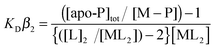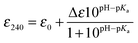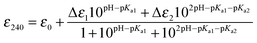DOI:
10.1039/C4MT00250D
(Paper)
Metallomics, 2015,
7, 145-155
Copper trafficking in the CsoR regulon of Streptomyces lividans†
Received
23rd September 2014
, Accepted 12th November 2014
First published on 12th November 2014
Abstract
In the actinobacterium Streptomyces lividans copper homeostasis is controlled through the action of the metalloregulator CsoR. Under copper stress, cuprous ions bind to apo-CsoR resulting in the transcriptional derepression of genes encoding for copper efflux systems involving CopZ-like copper chaperones and CopA-like P-type ATPases. Whether CsoR obtains copper via a protein–protein mediated trafficking mechanism is unknown. In this study we have characterised the copper trafficking properties of two S. lividans CopZ proteins (SLI_1317 and SLI_3079) under the transcriptional control of a CsoR (SLI_4375). Our findings indicate that both CopZ-proteins have cysteine residues in the Cu(I) binding MX1CX2X3C motif with acid–base properties that are modulated for a high cuprous ion affinity and favourable Cu(I)-exchange with a target. Using electrophoretic mobility shift assays transfer of Cu(I) is shown to occur in a unidirectional manner from the CopZ to the CsoR. This transfer proceeds via a shallow thermodynamic affinity gradient and is also kinetically favoured through the modulation of the acid–base properties of the cysteine residues in the Cys2His cuprous ion binding motif of CsoR. Using RNA-seq coupled with the mechanistic insights of Cu(I) transfer between CopZ and CsoR in vitro, we propose a copper trafficking pathway for the CsoR regulon that initially involves the buffering of cytosolic copper by three CopZ chaperones followed by transfer of Cu(I) to CsoR to illicit a transcriptional response.
Introduction
The intricate interplay between proteins that serve to sense, traffic and transport copper (Cu) is essential for maintaining the delicate balance between minimizing the toxicity of unbound Cu and to ensure that the metabolic needs of the cell for Cu are met. Cytosolic Cu metallochaperones play a key role in maintaining cellular Cu homeostasis through tightly binding the cuprous ion and trafficking it to target proteins or membrane transporters.1–6 A common fold utilized by both the Cu chaperone and transporter is the classic ferredoxin βαββαβ-fold, containing within a loop connecting β-sheet 1 to α-helix 1 a MX1CX2X3C metal binding motif that utilizes the two Cys residues for bis-cysteinate Cu(I) coordination.7,8 Atx1 from Saccharomyces cerevisiae was the first Cu(I) metallochaperone with a βαββαβ-fold to be discovered.9,10 Subsequently, other eukaryotic Atx1 members with Cu(I) trafficking roles have been discovered, such as HAH1 (or Atox1) from Homo sapiens. The role of HAH1 is to chaperone Cu(I) to the cytosolic N-terminal metal binding domains (MBD) of the ATP7A and ATP7B Cu-transporters, also known as the Menkes and Wilson proteins, respectively.11 These MBD have the homologous βαββαβ-fold as the Atx1 chaperone and a CXXC motif to bind the Cu(I) ion. The importance of pH and ionization properties of the Cys residues in the CXXC motif of both chaperone and acceptor have been reported to be a determining kinetic factor in eukaryotic Cu(I) trafficking.12
The Atx1 homologue in bacteria is CopZ, first identified in Enterococcus hirae as part of the copYZAB operon.13 In general, the copZ gene is part of an operon that includes a copA gene that encodes for a Cu-exporting P-type ATPase. Unlike their eukaryotic homologs, which can have up to six MBD, bacteria and archaea contain only one or two cytosolic MBD.14,15 Recent studies have indicated that the binding of Cu(I) to the MBD in bacterial systems serves to regulate the activity of the ATPase,16,17 with delivery of Cu(I) to a transmembrane metal binding platform by CopZ required for Cu(I) transport to occur.18,19 Metalloregulatory proteins regulate the expression of metallochaperones and effluxers in bacterial systems.20,21 In E. hirae transcription of the copYZAB operon is regulated by the Zn(II) bound repressor CopY.22 Under Cu stress CopY accepts two Cu(I) ions from CopZ with concomitant release of Zn(II) and derepression of the cop operon.13,23 In eubacteria where a copY gene is absent, a gene encoding for a copper sensitive operon repressor (CsoR) protein has been identified.24 This metalloregulator responds to elevated cytosolic Cu(I) levels through an allosterically induced structural change on binding Cu(I), leading to dissociation from its cognate DNA operator sequence(s),25–27 triggering the increased expression of an efflux system often involving a CopZ-like chaperone and a CopA-like ATPase.
The soil dwelling antibiotic producing Gram-positive bacterium Streptomyces lividans has been shown to have a distinct dependence on Cu ions for its morphological development.28–33 A CsoR (SLI_4375) has been identified in S. lividans and shown to regulate a 3-loci regulon consisting of two copZA-like operons and also its own csoR gene.34 Under Cu stress, the transcript levels of the copZ genes increase concomitantly with the copA genes and eventually the csoR gene.34 Whilst it is accepted that in bacteria containing a CsoR a CopZ can safely route Cu(I) to a CopA-like ATPase for efflux under Cu stress conditions, it has not escaped attention that there may be a role for CopZ in delivering Cu(I) to the DNA bound apo-CsoR,35 in a manner analogous to the delivery of Cu(I) to the CopY repressor in E. hirae.13,23
In the present study we have investigated the chaperoning characteristics of two S. livdians CopZ-like proteins (SLI_1317 and SLI_3079) both under the transcriptional control of CsoR (SLI_4375).36 This has included an evaluation of the pKa properties of the Cys residues in the MX1CX2X3C motif and a determination of the Cu(I) binding affinities. Two site-directed mutants of CopZ-1317 have been prepared (H22G and Y71F) enabling further insight into factors tuning the chaperone properties of this CopZ-pair. We have also investigated in vitro whether Cu(I) exchange can take place between the CopZ proteins and CsoR in the presence of DNA operators and reveal that unidirectional exchange from either CopZ to the CsoR does occur. Finally, from the re-analysis of our previous RNA-seq data using the S. lividans genome as input,36 a model of the CsoR regulon in S. lividans is discussed.
Results and discussion
Three out of the four copZ genes present in S. lividans are part of the CsoR regulon
From previous RNA-seq data and promoter probing experiments a model of the S. lividans regulon under the control of CsoR and its response to Cu stress has been proposed.34 Since then the S. lividans genome sequence has become available36 revealing that the cytosolic Cu(I) handling capacity is much larger than anticipated in the earlier study.34 It is now apparent that four copZ and five P-type ATPase (copA) encoding genes are present on the genome.36 All five P-type ATPases are of the ATPase-IB1_Cu class and four of them are immediately downstream and in an operon with one of the copZ genes. All copZ genes are preceded by a CsoR operator sequence, with one copZ gene, SLI_1317, having two CsoR binding sites. Re-analysis of the RNA-seq data34 with the S. lividans genome sequence as input (previously used S. coelicolor genome as input) indicates on the basis of transcription that CopZ-0895, 1063, 3079 and their cognate P-type ATPases 0896, 1064 and 3080 are the major players in Cu resistance in S. lividans (Table 1). The 1317/1318 couple maximally contributes 7% of total transcription, which may be a consequence of a strong repression because of the two CsoR operator sites present, whereas the expression level of the single P-type ATPase 0845 is even lower (Table 1). Addition of copper induces transcription of all genes with the exception of the single ATPase gene 0845 (Table 1). In the csoR mutant (ΔcsoR), despite all four copZ/ATPase couples having a CsoR operator binding site, only three of these couples are induced (Table 1). Thus the 0895–0896 operon, which is preceded by a predicted CsoR binding site appears not to be within the CsoR regulon. This raises the possibility of the presence of a second Cu-control mechanism using a repressor binding site very similar to that of CsoR. A second Cu-sensing repressor competing with CsoR for the binding sites could also explain why the induction level in the ΔcsoR mutant is lower than found in Cu induced cultures (Table 1). Furthermore, the high capacity for buffering and removing cytoplasmic Cu(I) may contain an answer to the observation that S. lividans does depend on higher Cu levels in the medium for full development compared to other Streptomyces species.29,30,36
Table 1 Transcription analysis of the copZA operons and csoR in S. lividans. RNA-seq data (obtained and reported earlier34) were analyzed with the S. lividans 1326 genome sequence as input.36 CsoR binding sites were identified with PREDetector61
| Annotation |
Gene |
CsoR binding site |
S. lividans 1326 (RPKM)a,b |
+Cu (RPKM)a |
ΔcsoR (RPKM)a |
Fold increase + Cuc |
Fold increase ΔcsoRc |
|
The transcriptomes of wild-type S. lividans 1326 and the ΔcsoR mutant strain were obtained without addition of Cu to the growth medium. The Cu induced transcriptome was obtained after a 2 hour exposure to 400 μM Cu(II).
