The intracellular metabolism of isoflavones in endothelial cells
Natalia
Toro-Funes
ab,
Francisco Javier
Morales-Gutiérrez
a,
M. Teresa
Veciana-Nogués
a,
M. Carmen
Vidal-Carou
a,
Jeremy P. E.
Spencer
b and
Ana
Rodriguez-Mateos
*c
aDepartment of Nutrition and Food Science-XaRTA, Faculty of Pharmacy, Campus de l'Alimentació Torribera, University of Barcelona, Avda. Prat de la Riba 171, 08921-Santa Coloma de Gramenet, Barcelona, Spain
bMolecular Nutrition Group, School of Chemistry, Food and Pharmacy, University of Reading, Reading, RG2 6AP, UK
cDivision of Cardiology, Pulmonology, and Vascular Medicine, Medical Faculty, University of Duesseldorf, D-40225 Duesseldorf, Germany. E-mail: ana.rodriguez-mateos@med.uni-duesseldorf.de; Tel: +49 2118115893
First published on 13th November 2014
Abstract
Data from epidemiological and human intervention studies have highlighted potential cardiovascular benefits of soy isoflavone-containing foods. In humans, genistein and daidzein are extensively metabolized after absorption into glucuronides and sulfate metabolites. However, limited data exist on isoflavone cellular metabolism, in particular in endothelial cells. We investigated the uptake and cellular metabolism of genistein, daidzein and its major in vivo microbial metabolite, equol, in human endothelial (HUVEC), liver (HepG2) and intestinal epithelial cells (Caco-2 monolayer). Our results indicate that genistein and daidzein are taken up by endothelial cells, and metabolized into methoxy-genistein-glucuronides, methoxy-genistein-sulfates and methoxy-daidzein-glucuronides. In contrast, equol was taken up but not metabolized. In HepG2 and Caco-2 cells, glucuronide and sulfate conjugates of genistein and daidzein and a sulfate conjugate of equol were formed. Our findings suggest that endothelial cell metabolism needs to be taken into account when investigating the cardioprotective mechanisms of action of isoflavones.
Introduction
Soy isoflavones have recently received much attention because of their potential health benefits, particularly on the prevention of different types of cancer, osteoporosis and cardiovascular diseases (CVD).1 However, there is also controversy regarding the safety and efficacy of isoflavones and potential adverse effects have also been reported.2,3 Data from human intervention studies suggest isoflavones may have beneficial effects on prognostically validated surrogate markers of CVD, such as blood pressure, endothelial function, or arterial stiffness.4–8 Moreover, several studies have shown that isoflavones exert favorable effects on other biomarkers of CVD, such as plasminogen activator inhibitor-1, endothelin 1, VCAM-1 or NO.9–13 Some studies have shown improvements in plasma lipids and lipoproteins after isoflavone consumption, including lowering blood triglycerides, total and LDL cholesterol levels, increasing HDL cholesterol and the ratio of HDL/LDL cholesterol,14–16 although some mixed data exist with a recent study and a meta-analysis concluding that there is not enough evidence to support positive effects of isoflavones on blood lipids.17,18The most abundant soy isoflavones in the diet are genistein (Ge) and daidzein (De) (Fig. 1).19 They are ingested mainly in the glucoside form, and undergo extensive hydrolysis by intestinal and bacterial β-glucosidases that release the main aglycones.20 De is converted into equol (Eq) (Fig. 1) due to the action of the intestinal microbiota before absorption.21 The isoflavone aglycones are converted into glucuronide metabolites by UDT-glucuronosyltransferases (UGT), and to a lesser extent to sulfate esters catalyzed by sulfotransferases (SULT) at either or both 4′ or 7 positions on the isoflavone ring by phase II enzymes during transfer across the small intestine and liver.22 These phase II metabolites are excreted in the bile and are deconjugated in the lower bowel allowing them to be reabsorbed again, creating an enterohepatic circulation.22 Studies showed that the conjugated metabolites of Ge and De are mainly found in human plasma as mono- and diglucuronides, mono- and disulfates, and sulfo-glucuronides.23 Trace amounts of mono- and dimethoxylated conjugates have also been found in urine.24,25
Most of the research investigating the mechanisms of action of flavonoids in the vascular system has tested the bioactivity of flavonoids and their metabolites in human and animal cell models. However, whether flavonoids and metabolites are taken up and metabolized further by cells is at present unclear. In order to assess the potential risks and benefits of soy isoflavones and the mechanisms by which health effects occur, it is important to have a more complete understanding of isoflavone intracellular metabolism. The metabolism of isoflavones has been reported in cell models of the gastrointestinal tract, such as enterocytes and Caco-2 cell monolayers,26–28 or in hepatic models,29,30 but little attention has been given to the potential intracellular metabolism of isoflavones in other human cells, such as human umbilical vein endothelial cells (HUVEC), despite being a widely used in vitro model for assessing mechanisms of action of isoflavones in the vascular endothelium.11,31 Therefore, the aim of this work is to determine the cellular uptake and intracellular metabolism of the isoflavones Ge, De and Eq in endothelial cells, using HepG2 hepatocytes (liver cell model) and Caco-2 cell monolayers (small intestine model) as positive controls.
Experimental
Cell culture
Human umbilical vein endothelial cells (HUVEC) were obtained from Lonza (Basingstoke, UK) and used at passages 2 or 3. The cells were cultured at 37 °C with 5% CO2 in a humidified atmosphere, and supplemented with endothelial culture medium consisted of: 2% foetal bovine serum (10 ml) plus supplements [hEGF (0.5 ml), Hydrocortisone (0.2 ml), GA-1000 (Gentamicin, Amphotericin-B) (0.5 ml), VEGF (0.5 ml), hFGF-B (2 ml), R3-IGF-1 (0.5 ml), ascorbic acid (0.5 ml), heparin (0.5 ml) to 500 ml endothelial cell basal medium without phenol red (Lonza, UK). Foetal bovine serum was heat inactivated by incubation at 56 °C for 30 minutes. Cells were kept at 37 °C in a humidified atmosphere containing 5% CO2. Cells were seeded at a density of 0.5–1 × 106 cells per dish in Petri dishes (diameter 100 mm).Liver hepatoma cells HepG2 cells (ATCC, Manassas, VA, US) were cultured at 37 °C in an atmosphere of 5% CO2 relative humidity between passages 19–21 in Dulbecco's modified Eagle's medium F-12 with glutamine (500 ml), with 10% FBS heat inactivated (50 ml) and 1% of penicillin/streptomycin solution (5 ml (PAA, UK)). Cells were seeded at a density of 0.5–1 × 106 cells per dish in Petri dishes (diameter 100 mm).
Human colon adenocarcinoma cells (Caco-2, ECACC Salisbury, Wiltshire, UK) were cultured between passages 15–19 in a humidified atmosphere of 5% CO2/95% air in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% heat-inactivated bovine serum, L-glutamine (2 mM), non-essential amino acids (1%), penicillin (100 U ml−1), and streptomycin (100 lg ml−1) (all from PAA, UK). Culture medium was changed every 2–3 days and the culture was split approximately every 7 days. For subculturing, the cells were removed enzymatically (0.25% trypsin-EDTA, 1 min, 37 °C), split 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and subcultured in 6 well plates (PAA, UK). For transport experiments, 2.5 × 105 cells were seeded in Transwell-clear, tissue culture treated polyester membrane filter inserts (pore size 0.4 mm, diameter 24 mm, PAA, UK) in 6-wells plates. Cells were allowed to grow and differentiate to confluent monolayers for about 20–22 days. The medium was changed twice a week. The apical and basolateral compartments contained 1.2 and 2 ml of culture medium, respectively. The integrity of the monolayers was checked by measuring transepithelial electrical resistance across the layer (TEER) values, using a Millicell-ers epithelial voltohmmeter (Millipore Co., Bedford, MA). Experiments were conducted only in cell monolayers that showed a TEER value between 400 and 1000 Ω cm−2.
