rev. Lett., 2000, 85, 4474 CrossRef CAS.
- L. M. Haupert and G. J. Simpson, Annu. Rev. Phys. Chem., 2009, 60, 345 CrossRef CAS PubMed.
- L. Fu, J. Liu and E. C. Y. Yan, J. Am. Chem. Soc., 2011, 133, 8094 CrossRef CAS PubMed.
- J. Zhao, R. D. Hu, M. F. M. Sciacca, J. R. Brender, H. Chen, A. Ramamoorthy and J. Zheng, Phys. Chem. Chem. Phys., 2014, 16, 2368 RSC.
- S. R. Walter and F. M. Geiger, J. Phys. Chem. Lett., 2010, 1, 9 CrossRef CAS.
- N. Khare, C. M. Eggleston, D. M. Lovelace and S. W. Boese, J. Colloid Interface Sci., 2006, 303, 404 CrossRef CAS PubMed.
- M. A. Belkin and Y. R. Shen, Phys. Rev. Lett., 2003, 91, 4474 CrossRef.
- E. C. Y. Yan, L. Fu, Z. Wang and W. Liu, Chem. Rev., 2014, 114, 8471 CrossRef CAS PubMed.
- K. T. Nguyen, J. T. King and Z. Chen, J. Phys. Chem. B, 2010, 114, 8291 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4cc06916a |
| This journal is © The Royal Society of Chemistry 2015 |