The selected expression measure is the RPKM and defined as the reads/kb of exon/million mapped reads i.e. dividing the total number of exon reads (in this case one exon per reference sequence) by the number of mapped reads (in millions) times the exon length (in kb).62
Compared to the wild-type strain in the absence of Cu.
|
| P-type ATPase-IB1-Cu |
SLI_0845 |
No |
2 |
0 |
2 |
0 |
1 |
| CopZ |
SLI_0895 |
|
39 |
253 |
33 |
6 |
1 |
| P-type ATPase-IB1-Cu |
SLI_0896 |
Yes |
7 |
24 |
7 |
3 |
1 |
| CopZ |
SLI_1063 |
|
33 |
128 |
67 |
4 |
2 |
| P-type ATPase-IB1-Cu |
SLI_1064 |
Yes |
18 |
65 |
40 |
4 |
2 |
| CopZ |
SLI_1317 |
Yes, two |
0 |
9 |
5 |
≫ |
≫ |
| P-type ATPase-IB1-Cu |
SLI_1318 |
3 |
3 |
3 |
1 |
1 |
| CopZ |
SLI_3079 |
|
16 |
142 |
80 |
9 |
5 |
| P-type ATPase-IB1-Cu |
SLI_380 |
Yes |
29 |
54 |
23 |
2 |
1 |
| CsoR |
SLI_4375 |
Yes |
170 |
212 |
0 |
1 |
0 |
CopZ-1317 and -3079 are monomers in solution and have a βαββαβ-fold
To address the possibility of Cu interplay between CopZ and CsoR in S. lividans two of the three CopZ proteins under the transcriptional control of CsoR as revealed in Table 1 were chosen for investigation. These were CopZ-1317, which has low expression and CopZ-3079, which has high expression (Table 1). A sequence alignment of CopZ proteins and eukaryotic Atx1 homologs is shown in Fig. 1A. Amongst the S. lividans CopZ proteins, 0895 and 1063 have Cys rich N-terminal extensions, which contain CCXXXXXC and CCXXC motifs, respectively. The MX1CX2X3C motif, characteristic of the Atx1 family, in which the Cys residues provide thiolate ligands to Cu(I), is conserved in all sequences, with variations apparent at the X positions between the eukaryotic and prokaryotic species, and a variation in the X1 and X2 positions between the prokaryotic proteins (Fig. 1A). CopZ-1317 and CopZ-3709 contain 77 and 80 amino acids, respectively, and their genes were successfully amplified from S. lividans and sub-cloned into an Escherichia coli expression vector. Using analytical gel filtration chromatography the His-tagged cleaved proteins eluted at 12.3 ml (CopZ-1317) and 12.5 ml (CopZ-3079). These retention volumes are consistent with mass of ∼10 kDa, based on the column calibration curve, and therefore monomer species in solution (Fig. 1B). The purified proteins gave masses determined from denaturing mass spectrometry of 8062 Da for CopZ-1317 (expected 8063 Da) and 8199 Da for CopZ-3079 (expected 8200 Da). The far UV-CD spectra for the apo-proteins clearly indicate that each apo-CopZ is folded (Fig. 1C). Dichroweb analysis of the individual spectrum gave a secondary structure content in agreement to that predicted from Jpred337 (Fig. 1A), providing good experimental support in the absence of tertiary structures that these S. lividans proteins have a βαββαβ-fold, distinctive of Atx1 family members.
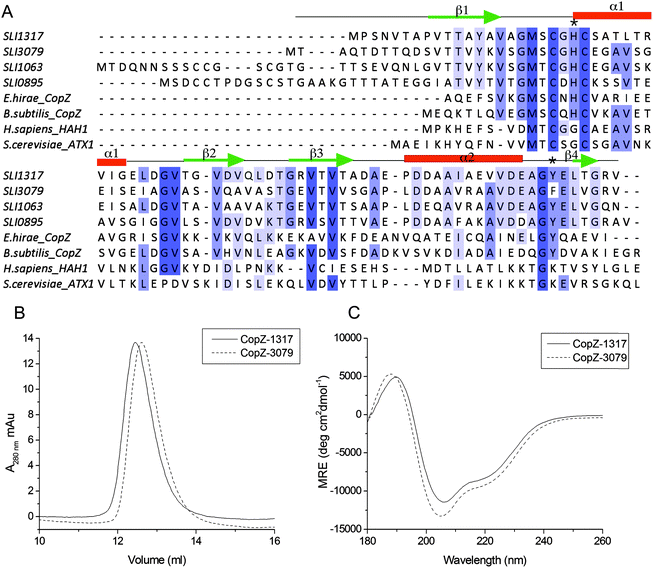 |
| | Fig. 1 Sequence alignments and preliminary characterization of CopZ-1317 and CopZ-3079. (A) Clustal Omega sequence alignments of the four S. lividans CopZ proteins (SLI), E. hirae and B. subtilis CopZ, and the H. sapiens HAH1 and S. cerevisiae Atx1 proteins. Completely and partially conserved residues are boxed in dark to light blue, respectively. The Jpred337 secondary structure prediction for CopZ-1317 and CopZ-3079 pair is indicated and the * indicates the position of H22 and Y71 (CopZ-1317 numbering). (B) Analytical gel filtration chromatography profile for apo-1317 and apo-3079 in 10 mM MOPS pH 7.5, 150 mM NaCl, 2 mM DTT. (C) Far UV-CD spectra at 20 °C, pH 7.0 with protein concentrations of 20 μM. Dichroweb analysis of the individual spectrum predicts for CopZ-1317; 20% α-helix, 22% β-sheet, 20% turns and 38% unordered; for CopZ-3079 23% α-helix, 18% β-sheet, 20% turns and 39% unordered. | |
CopZ-3079 has the higher affinity for Cu(I)
Prior to determining the Cu(I) affinity, the accessibility of the two Cys thiols in the MX1CX2X3C motif was assessed following anaerobic treatment with DTT. It should be noted that unlike 0895 and 1063, CopZ-1317 and CopZ-3079 have no other Cys residues present other than those in the MX1CX2X3C motif (Fig. 1A). Both proteins readily reduced DTNB to consistently give an average protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) thiol ratio of 1
thiol ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, indicative of a high level of Cys solvent exposure, ideal for capturing and binding Cu(I). Cu(I) binding affinities (KD(CuI)) were determined using a robust assay that employs the specific Cu(I) bidentate chelators BCA or BCS.38 At pH 7.5, addition of either apo-CopZ protein into [CuI(BCA)2]3− leads to a linear decrease in absorbance at 562 nm until a ratio of ∼1 CopZ/Cu(I) is reached (Fig. 2A and B). This indicates that under concentrations employed, BCA cannot compete with either CopZ protein for Cu(I), and infers that 1 equivalent of Cu(I) is bound per CopZ monomer. Competition for Cu(I) between the apo-proteins and [CuI(BCS)2]3− was observed, with analysis of the data plotted in Fig. 2C and D using eqn (2) and a β2 of 1019.8 M−2 for [CuI(BCS)2]3− giving KD(CuI) values of 2.1 × 10−17 M for CopZ-1317 and 3.7 × 10−18 M for CopZ-3079. Triplicate data sets were obtained for both proteins with varying [CuI]total or [BCS] and average KD(CuI) values are reported in Table 2.
2, indicative of a high level of Cys solvent exposure, ideal for capturing and binding Cu(I). Cu(I) binding affinities (KD(CuI)) were determined using a robust assay that employs the specific Cu(I) bidentate chelators BCA or BCS.38 At pH 7.5, addition of either apo-CopZ protein into [CuI(BCA)2]3− leads to a linear decrease in absorbance at 562 nm until a ratio of ∼1 CopZ/Cu(I) is reached (Fig. 2A and B). This indicates that under concentrations employed, BCA cannot compete with either CopZ protein for Cu(I), and infers that 1 equivalent of Cu(I) is bound per CopZ monomer. Competition for Cu(I) between the apo-proteins and [CuI(BCS)2]3− was observed, with analysis of the data plotted in Fig. 2C and D using eqn (2) and a β2 of 1019.8 M−2 for [CuI(BCS)2]3− giving KD(CuI) values of 2.1 × 10−17 M for CopZ-1317 and 3.7 × 10−18 M for CopZ-3079. Triplicate data sets were obtained for both proteins with varying [CuI]total or [BCS] and average KD(CuI) values are reported in Table 2.