3, and subcultured in 6 well plates (PAA, UK). For transport experiments, 2.5 × 105 cells were seeded in Transwell-clear, tissue culture treated polyester membrane filter inserts (pore size 0.4 mm, diameter 24 mm, PAA, UK) in 6-wells plates. Cells were allowed to grow and differentiate to confluent monolayers for about 20–22 days. The medium was changed twice a week. The apical and basolateral compartments contained 1.2 and 2 ml of culture medium, respectively. The integrity of the monolayers was checked by measuring transepithelial electrical resistance across the layer (TEER) values, using a Millicell-ers epithelial voltohmmeter (Millipore Co., Bedford, MA). Experiments were conducted only in cell monolayers that showed a TEER value between 400 and 1000 Ω cm−2.
Assessment of cellular uptake/association and metabolism
HUVEC and HepG2 were grown in petri dishes to a confluence of 80–90%. Prior to experiments, old medium was removed and cells were washed with PBS, pH 7.4. Appropriate amounts of Ge, De and Eq (0, 0.1, 1, 10 and 100 μM) were added to 7 ml of the growth medium and cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 for 2 h. After incubation, 1 mL of the medium was removed and immediately frozen in liquid nitrogen and kept at −80 °C afterwards. Cells were washed twice with ice-cold PBS, and 200 μL of ice-cold 0.1 M HCl was added. Cells were scraped, the cell homogenates sonicated 3 times for 30 s, centrifuged at 800g for 10 min and the supernatant was collected and transferred to −80 °C storage.For Caco-2 cells experiments, the medium was removed and cells were washed with PBS, pH 7.4. Test compounds (conc. as above) were added to the apical side in 1.2 mL of transport buffer consisting in PBS, 1% non-essential amino acids and 1 mM of ascorbic acid. Transport buffer (2 mL) was also added to the basolateral side. Incubation was performed for 2 h at 37 °C at a humidified atmosphere of 5% CO2. Then, the apical and basolateral buffer were collected and immediately stored at −80 °C. In addition, cell filters were washed with ice-cold PBS, prior to addition of 500 μL of 0.1 M HCl, and cell homogenate collection, which were again sonicated 3 times for 30 s and centrifuged at 800g for 10 min, prior to storage at −80 °C.
Uptake data is expressed as μM in supernatant and cell lysates per petri dish, or pmol per mg protein in cell lysate. Uptake refers to both cytosolic accumulation and membrane/cell-associated. Recoveries were calculated respect to the amount of compound recovered after incubation without cells (control).
Stability test of genistein, daidzein and equol in cell culture medium
Each isoflavone (10 μM) was dissolved in endothelial culture medium phenol red free and kept in 6-well plates at 37 °C with 5% CO2 in a humidified atmosphere. Samples were taken at pre-determined time points (0, 0.4, 1, 6 and 24 h). All the tests were done in triplicate. The amount of each compound remained in endothelial medium was determined and the residue amount of each compound was plotted against time to obtain their stability profiles.Sample preparation and analysis of isoflavones
Supernatants and cell lysates were evaporated to dryness using a speedvac concentrator (Savant) and dissolved with 0.1 ml of ultra pure water for chromatographic analysis.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. Resulting supernatant was evaporated to dryness using a speedvac concentrator (Savant) and dissolved with 0.1 ml of ultra pure water for chromatographic analysis.
000g for 10 min. Resulting supernatant was evaporated to dryness using a speedvac concentrator (Savant) and dissolved with 0.1 ml of ultra pure water for chromatographic analysis.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resolution at m/z 400. Operation parameters were set as follow: source voltage, 3.5 kV; sheath gas, 50 (arbitrary units); auxiliary gas, 20 (arbitrary units); sweep gas, 0 (arbitrary units); and capillary temperature, 275 °C. Default values were used for most other acquisition parameters (Fourier transform (FT) Automatic gain control (AGC) target 5 × 105 for MS mode and 5 × 104 for MSn mode). Samples were analysed in full MS mode with the Orbitrap resolution set at 30
000 resolution at m/z 400. Operation parameters were set as follow: source voltage, 3.5 kV; sheath gas, 50 (arbitrary units); auxiliary gas, 20 (arbitrary units); sweep gas, 0 (arbitrary units); and capillary temperature, 275 °C. Default values were used for most other acquisition parameters (Fourier transform (FT) Automatic gain control (AGC) target 5 × 105 for MS mode and 5 × 104 for MSn mode). Samples were analysed in full MS mode with the Orbitrap resolution set at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at m/z 400. The maximum injection time was set to 100 ms with two micro scans for MS mode. The mass range was from 100 to 1000 m/z. Data analyses were performed using XCalibur software. We used an Accela liquid chromatograph (Thermo Scientific, Hemel Hempstead, UK) equipped with a quaternary pump, a photodiode array detector (PDA) and a thermostated autosampler. An Acquity UPLC™ EH C18 1.7 μm column (2.1 × 50 mm) (Waters corp., Milford, MA, USA) with solvent A (ultra pure water with 0.1% formic acid) and B (acetonitrile with 0.1% formic acid) was used. Linear gradient conditions were as follows: 0 min – 10% B; 1 min – 12% B; 3 min – 22% B; 4 min – 23% B; 5 min – 35% B; 6 min – 50%; 8 min – 50% B; 10 min – 10% B at flow of 0.6 ml min−1. The injection volume was 2 μl.
000 at m/z 400. The maximum injection time was set to 100 ms with two micro scans for MS mode. The mass range was from 100 to 1000 m/z. Data analyses were performed using XCalibur software. We used an Accela liquid chromatograph (Thermo Scientific, Hemel Hempstead, UK) equipped with a quaternary pump, a photodiode array detector (PDA) and a thermostated autosampler. An Acquity UPLC™ EH C18 1.7 μm column (2.1 × 50 mm) (Waters corp., Milford, MA, USA) with solvent A (ultra pure water with 0.1% formic acid) and B (acetonitrile with 0.1% formic acid) was used. Linear gradient conditions were as follows: 0 min – 10% B; 1 min – 12% B; 3 min – 22% B; 4 min – 23% B; 5 min – 35% B; 6 min – 50%; 8 min – 50% B; 10 min – 10% B at flow of 0.6 ml min−1. The injection volume was 2 μl.
Due to the absence of commercial standards, metabolites were tentatively identified from the accurate exact mass data provided by the LC-MS analysis. Deviation from the calculated mass (5 ppm) and the isotopic pattern score were used to confirm the accuracy of possible molecular formulas. Deconjugation experiments with β-glucuronidase/sulfatase were also conducted to confirm the identity of metabolites, as described above. The concentration of Ge, De and Eq was determined using an external calibration curve produced with the use of authentic standards, while their metabolites were quantified as their corresponding aglycone.
