Retracted Article: Synthesis of diacylglycerol analogs bearing photoaffinity tags for labelling mammalian diacylglycerol kinase†
Sammy Eni Eni*,
Meng Rowland and
Michael D. Best
Department of Chemistry, University of Tennessee, 1420 Circle Dr., Knoxville, TN 37996-1600, USA. E-mail: senieni@vols.utk.edu
First published on 2nd March 2015
Abstract
Signaling lipids such as diacylglycerol (DAG), phosphatidic acid, and the phosphatidylinositol polyphosphates are site-specific ligands for protein binding partners. Herein, we report the apotheoses of our initial approach to the development of diverse probes which are vital for understanding lipid–protein interactions. When incorporated into liposomes, these probes reduce mammalian diacylglycerol kinase's (DGK) enzymatic activity in a concentration, time, and light dependent manner.
A small family of unique lipids present within plasma and organelle membranes act as critical regulatory biomolecules that control a variety of critical cellular pathways.1,2 Although a number of mechanisms exist for the regulation of protein function by lipids, a common motif involves the actions of lipids such as diacylglycerol (DAG),3,4 phosphatidic acid,5,6 and the phosphatidylinositol polyphosphates (PIPns)2 as site-specific ligands for protein binding partners. These binding interactions anchor peripheral proteins on the surfaces of cellular membranes and modulate the function of the bound protein either directly or through interaction with other proteins at the membrane interface. In addition, the generation of these signalling lipids is strictly regulated in a spatial and temporal manner, as they exist at low physiological concentrations, but become upregulated in response to biological processes, and thus their presentation within the cell explicitly controls the localization of receptors.7 As a result of the prominence of protein–lipid binding events as the basic interactions that feed information into critical cellular pathways, aberrant lipid activities often correlate to disabling diseases. For instance, the activities of DAG have been linked to a litany of noteworthy pathophysiological events including cancer and diabetes.8
Elucidation of the molecular level details of protein–lipid binding interactions has proven to be a challenging task, due in part to the complex nature of the membrane environment as well as the protein targets, which often possess multiple binding domains that display differential binding properties towards different lipids. For example, the most prominent family of DAG-binding proteins is the protein kinase Cs,9 which includes multiple isozymes possessing C1 binding domains that exhibit varying DAG-binding affinity.10,11 To overcome the challenges of studying protein–lipid binding interactions, there has been substantial interest in the development of functionalized lipid probes that are effective for the characterization of protein binding properties, which are typically synthetic lipid probes bearing appended reporter tags.9,12–15 Probes bearing photoaffinity tags are of particularly interest as they allow for covalent cross-linking to protein targets, which enables studies such as the mapping of protein binding domains and discovery of protein targets.16,17 In examples involving photoaffinity-labeled lipid probes, Nemoz and co-workers developed PA probes bearing phenylazide photoaffinity lables within the acyl chains that was used in the covalent labeling of type 4 cAMP-phosphodiesterases (PDE4).16 In addition, a range of photoactivatable PIPn probes have been developed and applied to characterize binding interactions.8,18–20
Previously, we reported a modular approach to the production of derivatized DAG probes17 that allowed for the efficient incorporation of a range of reporter groups in the final step of synthesis using click chemistry. This included the production of an analog bearing a benzophenone tag linked at the sn-1 position of the headgroup for photoaffinity labeling of DAG-binding proteins. Following synthesis, each of these probes was found to retain PKC-binding using an in vitro assay, indicating that modification of the lipid headgroup, but away from the hydroxyl group, was tolerated by protein targets. Headgroup derivatization was specifically targeted to enhance labeling by placing the tag in close proximity to bound peripheral proteins. While this initial approach was effective for producing active probes, the synthesis was complicated by significant problems with acyl chain migration that was only circumvented through careful protecting group manipulations. In addition, since cross-linking is affected by the presentation of the photoaffinity tag, which is difficult to predict, it is beneficial to produce multiple probes with varying tag displays. Thus, herein we report a strategy for the production of multiple photoactivatable DAG probes with varying photoaffinity tag presentation that promotes protein–lipid interactions and circumvents our previously encountered acyl migration enigma.