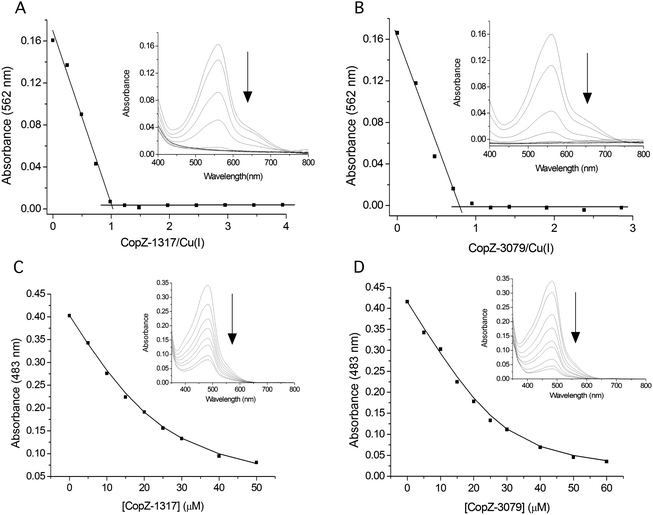 |
| | Fig. 2 Determining the Cu(I) stoichiometry and affinity using the chromogenic affinity probes BCA and BCS. (A) and (B) the absorbance at 562 nm in the visible spectrum of [CuI(BCA)2]3− (insets) decreases to zero upon increasing additions of each apo-CopZ, with plots of absorbance change at 562 nm as a function of [protein/Cu(I)] indicating an ∼1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry based on the intersection of the lines at the start and end of the titration. (C) and (D) Under the copper-limiting conditions imposed by [CuI(BCS)2]3− the absorbance at 483 nm in the visible spectrum of [CuI(BCS)2]3− (inset) decreases upon addition of each apo-CopZ and the KD(CuI) determined using eqn (2). The lines through the data points represents a best fit to the data using a KD(CuI) of 2.1 × 10−17 M and 3.7 × 10−18 M for CopZ-1317 and CopZ-3079, respectively. 1 stoichiometry based on the intersection of the lines at the start and end of the titration. (C) and (D) Under the copper-limiting conditions imposed by [CuI(BCS)2]3− the absorbance at 483 nm in the visible spectrum of [CuI(BCS)2]3− (inset) decreases upon addition of each apo-CopZ and the KD(CuI) determined using eqn (2). The lines through the data points represents a best fit to the data using a KD(CuI) of 2.1 × 10−17 M and 3.7 × 10−18 M for CopZ-1317 and CopZ-3079, respectively. | |
Table 2 Cu(I) affinity constants determined by BCS at pH 7.5, pKa and Δε240 values for the wild type S. lividans CopZ pair, the H22G and Y71F mutants of CopZ-1317 and CsoR. Values obtained for other species relevant to this work and discussed in the text are also reported
| Protein |
pKa |
Δε240 mM−1 cm−1 |
K
D(CuI) M |
|
B. subtilis CopZ KD(CuI) determined at pH 7.5 and pKa values determined using Badan.39
H. sapiens HAH1 and the MBD of the Menkes protein (MNK1) KD(CuI) determined at pH 7.0.12
Synechocystis PCC 6803 Atx1 KD(CuI) determined at pH 7.5.40
S. cerevisiae Atx1 KD(CuI) determined at pH 7.0.41
S. lividans CsoR KD(CuI) determined at pH 7.5.34
In addition to the Cys residues in the MXCXXC motif a third Cys residue is present in HAH1, which based on the Δε(1)240 value suggests ionizes in the same pH region as CysN (pKa1).12
|
| CopZ-1317 |
7.5 (0.2) |
4.4 (0.1) |
2.0 (0.2) × 10−17 |
| CopZ-3079 |
7.8 (0.2) |
3.5 (0.25) |
3.9 (0.5) × 10−18 |
| 1317-H22G |
8.0 (0.2) |
3.7 (0.1) |
6.0 (0.1) × 10−17 |
| 1317-Y71F |
7.8 (0.1) |
3.4 (0.1) |
1.7 (0.8) × 10−17 |
| |
pKa1 |
pKa2 |
Δε(1)240 |
Δε(2)240 |
|
|
Bs-CopZa |
6.1 (0.1) |
< 4 |
— |
— |
∼10−18 |
| HAH1b |
8.9 (0.1) |
5.5 (0.1) |
7.9 (0.5)f |
3.6 (0.2) |
1.8 × 10−18 |
| HAH1-K60Ab |
8.9 (0.1) |
7.0 (0.2) |
7.7 (0.5)f |
3.6 (0.4) |
5.5 × 10−18 |
|
Syn-Atx1c |
nd |
nd |
nd |
nd |
7.0 × 10−19 |
|
Sc-Atx1d |
nd |
nd |
nd |
nd |
2.0 × 10−18 |
| MNK1b |
9.2 (0.2) |
7.0 (0.1) |
4.0 (0.6) |
3.3 (0.2) |
2.8 × 10−18 |
| NmerA |
9.0 (0.5) |
6.4 (0.2) |
2.6 |
5.5 |
na |
| NmerA-Y62F |
9.3 (0.1) |
6.9 (0.1) |
3.3 |
2.8 |
na |
| CsoRe |
9.3 (0.3) |
6.7 (0.2) |
1.9 (0.3) |
2.6 (0.4) |
2.6 × 10−18 |
Recent studies using BCS at physiological pH have determined attomolar Cu(I) affinities (KD(CuI) 10−18 M) for Atx1 family members,12,39–41 and are thus comparable to the values reported here for the S. livdians CopZ pair (Table 2). It is noted however, that CopZ-3079 appears to have a 5-fold higher affinity for Cu(I) than CopZ-1317, and thus CopZ-3079 lies closer in terms of affinity for Cu(I) to HAH1, B. subtilis CopZ, and the Atx1 proteins from yeast and Synechocystis PCC 6803 (Table 2). The higher Cu(I) affinity determined for CopZ-3079 may provide a physiological advantage to the organism and would be consistent with this CopZ having a high capacity to buffer Cu(I) concentration in the cytosol during homeostasis. The lower Cu(I) affinity of CopZ-1317 does not have a direct physiological implication because the contribution of CopZ-1317 to total homeostasis capacity is close to zero and during Cu-stress is less than 2% on the basis of transcription levels (Table 1). Once Cu(I) buffering capacity is exceeded, increased Cu(I) concentrations may be sensed directly by the CsoR to trigger a transcriptional response, or alternatively, CopZ-3079 may act to chaperone Cu(I) to apo-CsoR bound to its operator sites (vide infra).
A single Cys ionization process is detected
The modulation of the ionization equilibria of the metal binding Cys thiols in metallochaperones possessing a MX1CX2X3C motif is important for facilitating metal ion binding and exchange of the bound metal ion between a cognate partner.12,33 The ionization properties of the Cys residues in the MX1CX2X3C motif for CopZ-1317 and CopZ-3079 were monitored by following absorbance changes at 240 nm (ΔA240) as a function of pH.42 Only data at pH 10.0 and below were considered as this is because both proteins contain Tyr residues that deprotonate at higher pH values and influence the absorbance at 240 nm.43 For CopZ-1317 and CopZ-3079 the ΔA240 over the pH range employed was best fit to eqn (3) that describes a single ionization process (Fig. 3), with pKa and Δε240 values reported in Table 2. The Δε values for both proteins (Table 2) are consistent with an absorbance change for the formation of a single Cys thiolate (Δε240 3–6 mM−1 cm−1).42 Therefore, assigning an equivalent pKa to both Cys residues is unlikely. To corroborate these results the reactivity of the Cys residues in the CopZ pair were assessed over the same pH range using the fluorescent alkylating agent Badan. Analysis of the data obtained with Badan was consistent again with a single Cys ionization process, with pKa values essentially identical to those determined from ΔA240 in Table 2 (see ESI† and Fig. S1). Our data, using two different probes, is also in line with the detection of a single Cys ionisation equilibrium in B. subtilis CopZ in the pH range 4 to 7, where a pKa of 6.1 was determined.39 However, using Badan in combination with mass spectrometry a second pKa estimated to be <4 was further determined for B. subtilis CopZ.39 This highly acidic value infers that the thiolate form of this Cys is stabilized to a greater extent (relative to the thiol form) than the Cys with a pKa of 6.1.
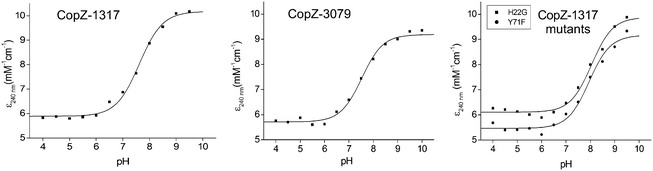 |
| | Fig. 3 Determination of Cys pKa values. Plots of ε240versus pH for CopZ-1317, CopZ-3079 and the H22G and Y71F mutants of CopZ-1317. The lines show fits of the data to eqn (3) and give pKa and Δε240 values reported in Table 2. | |
His22 is not responsible for a highly acidic Cys pKa
By comparing the acid–base properties of the CX1X2C motif in thioredoxins, Le Brun and co-workers noted39 that a very low pKa of 3.5 assigned to the N-terminal Cys (CysN) in the thioredoxin DsbA was attributed to an electrostatic interaction of a His residue at the X2 position stabilizing the thiolate form.44 A His residue is present at the equivalent position (X3) in the S. lividans CopZ pair as well as in B. subtilis and E. hirae CopZ, but is a Gly in the eukaryotic proteins (Fig. 1A). To assess whether this His residue is a contributing factor in modulating the pKa of a Cys to <4 in bacterial CopZ proteins, the H22G mutant of CopZ-1317 was prepared. The purified H22G CopZ-1317 mutant was folded as determined by far UV-CD spectroscopy (data not shown), with a BCA assay indicative of Cu(I) binding in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (Fig. S2A, ESI†). As for the wild type (WT) protein, the ΔA240 was monitored over the pH range 4 to 9.5 and the data fitted best to eqn (3) giving a pKa and a Δε240 consistent with the ionization of a single Cys thiolate (Fig. 3 and Table 2). Thus replacing His22 with an inert Gly residue does not result in the observation of a second ionization process, whereby a Cys with a very acidic pKa is no longer stabilized as a consequence of removing a potentially stabilizing thiolate interaction. However, the pKa value determined for the H22G mutant is ∼0.5 pH units higher than WT CopZ-1317, suggesting His22 does have a role in modulating the pKa of the observable Cys in the pH range studied (Table 2). Furthermore, removal of His22 leads to a ∼3-fold decrease in the affinity for Cu(I) as determined by using BCS (Table 1 and Fig. S2B, ESI†). An explanation for this Cu(I) affinity decrease is likely to concur with the decreased concentration of the thiolate form available (KD(CuI) determined at pH 7.5) supporting the notion that low Cys proton affinities increase Cu(I) affinity.
1 stoichiometry (Fig. S2A, ESI†). As for the wild type (WT) protein, the ΔA240 was monitored over the pH range 4 to 9.5 and the data fitted best to eqn (3) giving a pKa and a Δε240 consistent with the ionization of a single Cys thiolate (Fig. 3 and Table 2). Thus replacing His22 with an inert Gly residue does not result in the observation of a second ionization process, whereby a Cys with a very acidic pKa is no longer stabilized as a consequence of removing a potentially stabilizing thiolate interaction. However, the pKa value determined for the H22G mutant is ∼0.5 pH units higher than WT CopZ-1317, suggesting His22 does have a role in modulating the pKa of the observable Cys in the pH range studied (Table 2). Furthermore, removal of His22 leads to a ∼3-fold decrease in the affinity for Cu(I) as determined by using BCS (Table 1 and Fig. S2B, ESI†). An explanation for this Cu(I) affinity decrease is likely to concur with the decreased concentration of the thiolate form available (KD(CuI) determined at pH 7.5) supporting the notion that low Cys proton affinities increase Cu(I) affinity.