Results
Stability test of isoflavones at 37 °C
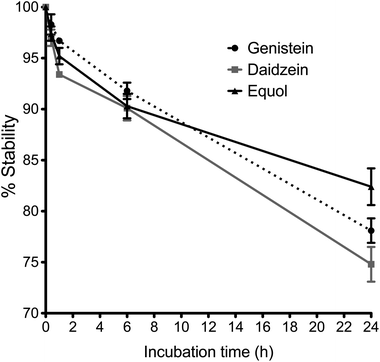
Stability profiles of Ge, De and Eq compounds in cell medium at 37 °C are shown in Fig. 2, as the percentage of initial concentration remaining with time. The concentration of Ge, De and Eq decreased by around 5 ± 2% after 2 hour of incubation, and by around 10 ± 2% following 6 hours incubation. After 24 hours, the 78 ± 1%, 75 ± 2% and 82 ± 2% of Ge, De and Eq, respectively, remained stable in endothelial medium.Identification of isoflavone conjugates in the cell models
The isoflavones and their metabolites identified in HUVEC, HepG2 cells and Caco-2 cell monolayers by LTQ-Orbitrap are shown in Table 1. No Ge, De, Eq or conjugates were detected in any of the control samples (0 μM Ge, De, or Eq) and after incubating cells with 0.1 μM of Ge, De, or Eq. The quantification limits (LOQ) of our method for Ge, De and Eq were 0.1, 0.1 and 0.2 nmol ml−1 for cell lysates, and 0.02, 0.03 and 0.05 nmol ml−1 for supernatant samples, respectively.| Compound | Cell model | [M + H]+ | Acc. mass | M.F. |
|---|---|---|---|---|
| a Abbreviations: Acc. mass, accurate mass; MF, molecular formula. | ||||
| Genistein | HUVEC/Hep-G2/Caco-2 | 271 | 271.0602 | C15H10O5 |
| Genistein-glucuronide | Hep-G2/Caco-2 | 447 | 447.0919 | C21H18O11 |
| Genistein-sulfate | Hep-G2/Caco-2 | 351 | 351.0176 | C15H10O8S |
| Methoxy-genistein-glucuronide | HUVEC | 477 | 477.1028 | C22H20O12 |
| Methoxy-genistein-sulfate | HUVEC | 381 | 366.0275 | C16H12O9S |
| Daidzein | HUVEC/Hep-G2/Caco-2 | 255 | 255.0655 | C15H10O4 |
| Daidzein-sulfate | Hep-G2/Caco-2 | 335 | 335.0229 | C15H10O7S |
| Daidzein-glucuronide | Hep-G2/Caco-2 | 431 | 431.0973 | C21H18O10 |
| Methoxy-daidzein-glucuronide | HUVEC | 461 | 461.1078 | C22H20O11 |
| Equol | HUVEC/Hep-G2/Caco-2 | 243 | 243.1018 | C15H14O3 |
| Equol-sulfate | Hep-G2 | 323 | 323.0584 | C15H14O6S |
The identity of glucuronides, sulfates, methoxy-glucuronides and methoxy-sulfates of isoflavones were further confirmed by treating samples with β-glucuronidase/sulfatase enzymes, which resulted in the appearance of their aglycones or methoxylated forms. Fig. 3 and 4 show representative chromatograms of HUVEC supernatants after incubation with Ge and De, before (Fig. 3a and 4a, respectively) and after enzymatic hydrolysis (Fig. 3b and 4b, respectively). Enzymatic hydrolysis led to the formation of methoxylated forms of Ge and De, which corresponded to [M − H]+ with m/z 301.0706 and m/z 285.0761, respectively. This confirmed that the methylation occur in the aglycone, not in the glucuronide or sulfate moiety. The increase in mass of 30 units between the glucuronide or sulfate metabolites and the methoxylated conjugates (Table 1) corresponds to the addition of a methoxy group, which can be explained by a hydroxylation step leading to the formation of a catechol moiety, followed by methylation by COMT, as it has been previously demonstrated.24,25
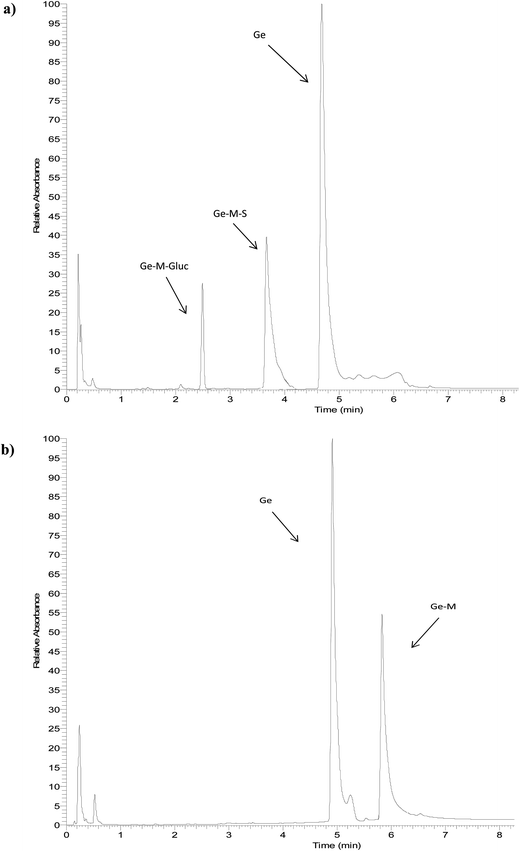 | ||
| Fig. 3 Representative UHPLC-UV chromatograms of HUVEC supernatant (a) incubated with genistein for 2 h and (b) after enzymatic hydrolysis with β-glucuronidase/sulfatase. | ||
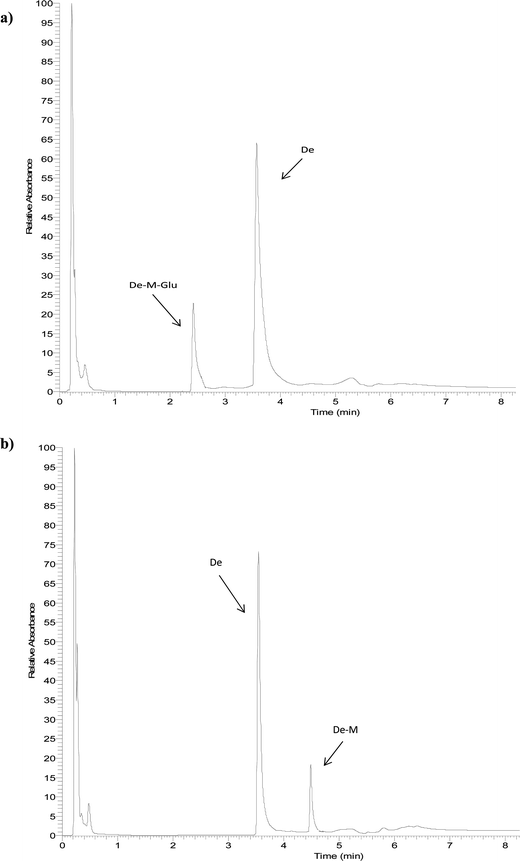 | ||
| Fig. 4 Representative UHPLC-UV chromatograms of supernatant of HUVEC incubated with daidzein for 2 h (a) and after enzymatic hydrolysis (b). | ||
Uptake and metabolism of isoflavones in HUVEC cells