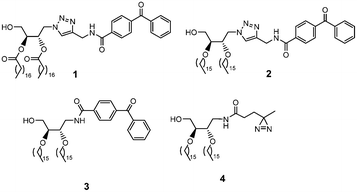
In photoaffinity labeling, the presentation of the cross-linking group is critical for successful labeling. As an example of the ramifications of tag placement, in the development of a PI(3,4,5)P3 activity probe for proteomic identification of protein targets, we found that the linker length between the lipid headgroup and the photoaffinity label significantly affected labeling.21 Since ideal tag presentation is difficult to predict, it is often advisable to generate multiple probes with varying tag display to discern the optimal approach. With the current strategy, in order to produce multiple photoaffinity probes with varying tag presentation, we took advantage of the versatile nature of the azide group to append photoactivatable groups using different linkage strategies. Specifically, the azide group was either directly functionalized via click chemistry or reduced to an amine for amide bond coupling, which would yield distinct linker lengths and geometries. In addition, different photoaffinity moieties were added, which would be expected to provide disparate results in cross-linking. In particular, we sought to decrease the size of the added cross-linking group to maximize the ability to fit within the confines of protein binding domains which led us to the synthesis of probed 1–4 in Scheme 1.
In the current studies, we additionally sought to overcome the issue of sn-2 to sn-3 acyl chain migration in the generation of DAG probes. Such migration in acylglycerols, which occurs under both acidic and basic conditions, generally complicates both synthesis and biological analysis.22 To circumvent this issue, the current DAG probe targets contain hydrophobic tails linked via ether bonds to lock these groups into place. This attachment was chosen due to prior evidence that ether-linked DAG analogs retain protein binding properties. Ether linkage also has the added benefits of blocking enzymatic removal of acyl chains, thus decreasing the routes by which the probes will be modified in biological samples. Finally, removal of the labile ester groups from these structures also expands the synthetic options available for probe synthesis and reporter group introduction.
Based on these designs, the targets of the current work include triazole-linked benzophenone analog 2, amide-linked benzophenone probe 3, and amide-linked diazirine 4, for which the synthetic routes are depicted in Scheme 2. The route commenced with diethyl-L-tartrate (5), for which the diol was protected through acetonide formation to 6, followed by reduction to bis-hydroxymethyl compound 7. Next mono-tosylation to 8 was followed by azide displacement to 9. Protection of the free hydroxyl group of 9 as a para-methoxybenzyl (PMB) ether to 10 was then followed by acetonide deprotection to produce diol 11. At this point, two hexadecyl chains were introduced through the Williamson ether synthesis at what will eventually become the sn-2 and sn-3 positions of the subsequent probes to generate 12. Finally, PMB deprotection was performed to set up late stage reporter tag introduction onto azido diether-DAG scaffold 13.
From precursor 13, tert-butyloxy (Boc) carbamate was added under hydrogen to 14. The Boc group was then deprotected, followed by amide bond coupling to append the benzophenone of probe 3. Additionally, we sought to further decrease the size of the photoaffinity label by instead introducing a diminutive diazirine group. To do so, diazirine-carboxylic acid was synthesized and then coupled to the DAG scaffold to produce probe 4.
DGKs are interfacial enzymes, catalyzing reactions at the two-dimensional interface between the membrane and the aqueous phase. Eukaryotic DGKs have been implicated in a number of physiological roles and human diseases. Very little is known about the structure of the catalytic domain, especially the lipid substrate binding site.
A photoaffinity labeling approach was carried out to identify the lipid binding site due to covalent attachment by the probes. As observed from Fig. 1, radiometric DGK activity assay was used to determine the enzyme's activity with probe 4 incorporated in liposomes in different concentrations pertaining to probe concentration, light exposure, and temperature. The results showed that probe 1 is used as a substrate to DGK and binds to DGK covalently as concentration of DGK increases which is also proportionate to time and light exposure. DGK activity studies were carried out with probe 1 which shows it deactivated DGK activity but not monotonically as probes 4 and 3 as shown in Fig. 2.
 | ||
| Fig. 1 DGK activity assay with probe 4 showing reduction of its activity in a concentration, temperature, and light dependent fashion. | ||
The results in Fig. 1 and 2 indicate a substantial difference in the activities of these probes to DGK. In these regard, we propose that the different photoaffinity tags might play a role in this variation in activity. Benzophenone, because of its bulky moiety and also requiring a high photo-activation energy, could account for its lower reactivity with DGK compared to the diazirine moiety which is smaller and requires a low photo-energy of activation.
The enzymatic activity of DGK was also measured on liposomes containing compound 3 which demonstrated that DGK's Michaelis constant (Km) for compound 3 must be high (Fig. 3A).
 | ||
| Fig. 3 DGK activity assay with probe 3 showing reduction of activity of DGK batches (601 E and 227 A). | ||
To further verify the cross-linking of the probes to the active site of DGK, probe 4 was used in which the band containing probe 4 and cross-linked DGK was evaluated using in-gel trypsin digestion following SDS-PAGE.