A thiolate stabilizing hydrogen bond interaction from a loop 5 residue is not operative
In contrast to the CopZ proteins, Badarau and Dennison report two well resolved ionization processes, with pKa values of 5.5 and 8.9 for HAH1.12 The acidic pKa was assigned to the C-terminal Cys (CysC) residue and the basic pKa to CysN of the MX1CX2X3C motif. This was inferred from structural information where the Sγ atom of CysC can participate in a hydrogen bond interaction with a side chain amino-group of a Lys residue located on loop 5 connecting α-helix 2 and β sheet 4 of HAH1 at sequence position 6010,44,45 (* in Fig. 1A). This polar interaction would therefore stabilize the thiolate form with respect to the N-terminal CysN, increasing the electrophilicity of the Cu(I) ligand and lowering the pKa. The K60A mutant of HAH1 confirmed these structural interpretations, resulting in an increased pKa of CysC to 7.0 and a subsequent 3-fold decrease in Cu(I) affinity relative to WT HAH1 (Table 2).12 Similarly, the N-terminal MBD (NmerA) of MerA, a Hg2+ reductase, has a Tyr residue in the equivalent position to the Lys60 in HAH1.43 For NmerA two ionization processes are reported over the pH range 3 to 10 with pKa values of 6.4 and 9.0 assigned to the CysC and CysN, respectively, on the basis that the CysC thiolate is stabilized through a side-chain hydrogen bonding interaction with the loop 5 Tyr residue.43 Subsequent mutation of the Tyr to a Phe increased the pKa of both Cys residues in NmerA, with the CysC retaining the lower pKa (Table 2).43
In CopZ-1317 a Tyr residue is found at the equivalent sequence position (Fig. 1A) and thus in analogy to HAH1 and NmerA may be expected to modulate the acidity of the pKa for CysC. To test whether this is the case the Y71F mutant of CopZ-1317 was prepared. The purified Y71F mutant was folded and a BCA titration was consistent with Cu(I) binding in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (Fig. S3A, ESI†). Using BCS the Cu(I) affinity for the mutant was essentially unchanged compared to the WT protein (Table 2 and Fig. S3B, ESI†). Likewise, the pKa remains within error of the WT and the Δε240 value is consistent with a single ionization process (Fig. 3 and Table 2). Therefore, replacing Tyr71 in CopZ-1317 with the non-hydrogen bonding Phe side chain does not lead to the detectability of a second (acidic) ionization process. Furthermore, a Phe residue is located at the equivalent sequence position in CopZ-3079 (Fig. 1A), for which a single ionization process is also detected (Fig. 3). Interestingly, a Phe is found in the equivalent position in eukaryotic transporter MBD.12 This apolar side chain serves to increase the pKa of CysC compared to the presence of a Lys residue in the HAH1 chaperone, and in the absence of a stabilizing hydrogen bond increases the nucleophilicity of CysC in the MBD acceptor.12
1 stoichiometry (Fig. S3A, ESI†). Using BCS the Cu(I) affinity for the mutant was essentially unchanged compared to the WT protein (Table 2 and Fig. S3B, ESI†). Likewise, the pKa remains within error of the WT and the Δε240 value is consistent with a single ionization process (Fig. 3 and Table 2). Therefore, replacing Tyr71 in CopZ-1317 with the non-hydrogen bonding Phe side chain does not lead to the detectability of a second (acidic) ionization process. Furthermore, a Phe residue is located at the equivalent sequence position in CopZ-3079 (Fig. 1A), for which a single ionization process is also detected (Fig. 3). Interestingly, a Phe is found in the equivalent position in eukaryotic transporter MBD.12 This apolar side chain serves to increase the pKa of CysC compared to the presence of a Lys residue in the HAH1 chaperone, and in the absence of a stabilizing hydrogen bond increases the nucleophilicity of CysC in the MBD acceptor.12
Bacterial CopZ members possess optimized Cys ligands for Cu(I) transfer
Based on our findings and those from B. subtilis CopZ,39 it appears that bacterial CopZ proteins have more acidic (low proton affinity) Cys pKa values than their eukaryotic counterparts (Table 2). Although our results are consistent with the detection of a single Cys ionization process for the S. lividans CopZ pair, we infer in analogy to B. subtilis CopZ39 that the second Cys in the MX1CX2X3C motif has a pKa< 4. Attempts to confirm this through the rational disruption of a hydrogen bonding side chain of a residue on loop 5 of the βαββαβ-fold in CopZ-1317 or through replacement of a His residue at the X3 position of the MX1CX2X3C motif did not however lead to a second observable ionization process. Therefore, other mechanisms contributing to thiolate stabilization of CysC in the bacterial proteins must be operative. In structures of Atx1 members, CysC is often positioned at the N-terminus of α-helix 1. The electrostatic effect of having a net partial charge at the N-terminus of the helix (macrodipole) can account for the stabilization of the thiolate. However, this contribution is usually modest, often contributing to a decrease in pKa of ∼−1.5 units relative to the intrinsic pKa of 9.1 for a Cys.45 Other factors that can stabilize a Cys thiolate are interactions with H2O molecules and cationic interactions with nearby amino acid side-chains. Hydrogen bond interactions alone can contribute to lowering the pKa of a Cys in the CXXC motif in thioredoxins by up to −3 units and cationic interactions in combination with the contribution from the N-terminus helix macrodipole by up to −5.6 units.45 Thus in bacterial CopZ proteins the highly acidic pKa of CysC must arise from summative contributions from the N-terminal macrodipole of α helix 1 and a hydrogen bond and/or a cationic interaction, other than His22, that differs to the loop5 Lys/Tyr interaction previously reported for HAH1 and NmerA.12,43 Less ambiguous in the bacterial proteins is the contribution of His22 at the X3 position of the MX1CX2X3C. Substitution to a Gly increases the pKa of the detectable Cys thiolate transition in CopZ-1317, which we assign to CysN, and may explain why CysN in bacterial proteins gives rise to a more acidic pKa (thiolate stabilized by His22) than its eukaryotic homologues where a non-cationic Gly residue is present in the X3 position (Table 2). In the Cu(I) bound form the CysN will now be a better leaving group (less nucleophilic) for Cu(I) than the corresponding CysN in HAH1 and favour Cu(I) release to an acceptor. Thus the additional hydrogen bond stabilization of CysN in the bacterial CopZ proteins creates a chaperone that is better optimized for Cu(I) transfer than their eukaryotic counterparts.
In vitro Cu(I) transfer from CopZ to CsoR is unidirectional
Other than the cytosolic MBD of CopA there is also the possibility that CopZ proteins can traffic Cu(I) to other cytosolic targets (not possessing a MX1CX2X3C motif), as has been illustrated for CopY in E. hirae (CXCXXXXCXC).23,46 To test if this is possible for a bacterial system involving a CsoR, electrophoretic mobility shift assay (EMSA) experiments were devised whereby the binding of apo-CsoR to each of its three cognate DNA operator sites was used to probe whether Cu(I) could be exchanged between proteins. CsoR binds Cu(I) via two Cys residues and a His residue in a trigonal coordination arrangement, with the two Cys ligands presented to the Cu(I) ion from two separate monomers of the CsoR tetramer assembly.34 The CsoR operator sequences for the individual targets have previously been determined.34 Two apo-CsoR tetramers bind the operator DNA,27,34 resulting in the formation of a low-mobility CsoR:DNA complex visualized by the retardation of the DNA in the EMSA (Fig. 4A and B). In contrast, incubation of the DNA operator sequences with Cu(I)-loaded CsoR results in the absence of a band shift and only the high-mobility band corresponding to the free operator DNA is observed (Fig. 4B).34 Cu(I)-loaded or apo-CopZ samples did not affect the mobility of the free DNA (Fig. 4A and B). Under anaerobic conditions a pre-mixed sample containing DNA and apo-CsoR was prepared followed by the addition of a 1 or 10-fold molar equivalent of a Cu(I)-loaded CopZ protein (Fig. 4A). From the EMSA, the low-mobility band corresponding to the apo-CsoR:DNA complex, is no longer observed and the high-mobility band is now prominent (Fig. 4A). This result was found for all three DNA operators (Fig. S4, ESI†) and can be interpreted as Cu(I) being readily transferred from both CopZ proteins to apo-CsoR, causing dissociation (derepression) of the operator DNA. The reverse experiments, again under anaerobic conditions, whereby the apo-CopZ proteins at 1 and 10-fold molar equivalent were added to pre-mixed samples containing Cu(I)-loaded CsoR and operator DNA were also performed. The EMSA shows only the low-mobility band (Fig. 4B), indicating that Cu(I) is not being transferred from Cu(I)-loaded CsoR to the apo-CopZ proteins. This was also the case with the other operator sequences (Fig. S4, ESI†) and suggests that Cu(I) transfer is a unidirectional process from the CopZ to CsoR.