When 1, 10 or 100 μM of Ge was incubated with HUVEC cells for 2 hours, Ge and its methoxy-glucuronide and methoxy-sulfate metabolites were detected in both supernatant and cells (Table 2), indicating that Ge is taken up and metabolized by endothelial cells. The amount of Ge (and metabolites) associated with cells represented 6–7% of the total initial Ge. The percentage of total conjugated Ge in the supernatant and cell lysate was around 14–18%, with 82–86% remaining in the unconjugated form. Total methoxy-genistein-glucuronide and methoxy-genistein-sulfate represented 25 and 55% of the metabolized aglycone, respectively (Table 2).| 1 μM | 10 μM | 100 μM | ||||
|---|---|---|---|---|---|---|
| Supernatant | Cell lysate (pmol per mg protein) | Supernatant | Cell lysate (pmol per mg protein) | Supernatant | Cell lysate (pmol per mg protein) | |
| a Abbreviations: Ge-Gluc, genistein-glucuronide; Ge-S, genistein-sulfate; M-Ge-Gluc, methoxy-genistein-glucuronide; M-Ge-S, methoxy-genistein-sulfate; De-Gluc, daidzein-glucuronide; M-De-Gluc, methoxy-daidzein-glucuronide; De-S, daidzein-sulfate; Eq-S, equol-sulfate; nd, not detected. | ||||||
| Genistein | 0.83 ± 0.05 | 0.05 ± 0.01 (60 ± 3) | 8.34 ± 0.09 | 0.48 ± 0.05 (571 ± 58) | 78.95 ± 4.85 | 0.61 ± 0.03 (726 ± 35) |
| Ge-Gluc | nd | nd | nd | nd | nd | nd |
| Ge-S | nd | nd | nd | nd | nd | nd |
| M-Ge-Gluc | 0.05 ± 0.01 | nd | 0.46 ± 0.02 | 0.08 ± 0.01 (95 ± 2) | 4.97 ± 0.39 | 1.62 ± 0.09 (1928 ± 106) |
| M-Ge-S | 0.09 ± 0.01 | 0.02 ± 0.01 (24 ± 1) | 0.68 ± 0.06 | 0.21 ± 0.02 (250 ± 19) | 7.85 ± 0.75 | 3.47 ± 0.25 (4131 ± 297) |
| Total | 0.97 ± 0.09 | 0.07 ± 0.02 (84 ± 3) | 0.95 ± 0.09 | 0.79 ± 0.06 (916 ± 57) | 91.77 ± 8.32 | 5.70 ± 0.52 (6785 ± 421) |
| % Recovery | 99% | 7% | 97% | 7% | 94% | 6% |
| Daidzein | 0.81 ± 0.04 | nd | 7.93 ± 0.56 | 0.38 ± 0.02 (452 ± 22) | 78.98 ± 6.05 | 1.46 ± 0.09 (1738 ± 104) |
| De-Gluc | nd | nd | nd | nd | nd | nd |
| M-De-Gluc | 0.08 ± 0.01 | nd | 0.84 ± 0.07 | 0.51 ± 0.04 (607 ± 48) | 10.61 ± 1.01 | 4.44 ± 0.55 (5286 ± 598) |
| De-S | nd | nd | nd | nd | nd | nd |
| Total | 0.89 ± 0.04 | 0 | 8.77 ± 0.58 | 0.89 ± 0.05 (1059 ± 56) | 89.59 ± 8.89 | 5.90 ± 0.84 (7024 ± 609) |
| % Recovery | 91% | 0% | 89% | 10% | 92% | 6% |
| Equol | 0.89 ± 0.07 | 0.12 ± 0.02 (143 ± 7) | 9.14 ± 0.84 | 0.08 ± 0.01 (95 ± 4) | 89.15 ± 3.85 | 10.84 ± 1.33 (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 905 ± 1311) 905 ± 1311) |
| Eq-S | nd | nd | nd | nd | nd | nd |
| Total | 0.89 ± 0.07 | 0.12 ± 0.02 (143 ± 7) | 9.14 ± 0.84 | 0.08 ± 0.01 (95 ± 4) | 89.15 ± 3.85 | 10.84 ± 1.33 (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 905 ± 1311) 905 ± 1311) |
| % Recovery | 95% | 13% | 96% | 8% | 95% | 12% |
When De was incubated with HUVEC cells, De and a methoxy-glucuronide metabolite were detected in the supernatant and cell lysate (Table 2), indicating that De is also taken up and metabolized by endothelial cells. Around 5–8% of De and its metabolite were found associated to cells at 10 and 100 μM. Neither De nor its metabolites were found associated to cells at the lower concentrations tested, probably because concentrations were lower than the limit of detection of our method. The methoxy-glucuronide metabolite represented around 10–15% of the initial De.
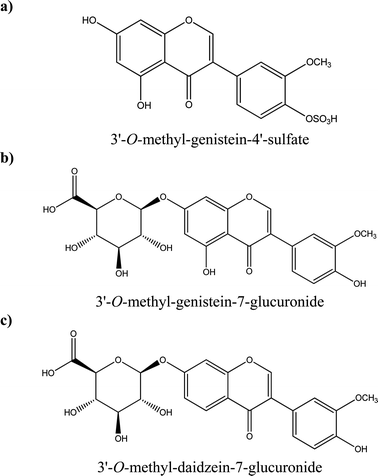
Some examples of tentative chemical structures of the isoflavone metabolites identified in HUVEC cells are shown in Fig. 5. The methoxy group could be mostly likely in position 3′ or 4′;24 whereas the glucuronide or sulfate moiety could be in position 3′, 4′, 5 or 7 for genistein and 3′,4′ and 7 for daidzein.
No Eq metabolites were detected in the supernatant or cell lysate at any concentration after the incubation of Eq in HUVEC. Around 90% of Eq was found in the supernatant, with around 8–12% of Eq found in the cell lysate at 1, 10 and 100 μM (Table 2).