Certain adjacent tryptic peptides were disproportionately absent from the cross-linked sample as compared to the control sample. For example, ARPGPGPGPGPER was found six times in the control peptide, APGPAAAPGHSFR was found eleven times and IPCTSVAPSLVRVPVAHCFGPR was found three times. Of these peptides, ARPGPGPGPGPER and IPCTSVAPSLVRVPVAHCFGPR were found in the cross-linked sample, but APGPAAAPGHSFR was not, even though it was the most abundant in the control sample. If peptides were covalently modified with compound 4, they will be disproportionately absent from the cross-linked sample, either because the mass of the cross-linked peptide had been shifted and thus no longer appeared as the unmodified mass, or because the cross-linked peptide remained in the gel matrix.
Two such disproportionately absent peptides in these samples were found and, even more encouragingly, both coincided with regions of the protein predicted by primary sequence homology to include DAG-binding sites as depicted in Fig. 4.
 | ||
| Fig. 4 Amino acid coverage of in-gel trypsin-digested DGK cross-linked with compound 4 with certain peptides disproportionately absent in the cross-linked sample as compared to control sample. | ||
Detected peptides are highlighted in yellow. Peptides disproportionately absent in the sample with compound 4 are marked in red. The three C1 domains and the catalytic domain of DGK, which are predicted to contain DAG-binding sites, are shaded in blue and green, respectively. Oxidized sites are marked in bright green. In addition to the sequencing, tandem MS was carried out on probe 4 that cross-linked DGK which produced a product ion whose molecular weight is consistent with loss of either alkyl chain as shown in Fig. 5. Cross-linking efficiency depended on the length of the linker between the photoactivatable cross-linker and the DAG-like moiety in the probe: probes with longer linkers are able to access the active site and be used as substrates. DAG does not protect DGK from cross-linking, possibly because DAG may be enhancing DGK binding to probe-containing liposomes.
When cross-linked DGK is subjected to LC/MS/MS, certain peptides are disproportionately missing from the cross-linked DGK as compared to DGK irradiated in the absence of photoaffinity probe. These missing peptides coincided with regions of the protein predicted by primary sequence homology to contain DAG-binding sites, and are candidates for sites of photoaffinity cross-linking. We were unable to find any peptides shifted by the total mass of the probe following cross-linking, but the probe fragments during tandem mass spectrometry. The photoactivable tags, benzophenone as well as diazirine, were presented at the aqueous interface to enforce proximity to bound proteins for easy cross-linking after irradiation.
The tag placement also greatly influences the accessibility for modification, in which presentation on the surface of the membrane is also rewarding. We have pursued a variety of schemes which introduces tags as a Y-shaped lysine linker.12 This method allowed us to vary the length of the linker between the head group and the photoactivable tag as this length has been known to affect the magnitude of cross-linking. The proximity of the benzophenone was found to be very vital for successful protein labeling. This further circumvented the problem of the photoaffinity tag being buried in the membrane core if a longer linker is used. Our results strongly indicated that shorter linkers enhanced labeling to peripheral proteins compared to longer linkers and also that small and easily photo-activated moieties like diazirine bind DGK readily than bulkier moieties like benzophenone.
Consequently, our synthesis of DAG probes 3 and 4 was geared towards the placement of the benzophenone close to the head group in the aqueous interface with a short hydrophobic moiety which proved potent in deactivating DGK activity as envisioned. Also included in these new probes 2, 3 and 4 unlike probe 1 is the azide tag as our secondary handle. This azide allows for selective detection and purification of proteins that have been successfully labeled by the probe. The azide group is specifically chosen due to its diminutive size, robust properties, and bio-orthogonal reactivity through the 1,3 dipolar cycloaddition (click chemistry).13
Conclusion
Herein, we report the apotheoses of our initial approach to the development of the diverse probes that are potent as functionalized derivatives of DAG. Probes 2 and 3 are composed of a benzophenone as the photoactivable tag while probe 4 has diazirine. These two unique tags on the DAG probes are essential for their correlation in reactivity pertaining to cross-linking of peripheral proteins. Assay studies indicate that these probes after incorporation on a liposome, reduce diacylglycerol kinase (DGK) activity in a concentration, time, and UV-dependent manner. This dependence varies among the probes and is observed considerably only when the linker connecting the photoaffinity moiety and the DAG-like moiety on the probe is sufficiently short (3 and 4), small and easily photo-activated. This therefore indicates that these DAG probes possess variable properties for the labelling of the DGK enzyme. Our future goal is to use and apply these probes in other complex proteome systems and even live cells, in order to characterize their interactions with peripheral proteins especially those upregulated in cancer cells.Notes and references
- W. Cho and R. V. Stahelin, Membrane–protein interactions in cell signaling and membrane trafficking, Annu. Rev. Biophys. Biomol. Struct., 2005, 34, 119–151 CrossRef CAS PubMed.