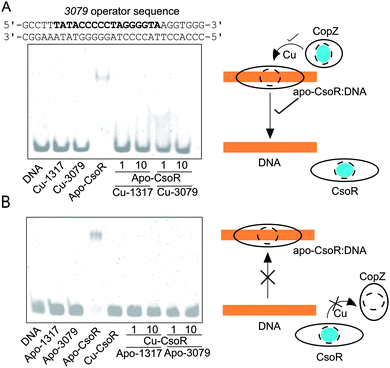 |
| | Fig. 4 Cu(I) transfer between CsoR and CopZ proteins probed by EMSA. Experiments were performed with three CsoR operator sequences (1317, 3079 and csoR), but is illustrated here only for the 3079 CsoR operator sequence (bold type). In (A) 1 or 10 molar equivalent of Cu(I)-loaded CopZ proteins were added to a pre-incubated sample of apo-CsoR and DNA. In (B) 1 or 10 molar equivalent of apo-CopZ proteins were added to a pre-incubated sample of Cu-CsoR and DNA. In (A) the cartoon indicates that Cu (blue sphere) is transferred from Cu-CopZ to apo-CsoR bound to DNA, leading to dissociation of the DNA, whilst in (B) Cu-CsoR cannot transfer Cu to apo-CopZ, and cannot therefore bind to the DNA and cause a band-shift. The components present in each lane of the gels are indicated with the concentrations as follows: 0.5 μM [DNA], 4 μM [apo-CsoR-monomer], 4 μM [Cu(I)-CsoR-monomer] (equivalent to 0.5 μM 2 × CsoR tetramer), 4 and 40 μM [apo-CopZ] or [Cu(I)-CopZ] (equivalent to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and a 1 1 and a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 CsoR-monomer 10 CsoR-monomer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CopZ). CopZ). | |
Mechanistic insight into Cu(I) transfer between CopZ and CsoR
Based on the KD(CuI) values reported in Table 2 shallow but favourable thermodynamic affinity gradients exist between the CopZ-pair and CsoR. This is consistent with the interpretation of the EMSA data, whereby Cu(I) transfer proceeds from CopZ to DNA-bound CsoR. However, the Cu(I) affinity for DNA-bound CsoR has not been directly determined but is expected to be of weaker affinity based on the thermodynamic linkage of the coupled equilibrium between CsoR and its DNA operator and Cu(I) ligands.20,47 This would yield an unfavourable affinity gradient for Cu(I) transfer from CopZ to DNA-bound CsoR and thus an alternative explanation of the EMSA results in Fig. 4A could be that transfer occurs with the ‘free’ apo-CsoR in this equilibrium. However, based on our experimental set-up this is unlikely. Biological systems are open systems that operate far from equilibrium and Cu(I) trafficking has been shown to occur in systems where there is an unfavourable thermodynamic gradient.40 This has highlighted that kinetic factors play an important role in Cu(I) trafficking pathways and that the tuning of the reactivity of the ligands involved in the transfer of Cu(I) from donor to the acceptor will be a considerable factor in determining whether transfer occurs. To address this the ionization properties of the Cu(I) coordinating Cys residues in CsoR were assessed by monitoring the ΔA240 in the pH range 5 to 9.5. Two ionization processes are apparent and despite the data being less well defined at alkaline pH (Fig. 5), eqn (4) fits the data well to give the Δε and pKa values reported in Table 2, with the pKa1 value considered a lower limit. Analysis of the apo-CsoR crystal structure reveals that the Cys thiol groups do not participate in hydrogen bond interactions with other amino acids and are readily accessible to solvent (inset Fig. 5). Cys75 is located on a loop region connecting α-helix 1 and α-helix 2 and is positioned towards the N-terminus of α-helix 2 with its side-chain modeled in two orientations, indicating increased motion in this region. Cys104 is located within α-helix 2 of another protomer but lies towards the C-terminal end of the α-helix and will therefore not experience a macrodipole. Thus the more acidic pKa in CsoR might be attributed to Cys75, whereby the thiolate is weakly stabilized by the α-helix 2 macrodipole. Importantly, in the absence of structural evidence for hydrogen bonds, both these Cys residues are potentially strong nucleophiles and at physiological pH the more acidic Cys will be predominately in the thiolate form (∼83% at pH 7.4). Therefore the enhanced electrophilicity of CysN (and possibly CysC) in the CopZ proteins coupled at physiological pH with the strongly nucleophilic CsoR Cys thiolate, all favour the lowering of the activation barrier for unidirectional Cu(I) transfer to proceed from CopZ to the CsoR regardless of whether the thermodynamic affinity is favourable.
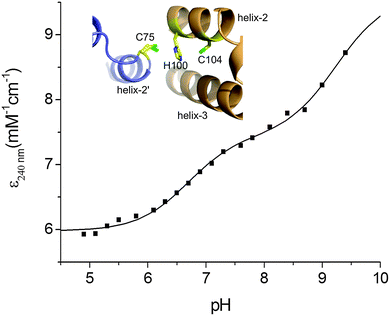 |
| | Fig. 5 Determination of Cys pKa values for CsoR. A plot of ε240 against pH with the line representing a fit of the data to eqn (4) to give pKa and Δε240 values reported in Table 2. Inset, a zoomed-in view of the Cu(I) ligands in the X-ray structure (PDB 4adz)34 of S. lividans apo-CsoR. | |
The role of CopZ within the S. lividans CsoR regulon
The large number of chaperone and transport proteins involved in cytosolic Cu handling in S. lividans is now included in a model (Fig. 6). In the present study we have elucidated at the molecular level some of the mechanistic detail of Cu(I) trafficking within this model. The higher Cu(I) affinity for CopZ-3079 over CopZ-1317 fits with a proposed role as a buffer to maintain homeostasis and reveals a physicochemical organism advantage to having basal expression levels of this CopZ–CopA couple. Possibly for similar reasons, although not determined here, the other two operons (0895–0896 and 1063–1064) are expressed during homeostasis creating a large pool of Cu(I) buffering capacity. At elevated Cu levels, our data are consistent with the CopZ proteins having Cu(I) binding Cys residues that are tuned to facilitate transfer to CsoR, which itself has a Cys optimized for ligand-exchange. Thus CopZ-3079/1063/0895 could first transfer Cu(I) to DNA-bound CsoR causing derepression of three copZ/copA efflux systems, with the resulting apo-CopZ's continuing to buffer Cu(I) and eventually being assisted by CopZ-1317 (steps 1 to 3 in Fig. 6). The Cu(I)-CopZ proteins then proceed to traffic their cargo to the metal binding platform of their cognate CopA-like transporters for export to the extracytoplasmic environment (step 4 in Fig. 6). Derepression of the csoR gene will only be partial as shown by the RNA-seq data, which we have subsequently shown is due to different thermodynamic binding properties of the csoR operator site27 (step 5 in Fig. 6). In the pathogenic bacterium Listeria monocytogenes a CopZ chaperone is also reported to buffer cytosolic Cu(I) concentration, but in vivo studies suggest that the CopZ is not strictly necessary to traffic Cu(I) to the CsoR to illicit a transcriptional response.48 Thus whilst our data in S. livdians favour the model outlined in Fig. 6 variations in CopZ function across bacterial species may be operative. Finally, once the cytoplasmic Cu(I) levels have been reduced the system needs to reset and return to homeostasis. This requires sufficient concentration of both apo-CopZ and apo-CsoR. For the CopZ proteins transfer of the metal load to their cognate ATPase will achieve this, but for CsoR the situation at present is less clear, but our data show that Cu-transfer from CsoR to a CopZ is disfavoured.
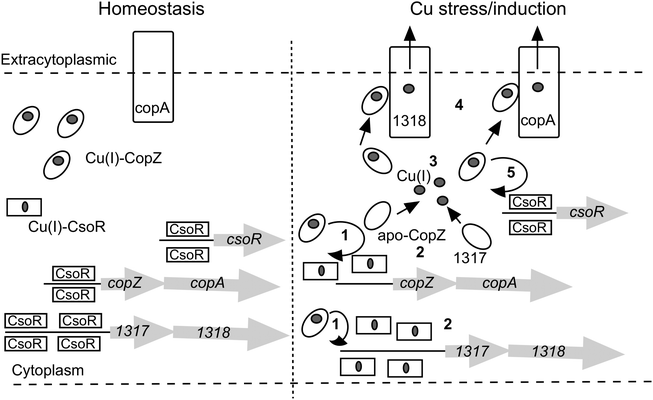 |
| | Fig. 6 A model of the CsoR regulon in S. lividans based on RNA-seq data presented in Table 1 and mechanistic implications from the present study. The copZ/copA arrows represent the highly expressed operons 1063/1064 and 3079/3080, with the Cu(I)-CopZ and CopA representative of these expressed genes. The copZ/copA couple 0895/0896 is not included in the model since the control mechanism is not known. However, it does contribute a significant amount of CopZ and CopA both under homeostasis and upon Cu stress/induction. | |
Experimental
Cloning and over-expression of CopZ-1317 and CopZ-3079
The SLI_1317 and SLI_3079 genes (234 and 243 base pairs, respectively) were amplified from S. lividans 1326 (S. lividans 66, stock number 1326 John Innes Centre) genomic DNA and cloned into the NdeI and HindIII sites of a pET28a vector (Novagen) to create N-terminal His6-tagged constructs for over-expression in Escherichia coli. CopZ-1317 and CopZ-3079 were over-expressed in the E. coli strain BL21 (DE3) following induction with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG; Melford). Cells were harvested after 16 h of growth at 25 °C and lysed using an EmulsiFlex-C5 cell disrupter (Avestin) followed by centrifugation at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min at 4 °C. The clarified supernatant was loaded to a Ni2+-NTA Sepharose column (GE Healthcare) equilibrated with Buffer A (50 mM Tris/HCl pH 8, 500 mM NaCl, 20 mM imidazole) and eluted with a linear imidazole gradient using Buffer B (Buffer A with 500 mM imidazole). Fractions were pooled and dialysed overnight at 4 °C against Buffer C (50 mM Tris/HCl pH 8, 150 mM NaCl, 2 mM EDTA, 2 mM DTT (Melford)). Following dialysis, the N-terminal His6-tag was removed by incubating the protein at room temperature overnight in the presence of 125 U of thrombin (Sigma). The protein–thrombin mixture was reapplied to the Ni2+-NTA Sepharose column (GE Healthcare) and the flow-through collected and concentrated at 4 °C using a centricon (vivaspin) with a 5 kDa cut-off, and loaded to a G75 Sephadex column (GE Healthcare) equilibrated with buffer C. Fractions eluting in the major peak of the G75 column were analysed by 15% SDS-PAGE and those deemed of good purity were concentrated and stored at −20 °C until required.
000 rpm for 20 min at 4 °C. The clarified supernatant was loaded to a Ni2+-NTA Sepharose column (GE Healthcare) equilibrated with Buffer A (50 mM Tris/HCl pH 8, 500 mM NaCl, 20 mM imidazole) and eluted with a linear imidazole gradient using Buffer B (Buffer A with 500 mM imidazole). Fractions were pooled and dialysed overnight at 4 °C against Buffer C (50 mM Tris/HCl pH 8, 150 mM NaCl, 2 mM EDTA, 2 mM DTT (Melford)). Following dialysis, the N-terminal His6-tag was removed by incubating the protein at room temperature overnight in the presence of 125 U of thrombin (Sigma). The protein–thrombin mixture was reapplied to the Ni2+-NTA Sepharose column (GE Healthcare) and the flow-through collected and concentrated at 4 °C using a centricon (vivaspin) with a 5 kDa cut-off, and loaded to a G75 Sephadex column (GE Healthcare) equilibrated with buffer C. Fractions eluting in the major peak of the G75 column were analysed by 15% SDS-PAGE and those deemed of good purity were concentrated and stored at −20 °C until required.