Uptake and metabolism of isoflavones in HepG2
Incubation of 1, 10, and 100 μM of Ge with HepG2 cells for 2 hours led to the detection of a genistein-glucuronide and a genistein-sulfate in supernatant and cells (Table 3). Around 50% of the initial Ge was metabolized at 1 and 10 μM, and around 20% at 100 μM. Practically all Ge associated to cells was metabolized, being the glucuronide conjugates the most abundant metabolites representing 60–70% of the total, and genistein-sulfate representing 30–40% (Table 3). At 1 μM, the aglycone was not detected in the supernatant neither in the cell lysate.| 1 μM | 10 μM | 100 μM | ||||
|---|---|---|---|---|---|---|
| Supernatant | Cell lysate (pmol per mg protein) | Supernatant | Cell lysate (pmol per mg protein) | Supernatant | Cell lysate (pmol per mg protein) | |
| a Abbreviations: Ge-Gluc, genistein-glucuronide; Ge-S, genistein-sulfate; M-Ge-Gluc, methoxy-genistein-glucuronide; M-Ge-S, methoxy-genistein-sulfate; De-Gluc, daidzein-glucuronide; M-De-Gluc, methoxy-daidzein-glucuronide; De-S, daidzein-sulfate; Eq-S, equol-sulfate; nd, not detected. | ||||||
| Genistein | nd | nd | 2.32 ± 0.25 | nd | 81.77 ± 6.11 | 0.91 ± 0.02 (276 ± 6) |
| Ge-Gluc | 0.28 ± 0.01 | 0.07 ± 0.01 (21 ± 3) | 2.14 ± 0.19 | 0.47 ± 0.02 (142 ± 6) | 8.54 ± 0.77 | 3.88 ± 0.20 (1176 ± 60) |
| Ge-S | 0.11 ± 0.01 | 0.04 ± 0.01 (12 ± 1) | 1.66 ± 0.14 | 0.16 ± 0.01 (48 ± 3) | 4.03 ± 0.32 | 1.22 ± 0.04 (370 ± 12) |
| M-Ge-Gluc | nd | nd | nd | nd | nd | nd |
| M-Ge-S | nd | nd | nd | nd | nd | nd |
| Total | 0.39 ± 0.01 | 0.11 ± 0.01 (33 ± 3) | 6.12 ± 0.74 | 0.63 ± 0.03 (190 ± 5) | 94.34 ± 9.28 | 6.01 ± 0.37 (1822 ± 71) |
| % Recovery | 40% | 11% | 62% | 6% | 96% | 6% |
| Daidzein | nd | nd | 5.09 ± 0.03 | nd | 78.63 ± 4.39 | 1.18 ± 0.03 (358 ± 9) |
| De-Gluc | 0.07 ± 0.01 | nd | 4.15 ± 0.02 | 0.63 ± 0.03 (191 ± 9) | 14.03 ± 1.22 | 4.45 ± 0.23 (1348 ± 69) |
| M-De-Gluc | nd | nd | nd | nd | nd | nd |
| De-S | nd | nd | nd | nd | nq | nq |
| Total | 0.07 ± 0.01 | 0 | 9.24 ± 0.06 | 0.63 ± 0.03 (191 ± 9) | 92.66 ± 7.73 | 5.63 ± 0.27 (1702 ± 669) |
| % Recovery | 7% | 0% | 94% | 6% | 94% | 6% |
| Equol | 0.93 ± 0.06 | 0.07 ± 0.01 (21 ± 1) | 8.72 ± 0.07 | 1.26 ± 0.09 (382 ± 27) | 96.71 ± 3.47 | 3.28 ± 0.12 (994 ± 32) |
| Eq-S | nd | nd | nq | nd | 4.11 ± 0.25 | nd |
| Total | 0.93 ± 0.06 | 0.07 ± 0.01 (21 ± 1) | 8.72 ± 0.07 | 1.26 ± 0.09 (382 ± 27) | 100.88 ± 5.26 | 3.28 ± 0.12 (994 ± 32) |
| % Recovery | 99% | 7% | 93% | 14% | 108% | 3% |
When incubation of HepG2 cells was carried out with De for 2 hours, De and daidzein-glucuronide and daidzein-sulfate metabolites were detected in both supernatant and cells. However, daidzein-sulfate could not be quantified because the level of the metabolite was lower than the limit of quantification (Table 3). At 1 μM, the aglycone De was not detected in the supernatant or cell lysate, and only the glucuronide metabolite was found in the supernatant (∼7% of total De).
After the incubation of Eq in HepG2 cells, Eq and equol-sulfate were detected in the supernatant and/or cells. However, the sulfate metabolite of equol could only be quantified in the supernatant at the highest concentration. Around 90% of Eq was found in the supernatant, and 3–13% associated to cells (Table 3).
Uptake, transport and metabolism of isoflavones in the Caco-2 monolayer model
After incubation with Ge for 2 hours, Ge-glucuronide and Ge-sulfate metabolites were detected in the apical side, basolateral side and cells. Ge-sulfate could not be quantified because the level of the metabolite present was lower than the limit of quantification. The glucuronide metabolite of Ge was observed to be associated with Caco-2 cells at all concentrations tested (Table 4). In addition, at the highest concentration Ge was also detected in the cell lysate. The glucuronide conjugate was excreted to the apical side (∼30–40%) and to the basolateral side (∼40–50%). Significant amounts of Ge were also detected in the basolateral side, suggesting that both the aglycone and glucuronide transverse the monolayer. At 1 and 10 μM, around 80% of the initial Ge was glucuronidated, whereas at 100 μM only 12% was found as Ge-glucuronide (Table 4).| 1 μM | 10 μM | 100 μM | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Apical | Basolateral | Cell lysate (pmol per mg protein) | Apical | Basolateral | Cell lysate (pmol per mg protein) | Apical | Basolateral | Cell lysate (pmol per mg protein) | |
| a Abbreviations: Ge-Gluc, genistein-glucuronide; Ge-S, genistein-sulfate; M-Ge-Gluc, methoxy-genistein-glucuronide; M-Ge-S, methoxy-genistein-sulfate; De-Gluc, daidzein-glucuronide; M-De-Gluc, methoxy-daidzein-glucuronide; De-S, daidzein-sulfate; Eq-S, equol-sulfate; nd, not detected. | |||||||||
| Genistein | 0.18 ± 0.01 | nd | nd | 2.13 ± 0.11 | 0.41 ± 0.02 | nd | 66.26 ± 2.33 | 14.05 ± 0.98 | 6.12 ± 0.04 (4451 ± 29) |
| Ge-Gluc | 0.34 ± 0.02 | 0.43 ± 0.03 | 0.03 ± 0.01 (22 ± 1) | 3.49 ± 0.22 | 4.48 ± 0.03 | 0.39 ± 0.01 (284 ± 8) | 3.87 ± 0.25 | 4.65 ± 0.03 | 3.77 ± 0.02 (2742 ± 15) |
| Ge-S | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| M-Ge-Gluc | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| M-Ge-S | nd | nd | nd | nd | nd | nd | nq | nq | nq |
| Total | 0.52 ± 0.02 | 0.43 ± 0.03 | 0.03 ± 0.01 (22 ± 1) | 5.62 ± 0.38 | 4.48 ± 0.06 | 0.39 ± 0.01 (284 ± 8) | 70.13 ± 3.60 | 18.70 ± 1.38 | 9.89 ± 0.07 (7193 ± 22) |
| % Recovery | 53% | 44% | 3% | 57% | 50% | 4% | 71% | 19% | 10% |
| Daidzein | 0.23 ± 0.01 | 0.11 ± 0.01 | nd | 3.19 ± 0.23 | 1.87 ± 0.07 | 0.41 ± 0.02 (298 ± 12) | 73.87 ± 4.52 | 10.49 ± 0.85 | 2.07 ± 0.01 (1505 ± 7) |
| De-Gluc | 0.34 ± 0.02 | 0.23 ± 0.01 | 0.03 ± 0.01 (22 ± 1) | 2.16 ± 0.15 | 1.59 ± 0.08 | 0.64 ± 0.04 (465 ± 30) | 3.58 ± 0.26 | 4.31 ± 0.02 | 2.46 ± 0.01 (1789 ± 8) |
| M-De-Gluc | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| De-S | nd | nd | nd | nd | nd | nd | nq | nq | nq |
| Total | 0.57 ± 0.02 | 0.34 ± 0.01 | 0.03 ± 0.01 (22 ± 1) | 5.35 ± 0.47 | 3.46 ± 0.11 | 1.05 ± 0.05 (763 ± 32) | 77.45 ± 6.54 | 14.80 ± 0.90 | 4.53 ± 0.01 (3294 ± 9) |
| % Recovery | 58% | 35% | 3% | 55% | 36% | 6% | 78% | 15% | 5% |
| Equol | 0.71 ± 0.01 | 0.22 ± 0.01 | 0.08 ± 0.01 (58 ± 2) | 6.94 ± 0.44 | 2.22 ± 0.16 | 0.82 ± 0.05 (596 ± 36) | 62.65 ± 3.77 | 22.46 ± 1.25 | 14.89 ± 1.00 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 829 ± 707) 829 ± 707) |
| Eq-S | nd | nd | nd | nd | nd | nd | nd | nd | |
| Total | 0.71 ± 0.01 | 0.22 ± 0.01 | 0.08 ± 0.01 (58 ± 2) | 6.94 ± 0.44 | 2.22 ± 0.16 | 0.82 ± 0.05 (596 ± 36) | 62.65 ± 3.77 | 22.46 ± 1.25 | 14.89 ± 1.00 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 829 ± 707) 829 ± 707) |
| % Recovery | 75% | 23% | 8% | 74% | 23% | 8% | 67% | 23% | 16% |
Incubation of Caco-2 cell monolayers with De led to the detection of De, a glucuronide and a sulfate metabolite in the cell lysate (Table 4). De-sulfate was detected only at the higher concentration tested and could not be quantified because levels were lower than the limit of quantification. When De was incubated at 1 and 10 μM, around 50% of the glucuronide conjugate of De was found in the apical side, and at the higher concentration tested, around 35%. Around 15% of the aglycone and 40% of its metabolite were found in the basolateral side. De-glucuronide represented around 50% of the initial De at 1 and 10 μM, and 10% at the higher concentration tested (Table 4).