- M. A. Lemmon, Membrane recognition by phospholipid-binding domains, Nat. Rev. Mol. Cell Biol., 2008, 9(2), 99–111 CrossRef CAS PubMed.
- S. Carrasco and I. Merida, Diacylglycerol, when simplicity becomes complex, Trends Biochem. Sci., 2007, 32(1), 27–36 CrossRef CAS PubMed.
- J. C. Gomez-Fernandez and S. Corbalan-Garcia, Diacylglycerols, multivalent membrane modulators, Chem. Phys. Lipids, 2007, 148, 1–25 CrossRef CAS PubMed.
- C. Testerink and T. Munnik, Phosphatidic acid: a multifunctional stress signaling lipid in plants, Trends Plant Sci., 2005, 10(8), 368–375 CrossRef CAS PubMed.
- M. D. Best, H. L. Zhang and G. D. Prestwich, Nat. Prod. Rep., 2010, 27(10), 1403–1430 RSC.
- H. Sprong, P. van der Sluijs and G. van Meer, How proteins move lipids and lipids move proteins, Nat. Rev. Mol. Cell Biol., 2001, 2(9), 698 Search PubMed.
- M. P. Wymann and R. Schneiter, Lipid signalling in disease, Nat. Rev. Mol. Cell Biol., 2008, 9(2), 162–176 CrossRef CAS PubMed.
- A. C. Newton, Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions, Chem. Rev., 2001, 101(8), 2353–2364 CrossRef CAS PubMed.
- B. Ananthanarayanan, R. V. Stahelin, M. A. Digman and W. H. Cho, Activation mechanisms of conventional protein kinase C isoforms are determined by the ligand affinity and conformational flexibility of their C1 domains, J. Biol. Chem., 2003, 278(47), 46886–46894 CrossRef CAS PubMed.
- R. V. Stahelin, M. A. Digman, M. Medkova, B. Ananthanarayanan, J. D. Rafter, H. R. Melowic and W. H. Cho, Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase C delta, J. Biol. Chem., 2004, 279(28), 29501–29512 CrossRef CAS PubMed.
- G. D. Prestwich, Touching all the bases: synthesis of inositol polyphosphate and phosphoinositide affinity probes from glucose, Acc. Chem. Res., 1996, 29(10), 503–513 CrossRef CAS.
- M. D. Best, M. M. Rowland and H. E. Bostic, Exploiting bioorthogonal chemistry to elucidate protein–lipid binding interactions and other biological roles of phospholipids, Acc. Chem. Res., 2011, 44, 686–698 CrossRef CAS PubMed.
- G. Dorman and G. D. Prestwich, Using photolabile ligands in drug discovery and development, Trends Biotechnol., 2000, 18(2), 64–77 CrossRef CAS.
- Y. Hatanaka and Y. Sadakane, Photoaffinity labeling in drug discovery and developments: Chemical gateway for entering proteomic frontier, Curr. Top. Med. Chem., 2002, 2, 271–288 CrossRef CAS.
- G. Nemoz, C. Sette and M. Conti, Selective activation of rolipram-sensitive, cAMP-specific phosphodiesterase isoforms by phosphatidic acid, Mol. Pharmacol., 1997, 51(2), 242–249 CAS.
- M. D. Smith, D. H. Gong, C. G. Sudhahar, J. C. Reno, R. V. Stahelin and M. D. Best, Synthesis and convenient functionalization of azide-labeled diacylglycerol analogues for modular access to biologically active lipid probes, Bioconjugate Chem., 2008, 19(9), 1855–1863 CrossRef CAS PubMed.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Click chemistry: diverse chemical function from a few good reactions, Angew. Chem., Int. Ed., 2001, 40(11), 2004–2021 CrossRef CAS.
- M. D. Best, Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules, Biochemistry, 2009, 48(28), 6571–6584 CrossRef CAS PubMed.
- E. M. Sletten and C. R. Bertozzi, Bioorthogonal chemistry: fishing for selectivity in a sea of functionality, Angew. Chem., Int. Ed., 2009, 48(38), 6974–6998 CrossRef CAS PubMed.
- M. M. Rowland, H. E. Bostic, D. Gong, A. E. Speers, N. Lucas, W. Cho, B. F. Cravatt and M. D. Best, Phosphatidylinositol 3,4,5-trisphosphate activity probes for the labeling and proteomic characterization of protein binding partners, Biochemistry, 2011, 50, 11143–11161 CrossRef CAS PubMed.
- D. R. Kodali, A. Tercyak, D. A. Fahey and D. M. Small, Acyl migration in 1,2-dipalmitoyl-sn-glycerol, Chem. Phys. Lipids, 1990, 52(3–4), 163–170 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra16730a |
| This journal is © The Royal Society of Chemistry 2015 |