Site-directed mutagenesis
The Quikchange (Stratagene) site-directed mutagenesis method was used to create the H22G and Y71F mutants of CopZ-1317. The mutagenic primers and PCR procedure are reported in ESI.†
Over-expression and purification of CsoR
CsoR (SLI_4375) was over-expressed in E. coli and purified as previously reported.34
Preparation of reduced proteins
Proteins were reduced in an anaerobic chamber (DW Scientific [O2] < 2 ppm) with 5 mM DTT and desalted (twice) using a PD-10 column (GE-Healthcare) into the desired buffer. Such high concentrations of DTT were used to ensure that cysteines were fully reduced prior to desalting. Free thiol content was determined by the reduction of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) monitored at 412 nm (ε = 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 M−1 cm−1).49
500 M−1 cm−1).49
CD and UV-visible spectroscopy
A Varian Cary 50 UV-visible spectrophotometer thermostated at 20 °C was used for all absorbance spectrum measurements. Apo-protein concentrations were determined at 280 nm using extinction coefficients (ε) of 3105 M−1 cm−1, 1615 M−1 cm−1 and 3105 M−1 cm−1 for CopZ-1317, CopZ-3079 and CsoR-monomer, respectively.50 CopZ samples (20 μM) for circular dichroism (CD) analysis were exchanged into 10 mM potassium phosphate pH 7, 50 mM potassium fluoride and far UV-CD spectra were recorded between 260 and 175 nm at 20 °C on an Applied Photophysics Chirascan CD spectrophotometer (Leatherhead, UK) equipped with a thermostatic cell holder controlled with a Peltier system. CD spectra were analysed using DichroWeb51,52 with the programs CDSSTR53–55 and Contin-LL56 and the databases 4, 755,57 and SP175.58
Analytical gel filtration
A 10/300 GL G-75 Superdex column (GE-Healthcare) was equilibrated in degassed buffer (10 mM MOPS pH 7.5, 150 mM NaCl) with 2 mM DTT. Reduced apo-CopZ proteins were injected onto the column and the elution profile monitored at 280 nm.
Competition assays using BCA and BCS
CuCl (Sigma) was dissolved under anaerobic conditions in 10 mM HCl and 500 mM NaCl and diluted with 10 mM MOPS pH 7.5, 150 mM NaCl. The Cu(I) concentration was determined spectrophotometrically by step-wise addition to a known concentration of the Cu(I) specific bidentate chelator bicinchoninic acid (BCA) using an extinction coefficient at 562 nm of ε = 7900 M−1 cm−1 for [CuI(BCA)2]3−.59 Competition assays were set up anaerobically with either BCA or bathocuprione disulfonate (BCS) (Sigma). Increasing protein concentrations (0–80 μM) were added to solutions of [CuIL2]3− of defined molar ratio L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu(I) ≥ 3 creating a series of individual solutions with constant [CuI] and [L] and varying [protein]. Samples were left for ∼1 h and the transfer of Cu(I) from the [CuIL2]3− complex to the protein determined by measuring the absorbance of the [CuIL2]3− complex spectrophotometrically for L = BCA at 562 nm (ε = 7900 M−1 cm−1)59 and L = BCS at 483 nm (13
Cu(I) ≥ 3 creating a series of individual solutions with constant [CuI] and [L] and varying [protein]. Samples were left for ∼1 h and the transfer of Cu(I) from the [CuIL2]3− complex to the protein determined by measuring the absorbance of the [CuIL2]3− complex spectrophotometrically for L = BCA at 562 nm (ε = 7900 M−1 cm−1)59 and L = BCS at 483 nm (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1).60 By interchanging L, assays favoring competitive or non-competitive Cu(I) binding were set-up, which for the latter led to an estimate of the binding stoichiometry. The dissociation constant for Cu(I) (KD(CuI)) was determined from competitive assays by assuming the following reaction (1)
000 M−1 cm−1).60 By interchanging L, assays favoring competitive or non-competitive Cu(I) binding were set-up, which for the latter led to an estimate of the binding stoichiometry. The dissociation constant for Cu(I) (KD(CuI)) was determined from competitive assays by assuming the following reaction (1)| | | apo-P + CuL2 ↔ Cu(I)–P + 2L | (1) |
where P is protein and by using eqn (2),| | 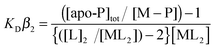 | (2) |
where [L] is the total ligand concentration (BCA or BCS) and the overall formation constant (β2) is 1017.2 M−2 for [CuI(BCA)2]3− and 1019.8 M−2 for [CuI(BCS)2]3−.41,59 Assays were performed in triplicate and the KD(CuI) value for a series was initially calculated for each individual solution and then averaged. Conditions for the plots shown in Fig. 2 for CopZ-1317 with BCA were; 5–80 μM [P], 20 μM [CuI] and 80 μM [BCA]; with BCS, 5–50 μM [P], 31 μM [CuI], and 80 μM [BCS]. Conditions for CopZ-3079 with BCA were; 5–60 μM [P], 21 μM [CuI] and 80 μM [BCS]; with BCS 5–60 μM [P], 32 μM [CuI] and 120 μM [BCS].
Determination of Cys pKa values
Reduced apo-proteins (∼400 μM) were prepared in an anaerobic chamber in 5 mM MOPS pH 7.0, 25 mM KCl, together with a series of mixed buffer systems containing 10 mM each of potassium acetate, MES, MOPS, Tris and 200 mM KCl with the pH of each solution individually adjusted in increments of 0.5 from pH values of 4 to 10.0. Reduced protein was added to each buffered solution to give a final concentration of 40 μM and left for 1 h before the absorbance at 240 nm was measured spectrophotometrically in a sealed quartz cuvette (Hellma) and plotted as a function of pH. Models describing one (eqn (3)) or two (eqn (4)) non-interacting macroscopic ionisations were used to determine pKa values of the Cys residues,| | 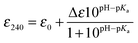 | (3) |
| | 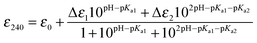 | (4) |
where ε0 is the extinction of the thiol form and Δε is the difference between the extinction coefficient of a thiol and thiolate form. The pKa values reported are an average of multiple data sets and the error reported is the standard deviation.
Electrophoretic mobility shift assays (EMSA)
DNA oligomers (Sigma) were prepared in 10 mM HEPES pH 7.5, 150 mM NaCl. Concentrations of individual oligonucleotides were determined using appropriate extinction coefficients at 260 nm on a Nanodrop 2000 (Thermo Scientific). Equal concentrations of complementary strands were annealed by heating to 96 °C in a water bath for 5 min and then left to cool to room temperature over-night. Reduced proteins were prepared in an anaerobic chamber in 10 mM HEPES pH 7.5, 150 mM NaCl. For Cu(I) loaded samples, CuCl solution was added to the respective proteins and allowed to incubate for 30 min followed by removal of any excess Cu(I) by a PD-10 column (GE-Healthcare). 0.5 μM of a DNA duplex was incubated with the desired concentration of protein (4 μM CsoR monomer and 4 or 40 μM CopZ) in 10 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT. All samples were incubated at room temperature under anaerobic conditions for 1 h and then loaded (20 μl) to a pre-run 6% Tris-Borate EDTA (TBE) polyacrylamide gel. Gels were stained for 30 min in an ethidium bromide solution followed by imaging.
Acknowledgements
This work was supported by a University of Essex Silberrad PhD scholarship to AKC.
References
- R. A. Pufahl, C. P. Singer, K. L. Peariso, S. J. Lin, P. J. Schmidt, C. J. Fahrni, V. C. Culotta, J. E. Penner-Hahn and T. V. O'Halloran, Science, 1997, 278, 853–856 CrossRef CAS.
- T. V. O'Halloran and V. C. Culotta, J. Biol. Chem., 2000, 275, 25057–25060 CrossRef PubMed.
- D. L. Huffman and T. V. O'Halloran, Annu. Rev. Biochem., 2001, 70, 677–701 CrossRef CAS PubMed.
- L. Banci, I. Bertini, K. S. McGreevy and A. Rosato, Nat. Prod. Rep., 2010, 27, 695–710 RSC.
- N. J. Robinson and D. R. Winge, Annu. Rev. Biochem., 2010, 79, 537–562 CrossRef CAS PubMed.
- M. D. Harrison, C. E. Jones, M. Solioz and C. T. Dameron, Trends Biochem. Sci., 2000, 25, 29–32 CrossRef CAS.
- A. K. Wernimont, D. L. Huffman, A. L. Lamb, T. V. O'Halloran and A. C. Rosenzweig, Nat. Struct. Biol., 2000, 7, 766–771 CrossRef CAS PubMed.
- F. Arnesano, L. Banci, I. Bertini, D. L. Huffman and T. V. O'Halloran, Biochemistry, 2001, 40, 1528–1539 CrossRef CAS PubMed.
- S. J. Lin, R. A. Pufahl, A. Dancis, T. V. O'Halloran and V. C. Culotta, J. Biol. Chem., 1997, 272, 9215–9220 CrossRef CAS PubMed.
- A. C. Rosenzweig, D. L. Huffman, M. Y. Hou, A. K. Wernimont, R. A. Pufahl and T. V. O'Halloran, Structure, 1999, 7, 605–617 CrossRef CAS.
- J. H. Kaplan and S. Lutsenko, J. Biol. Chem., 2009, 284, 25461–25465 CrossRef CAS PubMed.