No Eq conjugates were detected in the apical side, basolateral side or cell lysate at any concentration after the incubation of Eq in Caco-2 cell monolayers (Table 4). Around 20% of Eq was transported to the basolateral side and 8–15% was found associated to cells.
Discussion
In this work, the uptake and intracellular metabolism of the isoflavones Ge, De and its microbial metabolite Eq in endothelial cells were investigated, and compared with cell models of liver and small intestine cells. We showed that Ge is metabolized into methoxy-genistein-glucuronide and methoxy-genistein-sulfate, and De is converted into methoxy-daidzein-glucuronide in HUVEC. This indicates that, beside the liver and the small intestine, endothelial cells can also metabolize isoflavones by the action of UDT-glucuronosyltransferases (UGT), sulfotransferases (SULT), and catechol methyl transferases (COMT). The isoflavone metabolites were found in the cell lysate and in the cell medium, suggesting that Ge and De are taken up by endothelial cells, methoxylated, glucuronidated, and sulfated before export to the medium. We cannot discard though that additional conjugation and/or deconjugation reactions might have occurred in the cell medium; however we did not see any of these reactions occurring in the cell medium when testing the stability of Ge, De and Eq, suggesting that the cell medium alone has no glucuronidase/sulfatase/UGT, SULT or COMT activity (Fig. 2).In vivo, glucuronidation and sulfation seem to be the major metabolic pathways in the metabolism of isoflavones, since glucuronides, sulfates, and sulfoglucuronides of Ge and De, and also freely circulating aglycones have been shown to be the major human plasma and urinary metabolites of isoflavones.23 However, little evidence exists regarding the formation of methoxylated isoflavone metabolites. Ge, De or Eq cannot be subjected to methylation by COMT, as they do not have catechol moieties. However, it has been demonstrated in human liver microsomes that Ge and De are converted to hydroxylated metabolites by cytochrome P450 enzymes.24 Because some of these aromatic hydroxylated products of Ge and De contain a catechol moiety, these metabolites could undergo further biotransformation by COMT in HUVEC, leading to the formation of Ge and De methoxylated sulfate and methoxylated glucuronide derivatives. Treatment of samples with glucuronidase and sulfatase led to the formation of methoxylated metabolites of Ge and De (Fig. 3 and 4), which confirms that the methoxy group is attached to the aglycone and not to the glucuronide or sulfate moiety, supporting the action of COMT in the cells. Indeed, COMT has been reported to be present in HUVEC cells,33 and we have recently identified the formation of O-methyl-glucuronide and O-methyl-sulfates derivatives of (−)-epicatechin in HUVEC.34 Kulling et al. and Heinonen et al. have identified mono- and dimethoxylated conjugates of Ge and De in trace amounts in human urine samples after soy supplementation,24,25 which suggests that methylation of isoflavones does occur in vitro but in vivo only to a very minor extent. As the metabolism of isoflavones in HUVEC cells in vitro is not the same than the observed in human in vivo, caution should be taken when reaching conclusions on the investigations of mechanisms of action of isoflavones in vitro.
It is presently unknown whether the major in vivo conjugates of isoflavones are taken up and further metabolized by endothelial cells. In our previous work, no uptake or metabolism of the major in vivo metabolites of (−)-epicatechin were observed after incubation with HUVEC cells.34 In agreement with this, it has been reported that glucuronide conjugates of epicatechin and quercetin were unable to enter dermal fibroblasts and cortical neurons.35–37 This suggests that the higher polarity of glucuronides and sulfates may limit their capacity to enter the cells and thus, being metabolized further. Further work is needed in order to confirm that this is the case for genistein and daidzein conjugates.
When the concentration of isoflavones in the three cell models investigated here was normalized per mg of protein, the uptake of genistein and daidzein in HUVECs was found to be higher than in HepG2 and Caco-2 monolayer (Tables 2–4), which is in agreement with our previous work with (−)-epicatechin,34 suggesting that endothelial cell metabolism may be of relevance in vivo.
Beside investigations on hepatic and small intestine cell models, some evidence exist regarding the intracellular metabolism of other flavonoids, such as flavanols, flavonols, and flavanones, in dermal fibroblasts, central nervous system and cancer cells,35–41 however very few reports on endothelial metabolism of flavonoids exist. We have recently shown that the flavonoid (−)-epicatechin was taken up and metabolized into 3′-O-methyl-(−)-epicatechin-7-β-D-glucuronide and 3′-O-methyl-(−)-epicatechin-7-sulfate conjugates after 1 hour of incubation,34 which is in agreement with the data presented here for isoflavones, where formation of methoxy-glucuronides and methoxy-sulfates took place in HUVEC cells. Anthocyanins, another subclass of flavonoids, have also been shown to be taken up and metabolized into methylated conjugates after incubation with EA.hy926 endothelial cells.42 To our knowledge, this is the first study to address the uptake and intracellular metabolism of isoflavones in endothelial cells (HUVEC).