- A. Badarau and C. Dennison, J. Am. Chem. Soc., 2011, 133, 2983–2988 CrossRef CAS PubMed.
- A. Odermatt and M. Solioz, J. Biol. Chem., 1995, 270, 4349–4354 CrossRef CAS PubMed.
- J. M. Arguello, J. Membr. Biol., 2003, 195, 93–108 CrossRef CAS PubMed.
- A. N. Barry, U. Shinde and S. Lutsenko, J. Biol. Inorg. Chem., 2010, 15, 47–59 CrossRef CAS PubMed.
- C. C. Wu, W. J. Rice and D. L. Stokes, Structure, 2008, 16, 976–985 CrossRef CAS PubMed.
- M. Gonzalez-Guerrero, D. Hong and J. M. Arguello, J. Biol. Chem., 2009, 284, 20804–20811 CrossRef CAS PubMed.
- P. Gourdon, X. Y. Liu, T. Skjorringe, J. P. Morth, L. B. Moller, B. P. Pedersen and P. Nissen, Nature, 2011, 475, 59–64 CrossRef CAS PubMed.
- T. Padilla-Benavides, C. J. McCann and J. M. Arguello, J. Biol. Chem., 2013, 288, 69–78 CrossRef CAS PubMed.
- D. P. Giedroc and A. I. Arunkumar, Dalton Trans., 2007, 3107–3120 RSC.
- Z. Ma, F. E. Jacobsen and D. P. Giedroc, Chem. Rev., 2009, 109, 4644–4681 CrossRef CAS PubMed.
- D. Strausak and M. Solioz, J. Biol. Chem., 1997, 272, 8932–8936 CrossRef CAS PubMed.
- P. Cobine, W. A. Wickramasinghe, M. D. Harrison, T. Weber, M. Solioz and C. T. Dameron, FEBS Lett., 1999, 445, 27–30 CrossRef CAS.
- T. Liu, A. Ramesh, Z. Ma, S. K. Ward, L. Zhang, G. N. George, A. M. Talaat, J. C. Sacchettini and D. P. Giedroc, Nat. Chem. Biol., 2007, 3, 60–68 CrossRef CAS PubMed.
- Z. Ma, D. M. Cowart, B. P. Ward, R. J. Arnold, R. D. DiMarchi, L. Zhang, G. N. George, R. A. Scott and D. P. Giedroc, J. Am. Chem. Soc., 2009, 131, 18044–18045 CrossRef CAS PubMed.
- F. M. Chang, H. J. Coyne, C. Cubillas, P. Vinuesa, X. Fang, Z. Ma, D. Ma, J. D. Helmann, A. Garcia-de Los Santos, Y. X. Wang, C. E. Dann, 3rd and D. P. Giedroc, J. Biol. Chem., 2014, 289, 19204–19217 CrossRef CAS PubMed.
- B. G. Tan, E. Vijgenboom and J. A. Worrall, Nucleic Acids Res., 2014, 42, 1326–1340 CrossRef CAS PubMed.
- K. Ueda, Y. Tomaru, K. Endoh and T. Beppu, J. Antibiot., 1997, 50, 693–695 CrossRef CAS.
- B. J. Keijser, G. P. van Wezel, G. W. Canters, T. Kieser and E. Vijgenboom, J. Mol. Microbiol. Biotechnol., 2000, 2, 565–574 CAS.
- J. A. Worrall and E. Vijgenboom, Nat. Prod. Rep., 2010, 27, 742–756 RSC.
- M. Fujimoto, A. Yamada, J. Kurosawa, A. Kawata, T. Beppu, H. Takano and K. Ueda, Microb. Biotechnol., 2012, 5, 477–488 CrossRef PubMed.
- K. L. Blundell, M. T. Wilson, D. A. Svistunenko, E. Vijgenboom and J. A. Worrall, Open Biol., 2013, 3, 120163 CrossRef PubMed.
- K. L. Blundell, M. A. Hough, E. Vijgenboom and J. A. Worrall, Biochem. J., 2014, 459, 525–538 CrossRef CAS PubMed.
- S. Dwarakanath, A. K. Chaplin, M. A. Hough, S. Rigali, E. Vijgenboom and J. A. Worrall, J. Biol. Chem., 2012, 287, 17833–17847 CrossRef CAS PubMed.
- Z. Ma, D. M. Cowart, R. A. Scott and D. P. Giedroc, Biochemistry, 2009, 48, 3325–3334 CrossRef CAS PubMed.
- P. Cruz-Morales, E. Vijgenboom, F. Iruegas-Bocardo, G. Girard, L. A. Yanez-Guerra, H. E. Ramos-Aboites, J. L. Pernodet, J. Anne, G. P. van Wezel and F. Barona-Gomez, Genome Biol. Evol., 2013, 5, 1165–1175 CrossRef PubMed.
- C. Cole, J. D. Barber and G. J. Barton, Nucleic Acids Res., 2008, 36, W197–201 CrossRef CAS PubMed.
- Z. Xiao and A. G. Wedd, Nat. Prod. Rep., 2010, 27, 768–789 RSC.
- L. Zhou, C. Singleton and N. E. Le Brun, Biochem. J., 2008, 413, 459–465 CrossRef CAS PubMed.
- A. Badarau and C. Dennison, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 13007–13012 CrossRef CAS PubMed.
- Z. Xiao, J. Brose, S. Schimo, S. M. Ackland, S. La Fontaine and A. G. Wedd, J. Biol. Chem., 2011, 286, 11047–11055 CrossRef CAS PubMed.
- R. E. Benesch and R. Benesch, J. Am. Chem. Soc., 1955, 77, 5877–5881 CrossRef CAS.
- R. Ledwidge, B. Hong, V. Dotsch and S. M. Miller, Biochemistry, 2010, 49, 8988–8998 CrossRef CAS PubMed.
- T. Kortemme, N. J. Darby and T. E. Creighton, Biochemistry, 1996, 35, 14503–14511 CrossRef CAS PubMed.
- T. K. Harris and G. J. Turner, IUBMB
Life, 2002, 53, 85–98 CAS.
- P. A. Cobine, G. N. George, C. E. Jones, W. A. Wickramasinghe, M. Solioz and C. T. Dameron, Biochemistry, 2002, 41, 5822–5829 CrossRef CAS PubMed.
- H. Reyes-Caballero, G. C. Campanello and D. P. Giedroc, Biophys. Chem., 2011, 156, 103–114 CrossRef CAS PubMed.
- D. Corbett, S. Schuler, S. Glenn, P. W. Andrew, J. S. Cavet and I. S. Roberts, Mol. Microbiol., 2011, 81, 457–472 CrossRef CAS PubMed.
- G. L. Ellman, Arch. Biochem. Biophys., 1959, 82, 70–77 CrossRef CAS.
- E. Gasteiger, A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel and A. Bairoch, Nucleic Acids Res., 2003, 31, 3784–3788 CrossRef CAS PubMed.
- L. Whitmore and B. A. Wallace, Nucleic Acids Res., 2004, 32, W668–W673 CrossRef CAS PubMed.
- L. Whitmore and B. A. Wallace, Biopolymers, 2008, 89, 392–400 CrossRef CAS PubMed.
- L. A. Compton and W. C. Johnson, Jr., Anal. Biochem., 1986, 155, 155–167 CrossRef CAS.
- P. Manavalan and W. C. Johnson, Jr., Anal. Biochem., 1987, 167, 76–85 CrossRef CAS.
- N. Sreerama, S. Y. Venyaminov and R. W. Woody, Anal. Biochem., 2000, 287, 243–251 CrossRef CAS PubMed.
- S. W. Provencher and J. Glockner, Biochemistry, 1981, 20, 33–37 CrossRef CAS.
- N. Sreerama and R. W. Woody, Anal. Biochem., 2000, 287, 252–260 CrossRef CAS PubMed.
- J. G. Lees, A. J. Miles, F. Wien and B. A. Wallace, Bioinformatics, 2006, 22, 1955–1962 CrossRef CAS PubMed.
- Z. Xiao, P. S. Donnelly, M. Zimmermann and A. G. Wedd, Inorg. Chem., 2008, 47, 4338–4347 CrossRef CAS PubMed.
- Z. Xiao, F. Loughlin, G. N. George, G. J. Howlett and A. G. Wedd, J. Am. Chem. Soc., 2004, 126, 3081–3090 CrossRef CAS PubMed.
- S. Hiard, R. Maree, S. Colson, P. A. Hoskisson, F. Titgemeyer, G. P. van Wezel, B. Joris, L. Wehenkel and S. Rigali, Biochem. Biophys. Res. Commun., 2007, 357, 861–864 CrossRef CAS PubMed.
- A. Mortazavi, B. A. Williams, K. McCue, L. Schaeffer and B. Wold, Nat. Methods, 2008, 5, 621–628 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4mt00250d |
|
| This journal is © The Royal Society of Chemistry 2015 |
Click here to see how this site uses Cookies. View our privacy policy here. 
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) thiol ratio of 1
thiol ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, indicative of a high level of Cys solvent exposure, ideal for capturing and binding Cu(I). Cu(I) binding affinities (KD(CuI)) were determined using a robust assay that employs the specific Cu(I) bidentate chelators BCA or BCS.38 At pH 7.5, addition of either apo-CopZ protein into [CuI(BCA)2]3− leads to a linear decrease in absorbance at 562 nm until a ratio of ∼1 CopZ/Cu(I) is reached (Fig. 2A and B). This indicates that under concentrations employed, BCA cannot compete with either CopZ protein for Cu(I), and infers that 1 equivalent of Cu(I) is bound per CopZ monomer. Competition for Cu(I) between the apo-proteins and [CuI(BCS)2]3− was observed, with analysis of the data plotted in Fig. 2C and D using eqn (2) and a β2 of 1019.8 M−2 for [CuI(BCS)2]3− giving KD(CuI) values of 2.1 × 10−17 M for CopZ-1317 and 3.7 × 10−18 M for CopZ-3079. Triplicate data sets were obtained for both proteins with varying [CuI]total or [BCS] and average KD(CuI) values are reported in Table 2.
2, indicative of a high level of Cys solvent exposure, ideal for capturing and binding Cu(I). Cu(I) binding affinities (KD(CuI)) were determined using a robust assay that employs the specific Cu(I) bidentate chelators BCA or BCS.38 At pH 7.5, addition of either apo-CopZ protein into [CuI(BCA)2]3− leads to a linear decrease in absorbance at 562 nm until a ratio of ∼1 CopZ/Cu(I) is reached (Fig. 2A and B). This indicates that under concentrations employed, BCA cannot compete with either CopZ protein for Cu(I), and infers that 1 equivalent of Cu(I) is bound per CopZ monomer. Competition for Cu(I) between the apo-proteins and [CuI(BCS)2]3− was observed, with analysis of the data plotted in Fig. 2C and D using eqn (2) and a β2 of 1019.8 M−2 for [CuI(BCS)2]3− giving KD(CuI) values of 2.1 × 10−17 M for CopZ-1317 and 3.7 × 10−18 M for CopZ-3079. Triplicate data sets were obtained for both proteins with varying [CuI]total or [BCS] and average KD(CuI) values are reported in Table 2.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry based on the intersection of the lines at the start and end of the titration. (C) and (D) Under the copper-limiting conditions imposed by [CuI(BCS)2]3− the absorbance at 483 nm in the visible spectrum of [CuI(BCS)2]3− (inset) decreases upon addition of each apo-CopZ and the KD(CuI) determined using eqn (2). The lines through the data points represents a best fit to the data using a KD(CuI) of 2.1 × 10−17 M and 3.7 × 10−18 M for CopZ-1317 and CopZ-3079, respectively.
1 stoichiometry based on the intersection of the lines at the start and end of the titration. (C) and (D) Under the copper-limiting conditions imposed by [CuI(BCS)2]3− the absorbance at 483 nm in the visible spectrum of [CuI(BCS)2]3− (inset) decreases upon addition of each apo-CopZ and the KD(CuI) determined using eqn (2). The lines through the data points represents a best fit to the data using a KD(CuI) of 2.1 × 10−17 M and 3.7 × 10−18 M for CopZ-1317 and CopZ-3079, respectively.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (Fig. S2A, ESI†). As for the wild type (WT) protein, the ΔA240 was monitored over the pH range 4 to 9.5 and the data fitted best to eqn (3) giving a pKa and a Δε240 consistent with the ionization of a single Cys thiolate (Fig. 3 and Table 2). Thus replacing His22 with an inert Gly residue does not result in the observation of a second ionization process, whereby a Cys with a very acidic pKa is no longer stabilized as a consequence of removing a potentially stabilizing thiolate interaction. However, the pKa value determined for the H22G mutant is ∼0.5 pH units higher than WT CopZ-1317, suggesting His22 does have a role in modulating the pKa of the observable Cys in the pH range studied (Table 2). Furthermore, removal of His22 leads to a ∼3-fold decrease in the affinity for Cu(I) as determined by using BCS (Table 1 and Fig. S2B, ESI†). An explanation for this Cu(I) affinity decrease is likely to concur with the decreased concentration of the thiolate form available (KD(CuI) determined at pH 7.5) supporting the notion that low Cys proton affinities increase Cu(I) affinity.
1 stoichiometry (Fig. S2A, ESI†). As for the wild type (WT) protein, the ΔA240 was monitored over the pH range 4 to 9.5 and the data fitted best to eqn (3) giving a pKa and a Δε240 consistent with the ionization of a single Cys thiolate (Fig. 3 and Table 2). Thus replacing His22 with an inert Gly residue does not result in the observation of a second ionization process, whereby a Cys with a very acidic pKa is no longer stabilized as a consequence of removing a potentially stabilizing thiolate interaction. However, the pKa value determined for the H22G mutant is ∼0.5 pH units higher than WT CopZ-1317, suggesting His22 does have a role in modulating the pKa of the observable Cys in the pH range studied (Table 2). Furthermore, removal of His22 leads to a ∼3-fold decrease in the affinity for Cu(I) as determined by using BCS (Table 1 and Fig. S2B, ESI†). An explanation for this Cu(I) affinity decrease is likely to concur with the decreased concentration of the thiolate form available (KD(CuI) determined at pH 7.5) supporting the notion that low Cys proton affinities increase Cu(I) affinity.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (Fig. S3A, ESI†). Using BCS the Cu(I) affinity for the mutant was essentially unchanged compared to the WT protein (Table 2 and Fig. S3B, ESI†). Likewise, the pKa remains within error of the WT and the Δε240 value is consistent with a single ionization process (Fig. 3 and Table 2). Therefore, replacing Tyr71 in CopZ-1317 with the non-hydrogen bonding Phe side chain does not lead to the detectability of a second (acidic) ionization process. Furthermore, a Phe residue is located at the equivalent sequence position in CopZ-3079 (Fig. 1A), for which a single ionization process is also detected (Fig. 3). Interestingly, a Phe is found in the equivalent position in eukaryotic transporter MBD.12 This apolar side chain serves to increase the pKa of CysC compared to the presence of a Lys residue in the HAH1 chaperone, and in the absence of a stabilizing hydrogen bond increases the nucleophilicity of CysC in the MBD acceptor.12
1 stoichiometry (Fig. S3A, ESI†). Using BCS the Cu(I) affinity for the mutant was essentially unchanged compared to the WT protein (Table 2 and Fig. S3B, ESI†). Likewise, the pKa remains within error of the WT and the Δε240 value is consistent with a single ionization process (Fig. 3 and Table 2). Therefore, replacing Tyr71 in CopZ-1317 with the non-hydrogen bonding Phe side chain does not lead to the detectability of a second (acidic) ionization process. Furthermore, a Phe residue is located at the equivalent sequence position in CopZ-3079 (Fig. 1A), for which a single ionization process is also detected (Fig. 3). Interestingly, a Phe is found in the equivalent position in eukaryotic transporter MBD.12 This apolar side chain serves to increase the pKa of CysC compared to the presence of a Lys residue in the HAH1 chaperone, and in the absence of a stabilizing hydrogen bond increases the nucleophilicity of CysC in the MBD acceptor.12

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min at 4 °C. The clarified supernatant was loaded to a Ni2+-NTA Sepharose column (GE Healthcare) equilibrated with Buffer A (50 mM Tris/HCl pH 8, 500 mM NaCl, 20 mM imidazole) and eluted with a linear imidazole gradient using Buffer B (Buffer A with 500 mM imidazole). Fractions were pooled and dialysed overnight at 4 °C against Buffer C (50 mM Tris/HCl pH 8, 150 mM NaCl, 2 mM EDTA, 2 mM DTT (Melford)). Following dialysis, the N-terminal His6-tag was removed by incubating the protein at room temperature overnight in the presence of 125 U of thrombin (Sigma). The protein–thrombin mixture was reapplied to the Ni2+-NTA Sepharose column (GE Healthcare) and the flow-through collected and concentrated at 4 °C using a centricon (vivaspin) with a 5 kDa cut-off, and loaded to a G75 Sephadex column (GE Healthcare) equilibrated with buffer C. Fractions eluting in the major peak of the G75 column were analysed by 15% SDS-PAGE and those deemed of good purity were concentrated and stored at −20 °C until required.
000 rpm for 20 min at 4 °C. The clarified supernatant was loaded to a Ni2+-NTA Sepharose column (GE Healthcare) equilibrated with Buffer A (50 mM Tris/HCl pH 8, 500 mM NaCl, 20 mM imidazole) and eluted with a linear imidazole gradient using Buffer B (Buffer A with 500 mM imidazole). Fractions were pooled and dialysed overnight at 4 °C against Buffer C (50 mM Tris/HCl pH 8, 150 mM NaCl, 2 mM EDTA, 2 mM DTT (Melford)). Following dialysis, the N-terminal His6-tag was removed by incubating the protein at room temperature overnight in the presence of 125 U of thrombin (Sigma). The protein–thrombin mixture was reapplied to the Ni2+-NTA Sepharose column (GE Healthcare) and the flow-through collected and concentrated at 4 °C using a centricon (vivaspin) with a 5 kDa cut-off, and loaded to a G75 Sephadex column (GE Healthcare) equilibrated with buffer C. Fractions eluting in the major peak of the G75 column were analysed by 15% SDS-PAGE and those deemed of good purity were concentrated and stored at −20 °C until required.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 M−1 cm−1).49
500 M−1 cm−1).49
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu(I) ≥ 3 creating a series of individual solutions with constant [CuI] and [L] and varying [protein]. Samples were left for ∼1 h and the transfer of Cu(I) from the [CuIL2]3− complex to the protein determined by measuring the absorbance of the [CuIL2]3− complex spectrophotometrically for L = BCA at 562 nm (ε = 7900 M−1 cm−1)59 and L = BCS at 483 nm (13
Cu(I) ≥ 3 creating a series of individual solutions with constant [CuI] and [L] and varying [protein]. Samples were left for ∼1 h and the transfer of Cu(I) from the [CuIL2]3− complex to the protein determined by measuring the absorbance of the [CuIL2]3− complex spectrophotometrically for L = BCA at 562 nm (ε = 7900 M−1 cm−1)59 and L = BCS at 483 nm (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1).60 By interchanging L, assays favoring competitive or non-competitive Cu(I) binding were set-up, which for the latter led to an estimate of the binding stoichiometry. The dissociation constant for Cu(I) (KD(CuI)) was determined from competitive assays by assuming the following reaction (1)
000 M−1 cm−1).60 By interchanging L, assays favoring competitive or non-competitive Cu(I) binding were set-up, which for the latter led to an estimate of the binding stoichiometry. The dissociation constant for Cu(I) (KD(CuI)) was determined from competitive assays by assuming the following reaction (1)