In the present work, different cell types led to different uptake and metabolism of isoflavones. The metabolism of tumour/transformed cell lines such as HepG2 and Caco-2 might not be exactly similar to that of primary hepatocytes and colonocytes, although they still represent a fair model for a comparative study to primary HUVEC cells. In the HepG2 cell model, glucuronide and sulfate conjugates of Ge and De were detected in both the supernatant and cell lysate, in agreement with previous work, where the 7-O-glucuronide conjugate was the major metabolite of De and Ge in hepatocytes, together with sulfate conjugates.29,30 In the Caco-2 monolayer, we observed that Ge and De were taken up by cells, glucuronidated (and, to a smaller extent, sulfated) and excreted to the apical side (∼30–40%) and to the basolateral side (∼40–50%) (Table 3). These results are also in agreement with previously reported data in Caco-2 cell monolayers.26,27
The microbial metabolite of daidzein, equol, was found to be associated to HUVEC, HepG2 and Caco-2 cells at significant concentrations (Tables 2–4). In HepG2 cells, a sulfate metabolite of Eq was excreted to the medium, suggesting that sulfation is the main metabolic pathway of equol in hepatocyte cell model. This is in accordance to an earlier study in which Eq was mainly sulfated (∼95% of initial Eq) in HepG2 cells after 4 days incubation.43 However, in our case conjugated equol represented around 4% of the initial Eq, and was only found at the higher concentrations tested, which may be due to the shorter incubation time with HepG2 cells (2 h vs. 4 days). In contrast, Schwen et al. reported glucuronidation as the primary pathway for the metabolism of Eq in human hepatocytes (∼73% of initial Eq), with lesser sulfation metabolism (∼22% of initial Eq).44 We did not find any Eq conjugates being formed in HUVEC and Caco-2 cell models. No data available about the metabolism of Eq in HUVEC was found in the literature. In Caco-2 monolayers, Eq was shown to be metabolized into phase II conjugates, determined using enzymatic treatment with glucuronidase and sulfatase, so it is unknown whether the metabolites formed were glucuronide or sulfate conjugates.45 Differences in the analytical methodology and incubation times may explain the differences observed between studies, an it is possible that the limit of detection of our method was not low enough to detect glucuronide conjugates, although the recoveries obtained respect to the initial Eq were around 100% (Tables 2–4). In humans, Eq circulates in plasma and is excreted in urine predominantly as a glucuronide conjugate, and to a lesser extent as a sulfate.21,25 Thus, according to the work presented here, equol in vitro metabolism does not reflect human in vivo metabolism.
Few reports have been published describing the biological activity of isoflavone conjugates, such as the weak estrogenic effect and the activation of human natural killer cells of Ge and De glucuronides,46 the inhibitory effect of daidzein-4′,7-disulfate on the sterol sulfatase in hamster liver microsomes,47 the stimulatory effect of daidzein-7-glucuronide-4′-sulfate on the growth of MCF-7 cells,48 and the hypotensive and vasodilator effects of De sulfates in rats.49 Rimbach et al. reported that sulfation of Ge decreased its antioxidant activity, anti-aggregatory effect and its impact on monocyte and endothelial function in vitro.50 Although the mechanisms by which isoflavones and their metabolites mediate their observed effects have not been fully established, methylation of Ge and De could have an important role in the metabolism of isoflavones in some target cells. Further studies are needed to understand the cellular bioactivity of isoflavones and their metabolites.
A limitation of this work is that the major isoflavone metabolites were quantified as their corresponding aglycone, due to the lack of authentic glucuronidated, sulfated and methoxylated standards. Enzymatic treatment with glucuronidase/sulfatase was not used for quantification as methoxylated compounds cannot be quantified with this method without authentic standards, and there are also limitations concerning quantification of glucuronides and sulfates, such as the inability to differentiate between glucuronide and sulfate concentrations, or batch to batch variations in the activity and specificity of the enzymes. In addition, the limits of quantification of the method were not low enough to quantify all the metabolites identified in the cell samples, in particular the sulfate conjugates of Ge and De. We cannot discard that additional metabolites present in the samples below our limits of detection were formed and not accounted for here.
Conclusion
Our data shows that the isoflavones Ge and De are taken up by endothelial cells and are metabolized by phase II enzymes into their methoxylated, glucuronide and sulfate conjugates. However, the microbial metabolite equol is taken up by endothelial cells but is not metabolized. These findings suggest that endothelial cell metabolism needs to be taken into account when investigating the mechanisms of action of isoflavones in the cardiovascular system. Further work in this area is warranted, as the uptake and metabolism of flavonoids in human cells will likely determine their biological actions.Acknowledgements
The authors would like to thank the Direcció General de Recerca of the Generalitat de Catalunya (2014–1438 SGR) for their support and the Ministerio de Educación y Ciencia (Spain) for a grant to Dr Natalia Toro-Funes.References
- H. B. Patisaul and W. Jefferson, Front. Neuroendocrin., 2010, 31, 400–421 CrossRef CAS PubMed.
- W. O. Song, O. K. Chun, I. Hwang, H. S. Shin, B. G. Kim, K. S. Kim, S. Y. Lee, D. Shin and S. G. Lee, J. Med. Food, 2010, 10, 571–580 CrossRef PubMed.
- A. C. Moreira, A. M. Silva, M. S. Santos and V. A. Sardão, J. Steroid Biochem., 2014, 143, 61–71 CrossRef CAS PubMed.
- H. J. Teede, B. P. McGrath, L. DeSilva, M. Cehun, A. Fassoulakis and P. J. Nestel, Arterioscler., Thromb., Vasc. Biol., 2003, 23, 1066–1071 CrossRef CAS PubMed.
- P. J. Nestel, T. Yamashita, T. Sasahara, S. Pomeroy, A. Dart, P. Komesaroff, A. Owen and M. Abbey, Arterioscler., Thromb., Vasc. Biol., 1997, 17, 3392–3398 CrossRef CAS.
- X. X. Liu, S. H. Li, J. Z. Chen, K. Sun, X. J. Wang, X. G. Wang and R. T. Hui, Nutr. Metab. Cardiovas., 2012, 22, 463–470 CrossRef CAS PubMed.
- F. Squadrito, H. Marini, A. Bitto, D. Altavilla, F. Polito, E. B. Adamo, R. D'Anna, V. Arcoraci, B. P. Burnett, L. Minutoli, A. Di Benedetto, G. Di Vieste, D. Cucinotta, C. de Gregorio, S. Russo, F. Corrado, A. Saitta, C. Irace, S. Corrao and G. Licata, J. Clin. Endocrinol. Metab., 2013, 98, 3366–3374 CrossRef CAS PubMed.
- M. Lilamand, E. Kelaiditi, S. Guyonnet, R. A. Incalzi, A. Raynaud-Simon, B. Vellas and M. Cesari, Nutr. Metab. Cardiovas., 2014, 24, 698–704 CrossRef CAS PubMed.
- F. Squadrito, D. Altavilla, N. Morabito, A. Crisafulli, R. D'Anna, F. Corrado, P. Ruggeri, G. M. Campo, G. Calapai, A. P. Caputi and G. Squadrito, Atherosclerosis, 2002, 163, 339–347 CrossRef CAS PubMed.
- G. Rimbach, C. Boesch-Saadatmandi, J. Frank, D. Fuchs, U. Wenzel, H. Daniel, W. L. Hall and P. D. Weinberg, Food Chem. Toxicol., 2008, 46, 1308–1319 CrossRef CAS PubMed.
- C. M. De Andrade, M. F. Silva de Sá and M. R. Torqueti-Toloi, Climateric, 2012, 15, 186–194 CrossRef CAS PubMed.
- H. Zhang, F. Zheng, J. Zhao, D. Guo and X. Chen, Arch. Med. Res., 2013, 44, 13–20 CrossRef CAS PubMed.
- A. B. Santhakumar, A. C. Bulmer and I. A. Singh, J. Hum. Nutr. Diet., 2014, 27, 1–21 CrossRef CAS PubMed.
- J. W. Anderson, B. M. Johnstone and N. Cook, N. Engl. J. Med., 1995, 333, 276–282 CrossRef CAS PubMed.
- K. Taku, K. Umegaki, Y. Sato, Y. Taki, K. Endoh and S. Watanabe, Am. J. Clin. Nutr., 2007, 85, 1148–1156 CAS.
- Z. M. Liu, S. C. Ho, Y. M. Chen, J. Liu and J. Woo, PLoS One, 2014, 9, e87861 Search PubMed.
- Y. Qin, K. Niu, Y. Zeng, P. Liu, L. Yi, T. Zhang, Q. Y. Zhang, J. D. Zhu and M. T. Mi, Cochrane Database Syst. Rev., 2013, 6, CD009518 Search PubMed.
- G. Mainini, M. Torella, M. C. Di Donna, E. Esposito, S. Ercolano, R. Correa, G. Cucinella, L. Stradella, A. Luisi, A. Basso, F. V. Cerreto, R. Cicatiello, M. Matteo and P. De Franciscis, Clin. Exp. Obstet. Gyn., 2012, 40, 337–341 Search PubMed.
- S. R. Shelnutt, C. O. Cimino, P. A. Wiggins, M. J. J. Ronis and T. M. Badger, Am. J. Clin. Nutr., 2002, 76, 588–594 CAS.
- K. Németh, G. W. Plumb, J. G. Berrin, N. Juge, R. Jacob, H. I. Naim, G. Williamson, D. L. Swallow and P. A. Kroon, Eur. J. Nutr., 2003, 42, 29–42 CrossRef PubMed.
- K. D. Setchell and C. Clerici, J. Nutr., 2010, 140, 1355S–1362S CrossRef CAS PubMed.
- S. Barnes, Lymphat. Res. Biol., 2010, 8, 89–98 CrossRef CAS PubMed.
- K. Hosoda, T. Furuta, A. Yokokawa and K. Ishii, Anal. Bioanal. Chem., 2010, 397, 1563–1572 CrossRef CAS PubMed.
- S. E. Kulling, D. M. Honig and M. Metzler, J. Agric. Food Chem., 2001, 49, 3024–3033 CrossRef CAS PubMed.
- S. M. Heinonen, A. Hoikkala, K. Wahala and H. Adlercreutz, J. Steroid Biochem., 2003, 87, 285–299 CrossRef CAS PubMed.
- K. Murota, S. Shimizu, S. Miyamoto, T. Izumi, A. Obata, M. Kikuchi and J. Terao, J. Nutr., 2002, 132, 1956–1961 CAS.
- J. Chen, H. Lin and M. Hu, Cancer Chemoth. Pharm., 2005, 55, 159–169 CrossRef CAS PubMed.
- B. Wu, K. Kulkarni, S. Basu, S. Zhang and M. Hu, J. Pharm. Sci., 2011, 100, 3655–3681 CrossRef CAS PubMed.
- J. Bursztyka, E. Perdua, J. Tulliez, L. Debrauwer, G. Delous and C. Canlet, Food Chem. Toxicol., 2008, 48, 939–948 CrossRef PubMed.
- Y. Chen, C. Huang, T. Zhou and G. Chen, Basic Clinical Pharmacol., 2008, 103, 553–559 CrossRef CAS PubMed.
- B. K. Chacko, R. T. Chandler, T. L. D'Alessandro, A. Mundhekar, N. H. Khoo, N. Botting, S. Barnes and R. P. Patel, J. Nutr., 2007, 137, 351–356 CAS.
- N. Toro-Funes, I. Odriozola-Serrano, J. Bosch-Fusté, M. L. Latorre-Moratalla, M. T. Veciana-Nogués, M. Izquierdo-Pulido and M. C. Vidal-Carou, Food Chem., 2012, 135, 2832–2838 CrossRef CAS PubMed.
- Y. Steffen, C. Gruber, T. Schewe and H. Sies, Arch. Biochem. Biophys., 2008, 469, 209–219 CrossRef CAS PubMed.
- A. Rodriguez-Mateos, N. Toro-Funes, T. Cifuentes-Gomez, M. Cortese-Krott, C. Heiss and J. P. Spencer, Arch. Biochem. Biophys., 2014, 559, 17–23 CrossRef CAS PubMed.
- J. P. Spencer, M. M. Abd-el-Mohsen and C. Rice-Evans, Arch. Biochem. Biophys., 2004, 423, 148–161 CrossRef CAS PubMed.
- J. P. Spencer, G. G. Kuhnle, R. J. Williams and C. Rice-Evans, Biochem. J., 2003, 372, 173–181 CrossRef CAS PubMed.
- J. P. Spencer, H. Schroeter, A. J. Crossthwaithe, G. Kuhnle, R. J. Willians and C. Rice-Evans, Free Radicals Biol. Med., 2001, 31, 1139–1146 CrossRef CAS PubMed.
- J. P. Spencer, H. Schroeter, G. Kuhnle, S. K. Srai, R. M. Tyrrell, U. Hahn and C. Rice-Evans, Biochem. J., 2001, 354, 493–500 CrossRef CAS PubMed.
- K. Vafeiadou, D. Vauzour, A. Rodriguez-Mateos, M. Whiteman, R. J. Williams and J. P. Spencer, Arch. Biochem. Biophys., 2008, 478, 195–200 CrossRef CAS PubMed.
- A. R. Proteggente, S. Basu-Modak, G. Kuhnle, M. J. Gordon, K. Youdim, R. Tyrrell and C. A. Rice-Evans, Photochem. Photobiol., 2003, 78, 256–261 CrossRef CAS PubMed.
- M. Salucci, L. A. Stivala, G. Maiani, R. Bugianesi and V. Vannini, Br. J. Cancer, 2002, 86, 1645–1651 CrossRef CAS PubMed.
- L. Ziberna, F. Tramer, S. Moze, U. Vrhovsek, F. Mattivi and S. Passamonti, Free Radicals Biol. Med., 2012, 52, 1750–1759 CrossRef CAS PubMed.
- M. Loukovaara, M. Carson and A. Palotie, Steroids, 1995, 60, 656–661 CrossRef CAS PubMed.
- R. J. Schwen, L. Nguyen and R. L. Jackson, Food Chem. Toxicol., 2012, 50, 2074–2083 CrossRef CAS PubMed.
- K. R. Walsh and M. L. Failla, J. Agric. Food Chem., 2009, 57, 8297–8302 CrossRef CAS PubMed.
- Y. Zhang, T. T. Song, J. E. Cunnick and P. A. Murphy, J. Nutr., 1999, 129, 399–405 CAS.
- C. K. Wong and W. M. Keung, Biochem. Biophys. Res. Commun., 1997, 233, 579–583 CrossRef CAS PubMed.
- J. Kinjo, R. Tsuchihashi, K. Morito, T. Hirose, T. Aomori, T. Nagao, H. Okabe, T. Nohara and Y. Masamune, Biol. Pharm. Bull., 2004, 27, 185–188 CAS.
- Y. X. Cao, X. J. Yang, J. Liu and K. X. Li, Basic Clinical Pharm. Toxicol., 2006, 99, 425–430 CrossRef CAS PubMed.
- G. Rimbach, P. D. Weinberg, S. de Pascual-Teresa, M. G. Alonso, B. A. Ewins, R. Turner, A. N. Minihane, N. Botting, B. Fairley, S. Matsugo, Y. Uchida and A. Cassidy, Biochim. Biophys. Acta, Gen. Subj., 2004, 1670, 229–237 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |