Open Access Article
Open Access ArticleEnzymatic modification of 5′-capped RNA with a 4-vinylbenzyl group provides a platform for photoclick and inverse electron-demand Diels–Alder reaction†
Josephin Marie
Holstein‡
a,
Daniela
Stummer‡
ab and
Andrea
Rentmeister
*ab
aWestfälische Wilhelms-Universität Münster, Institute of Biochemistry, 48149 Muenster, Germany. E-mail: a.rentmeister@uni-muenster.de
bCells-in-Motion Cluster of Excellence (EXC 1003 – CiM), University of Muenster, 48149 Muenster, Germany
First published on 26th November 2014
Abstract
Chemo-enzymatic strategies provide a highly selective means to label different classes of biomolecules in vitro, but also in vivo. In the field of RNA, efficient labeling of eukaryotic mRNA with small organic reporter molecules would provide a way to detect endogenous mRNA and is therefore highly attractive. Although more and more bioorthogonal reactions are being reported, they can only be applied to chemo-enzymatic strategies if a suitable (i.e., click compatible) modification can be introduced into the RNA of interest. We report enzymatic site-specific transfer of a 4-vinylbenzyl group to the 5′-cap typical of eukaryotic mRNAs. The 4-vinylbenzyl group gives access to mRNA labeling using the inverse electron-demand Diels–Alder reaction, which does not work with an enzymatically transferred allyl group. The 4-vinylbenzyl-modified 5′-cap can also be converted in a photoclick reaction generating a “turn-on” fluorophore. Both click reactions are bioorthogonal and the two step approach also works in eukaryotic cell lysate. Enzymatic transfer of the 4-vinylbenzyl group addresses the lack of flexibility often attributed to biotransformations and thus advances the potential of chemo-enzymatic approaches for labeling.
Introduction
Chemo-enzymatic strategies have proven valuable for labeling biomolecules, including proteins,1–3 DNA,4,5 RNA,6,7 and glycans8,9in vitro or even in vivo. The key step is the specific, enzyme-mediated incorporation of a click-compatible residue into the biomolecule of choice. In a second step, the non-natural moiety can be targeted by a suitable click reaction that covalently links the reporter to the target molecule. For the enzymatic modification, wildtype or engineered methyltransferases with promiscuous activity on analogs of the natural methyl donor S-adenosyl-L-methionine (AdoMet) have proven useful to site-specifically transfer reactive handles to the biomolecule of interest.3,4,6,10 This two-step approach has also been realized for RNA and represents a promising basis for novel RNA isolation and labeling techniques.6,7,11,12 The field of RNA labeling is currently emerging, because a large fraction of eukaryotic mRNAs can show a distinct localization inside a cell, e.g. in Drosophila oocytes,13 yeast,14 fungi,15,16 and neurons.17,18 Subcellular mRNA localization contributes to the regulation of gene expression,19 and impairments can cause severe defects such as the fragile X syndrome or multiple sclerosis.20–22 Labeling of mRNAs in living cells in combination with modern sensitive live cell imaging techniques could allow investigation of transport mechanisms, dynamics, but also misregulation of these mRNAs. Consequently, a diverse set of approaches has been developed, ranging from hybridization-based techniques,23–26via genetically encoded protein- or RNA-based probes27–31 to the above mentioned post-synthetic modification strategies.11,12 The MS2-system relies on the tight interaction between the MS2 binding site and the MS2 coat protein of the bacteriophage MS2 and has recently been advanced to function in mice.32,33 However, it relies on a very long tag appended to the RNA of interest which may affect RNA interactions and transport.Chemo-enzymatic approaches aim to circumvent this limitation by installing small organic reporter molecules site-specifically to the RNA of interest. Besides enzymatic incorporation of modified nucleotides,34–38 several RNA methyltransferases have been successfully used to install alkene, alkyne, and azido groups.6,7,11,12 For the purpose of RNA labeling, one advantage of methyltransferases over polymerases is their selectivity for one particular class or even sequence of RNA (e.g. tRNA, snoRNA, mRNA). Thus, the RNA class or even sequence of interest can be modified in a complex mixture—an indispensable feature if endogenous RNA shall be modified in the cellular context. The enzymatically transferred modifications were so far reacted in thiol-ene, copper-catalyzed and strain-promoted azide–alkyne cycloaddition (CuAAC and SPAAC) reactions.6,7,11,12 Out of these, only SPAAC is bioorthogonal and thus suitable for intracellular RNA modification, although side-reactions with intracellular thiols have been reported stressing the importance of fine-tuning the reactivity.39
It would be highly desirable to make more of the recently developed bioorthogonal reactions featuring high yields, selectivities, and minimal side reactions (such as tetrazine ligation, photoinduced tetrazole–alkene cycloaddition, strain-promoted alkyne–nitrone cycloaddition40–43) (for review see ref. 44) accessible to enzymatically modified biomolecules, especially RNA. Tetrazine ligation is based on the inverse electron-demand Diels–Alder (IEDDA) reaction and was found to be biocompatible and established for protein labeling in cells.45 This reaction has also been used for RNA labeling of chemically or enzymatically synthesized dienophile-containing RNA.34,46 Typically, strained alkenes are used as dienophiles34,45 (especially trans-cyclooctene, norbornene, cyclopropene, and bicyclononyne) because very high reaction rates can be obtained (k2 ∼ 2000 M−1 s−1).43 The photoinduced tetrazole–alkene cycloaddition combines the speed and specificity of a click reaction with the versatility of a photochemical process.44,47 This “photoclick” (PC) chemistry was successfully used to label proteins in live prokaryotic and mammalian cells.1,48 It is triggered by light and it is fluorogenic, enabling generation of a fluorophore with spatio-temporal control. Long-wavelength photoactivatable tetrazoles are currently being developed and will make application in cells more interesting.49
However, the PC reaction has not been reported with RNA. For IEDDA, enzymatic incorporation and subsequent reaction of appropriately modified nucleotides into DNA and RNA has been reported,34,46,50 but enzymatic transfer of residues suitable for IEDDA is not yet possible.
We report here a novel AdoMet-analog and its use for enzymatic transfer of a 4-vinylbenzyl residue to the 5′-cap of RNA. The 4-vinylbenzyl residue can be efficiently reacted in IEDDA and PC reactions where the previously reported modifications failed to react or showed only very minor reactivity. The two-step approach can be performed in eukaryotic cell lysate. Changing the enzymatically transferred dipolarophile/dienophile influences the reaction and provides a valuable alternative to adjusting the reagent which is more common in biomolecular labeling approaches.51–54 Different fluorescence properties of the products, including a significant increase in fluorescence when the 4-vinylbenzyl-modification is reacted, were observed. The fact that the PC reaction is triggered by light (allowing for spatial and temporal control), and the significant increase in fluorescence represent major advances, especially regarding future intracellular applications.
Experimental
Materials
The reagents 4-vinylbenzylchloride, 3-bromo-1-propene, benzylbromide and S-adenosyl-L-homocysteine, aniline, sodium nitrite, p-methoxybenzaldehyde and phenylsulfonylhydrazine were purchased from Sigma-Aldrich. Cap analog m7GpppA was obtained from NEB. Tetrazine-5-TAMRA (545/575) was obtained from Jena Bioscience. HPLC grade acetonitrile was purchased from VWR. All components were used without further purification.Membrane Filters (0.025 μm, CME) for dialysis of bioconversions were purchased from Roth. The UV handlamp 254/365 nm UV-6 S/L was from Herolab. Analytical HPLC was carried out on an Agilent 1260 Infinity HPLC equipped with a Diode Array Detector (190–640 nm) using a Nucleodur® C18 Pyramid reversed-phase column (5 μm, 125 × 10 mm, 4 mm ID) from Macherey-Nagel. Preparative HPLC purification was carried out on the same HPLC using Nucleodur® C18 Pyramid reversed-phase column (5 μm, 125 × 10 mm, 10 mm ID). Fluorescence images of PAA gels were recorded on a ChemoCam Imager ECL Type HR 16-3200 (Intas) or with a Canon EOS Rebel XT Digital SLR 350D DS126071 8.0MP camera. Proton nuclear magnetic resonance spectra (1H NMR) were recorded on an Agilent DD2 600 MHz instrument at 10 °C. Chemical shifts were reported as δ in units of parts per million (ppm) relative to the solvent signal deuterium oxide-d2 (δ 4.80 ppm, singlet). Coupling constants are expressed in Hz. ESI-Orbitrap-MS were recorded on a LTQ Orbitrap XL™ Hybrid Ion Trap-Orbitrap Mass Spectrometer from Thermo Scientific and MALDI-TOF/TOF-MS on a Bruker Autoflex™ Speed system.
Synthesis and characterization of 5′-[(R/S)-[(3S)-3-amino-3-carboxypropyl]prop-2-enylsulfonio]-5′-deoxyadenosine (AdoPropen) 2, 5′-[(R/S)-[(3S)-3-amino-3-carboxypro-pyl]benzylsulfonio]-5′-deoxyadenosine (AdoBenz) 12 and 5′-[(R/S)-[(3S)-3-amino-3-carboxypropyl]4-vinylbenzylsulfonio]-5′-deoxyadenosine (AdoViBe) 3
AdoPropen 2 was synthesized according to Dalhoff et al.5 By following this synthesis strategy also AdoBenz 12 and AdoViBe 3 were generated. Briefly, S-adenosyl-L-homocysteine 4 (10 mg, 26 μmol) was dissolved in a 1.5 mL of a mixture of formic and acetic acid (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and cooled to 0 °C with an ice bath. Benzylbromide (186 μL, 1.6 mmol) or 4-vinylbenzylchloride (400 μL, 2.8 mmol) was slowly added to the stirred solution. The reaction mixture was allowed to warm up to room temperature and stirred for 4 days. The reaction was quenched by adding the mixture into 10–20 mL cool water and the water phase was extracted with 5 mL diethyl ether for three times. After lyophilization the crude product was purified by reversed-phase HPLC using a Nucleodur® C18 Pyramid column (5 μm, 125 × 10 mm, 10 mm ID). Elution was performed at a flowrate of 5 mL min−1 using water containing 0.01% TFA as eluent A and a gradient of acetonitrile containing 0.01% TFA from 0 to 7% percent within 15 min. The obtained diastereomeric mixture (approximately 1
1) and cooled to 0 °C with an ice bath. Benzylbromide (186 μL, 1.6 mmol) or 4-vinylbenzylchloride (400 μL, 2.8 mmol) was slowly added to the stirred solution. The reaction mixture was allowed to warm up to room temperature and stirred for 4 days. The reaction was quenched by adding the mixture into 10–20 mL cool water and the water phase was extracted with 5 mL diethyl ether for three times. After lyophilization the crude product was purified by reversed-phase HPLC using a Nucleodur® C18 Pyramid column (5 μm, 125 × 10 mm, 10 mm ID). Elution was performed at a flowrate of 5 mL min−1 using water containing 0.01% TFA as eluent A and a gradient of acetonitrile containing 0.01% TFA from 0 to 7% percent within 15 min. The obtained diastereomeric mixture (approximately 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was concentrated by lyophilization and the dried product was redissolved in water. Aliquots were stored at −20 °C before use. Concentrations of the AdoMet-analogs were determined by UV absorption with ε260 = 15
1) was concentrated by lyophilization and the dried product was redissolved in water. Aliquots were stored at −20 °C before use. Concentrations of the AdoMet-analogs were determined by UV absorption with ε260 = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 L mol−1 cm−1.5
400 L mol−1 cm−1.5
Synthesis of 2-(4-methoxyphenyl)-5-phenyl-2H-tetrazole 10
Synthesis of 10 was performed according to Ito et al.55 Briefly, 2-methoxybenzaldehyde (0.05 mol) was mixed with benzenesulfonyl hydrazide (0.05 mol) and dissolved in hot ethanol (50 mL). After cooling to room temperature the reaction mixture was diluted with water. The separated phenylsulfonylhydrazone was filtered, washed with ethanol and air dried. Parallel a cooled solution of sodium nitrite (5 mmol) in 2 mL water was added to a solution of aniline (5 mmol) and 1.3 mL of concentrated hydrochloric acid in 8 mL of 50% ethanol below 5 °C. The obtained arenediazonium chloride was added dropwise over a period of 30 min to a stirred solution of the prepared phenylsulfonylhydrazone dissolved in 40 mL ethanol at 2–5 °C. The reaction mixture was extracted with chloroform. The chloroform layer was washed with diluted hydrochloric acid followed by water and dried. After removal of the solvent, the crude product was purified by silica gel chromatography using toluene as eluent. The resulting tetrazole was purified by recrystallization in ethanol and characterized by ESI-MS as well as 1H NMR.Recombinant production and purification of GlaTgs2-Var1, MTAN and LuxS
GlaTgs2-Var1, MTAN and LuxS were recombinantly produced and purified as previously described.11,12,56Synthesis of P1-adenosine(5′)-P3-[N2-prop-2-enyl,7-methylguanosine(5′)] triphosphate 5, P1-adenosine(5′)-P3-[N2-benzyl,7-methylguanosine(5′)] triphosphate 13 and P1-adenosine(5′)-P3-[N2-4-vinylbenzyl,7-methylguanosine(5′)] triphosphate 6
N 2-Allyl-modified m7GpppA 5 was obtained as described earlier.12 Enzymatic conversion of m7GpppA 1 (275 μM or 1 mM in bioconversions with AdoViBe) with AdoBenz 12 (400 μM) or AdoViBe 3 (385 μM or 1.39 mM) by GlaTgs2-Var1 (30–90 μM) was performed in the presence of 4 μM MTAN and 3 μM LuxS in PBS (pH 7.4) or reaction buffer (50 mM Tris–HCl, 100 mM MgCl2, 100 mM NH4OAc, pH 8.4) at 37 °C for 3–4 h. Transfer reactions were analyzed by reversed-phase HPLC after protein precipitation with 1/10 volume 1 M HClO4 on an analytical Nucleodur® pyramid column as described earlier.11,12,56,57Photoclick reaction with modified m7GpppA
The bioconversions described above were stopped by incubating samples for 25 min at 68 °C, followed by centrifugation and dialysis against PBS buffer for 20 min on CME-filters (13 mm, 0.025 μm). Of these bioconversions, 8 μL were mixed with 8 μL acetonitrile and 5.5 mM tetrazole 10 in a black 384-well plate. Samples were mixed by pipetting and irradiated for 5 min with a handheld UV-lamp at 254 nm. Afterwards samples were transferred in reaction vials and wells were washed with 2.5 μL acetonitrile, which was added to the reactions. The photoclick labeling was allowed to proceed at 4 °C for 16 h. Afterwards samples were applied on a 20% denaturating PAA gel (using 2× loading buffer without tracking dyes) and separated at 10 W for 50 min. Analysis was performed by irradiating gels with handheld UV-lamp at 365 nm and detecting in-gel-fluorescence with a Canon EOS Rebel XT Digital SLR 350D DS126071 8.0MP camera.Photoclick reaction with modified m7GpppA in lysate
For experiments performed in lysate of eukaryotic cells, NIH 3T3 cells (ca. 1.5 × 107) were pelleted, resuspended in 50 μL PBS buffer and lysed by sonication. The supernatant was added to bioconversions instead of buffer generating N2-allyl-modified m7GpppA 5. Of this reaction mixture 8 μL were used for photoclick reaction as described above.Quantum yield measurements
To determine the relative fluorescence quantum yields of pyrazolines, 7 and 8 were detected in PAA gels by using a handheld UV-lamp (λex = 365 nm). Gel slices of isolated pyrazolines were used to extract desired compounds using 0.3 M NaOAc solution. Absorbance and fluorescence properties were analyzed by using a TECAN reader (Infinite® M1000 Pro). Quantum yields were determined by comparing the integrated fluorescence intensity to that of POPOP (1,4-bis-(5-phenyloxazol-2-yl)-benzol) (dissolved in cyclohexane, Φ = 0.97) with λex = 365 nm. Calculations were made according to Fery-Forgues and Lavabre.58Inverse electron-demand Diels–Alder reaction (IEDDA) with 4-vinylbenzyl-modified m7GpppA
The in situ generated N2-4-vinylbenzyl-m7GpppA 6 was used for IEDDA without further purification. Reactions were incubated with commercially available tetrazine-5-TAMRA 11 (1 mM from a 10 mM stock in DMSO) for 6 h at 37 °C in darkness according to Niederwieser et al.9 Samples were analyzed for successful conversion after gel electrophoresis on a 20% denat. PAA gel by fluorescence scanning with the ChemoCam Imager ECL Type HR 16-3200 (Intas) using green LED, excitation filter green (520 nm/35 nm) and an emission filter green (609 nm/54 nm).IEDDA with modified m7GpppA in lysate
Eukaryotic cell lysate was prepared as described above and used instead of buffer in the enzymatically catalyzed reaction resulting in N2-4-vinylbenzyl-m7GpppA 6. Reactions were incubated with 1 mM tetrazine-5-TAMRA 11 and analyzed as described above.Chemo-enzymatic labeling of capped RNA by IEDDA
In vitro produced and capped 106 nt RNA was generated as previously described.12 Precipitated RNA was resuspended in GlaTgs2-Var1 (100 μM), MTAN (1.2 μM), LuxS (0.6 μM), all purified using an ÄKTA purifier™ system and HisTrap™ FF 1 columns (GE Healthcare), and AdoViBe 3 (860 μM). Volume was adjusted to 5 μL with PBS buffer and possible RNAses were inhibited by 0.5 μL RiboLock RNase Inhibitor (Thermo Scientific). Samples were incubated for 90 min at 37 °C. After RNA precipitation, tetrazine-5-TAMRA 11 (400 μM) was added and the solution was adjusted to a final volume of 5 μL with PBS buffer. After an incubation period of 3 hours at 37 °C in darkness, samples were again analyzed on 10% denaturing PAA gels by fluorescence scanning with the ChemoCam Imager ECL Type HR 16-3200 (Intas).Results
GlaTgs2-Var1 catalyzes transfer of aromatic residues to the 5′-cap
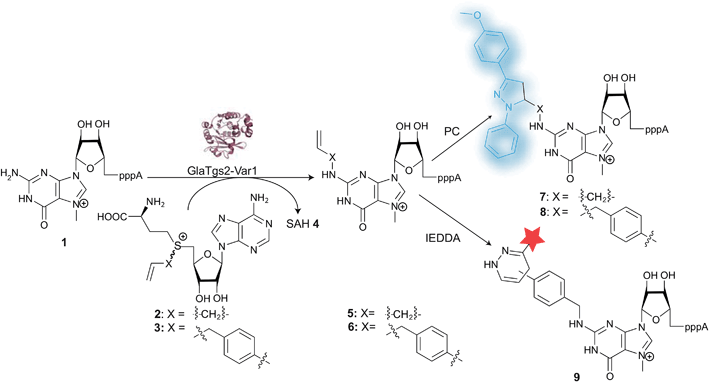
We previously reported on a variant of trimethylguanosine synthase 2 from Giardia lamblia (GlaTgs2-Var1) for transfer of allyl-, pentenynyl- or azido-butenyl group to 5′-capped RNA (e.g. eukaryotic mRNA) with yields of 30–90%, depending on the residue.11,12 These groups were transferred from analogs of the natural methyl group donor S-adenosyl-L-methionine (AdoMet) that have also been used with protein- and other DNA- or RNA-methyltransferases.3–7,59 AdoMet-analogs for the transfer of aromatic residues, however, have not been reported. Benzyl-substituted AdoMet analogs at the N6, 2′- and 3′-positions of AdoMet have only been made and tested to modulate the activity of methyltransferases—but not for methyl transfer.60,61 To test whether aromatic residues can also be enzymatically transferred, we synthesized a benzyl-analog of AdoMet, namely 5′-[(R/S)-[(3S)-3-amino-3-carboxy-propyl]benzylsulfonio]-5′-deoxyadenosine (AdoBenz, 12) and used it in bioconversions with GlaTgs2-Var1. Enzymatic transfer of the benzyl group to the minimal capped mRNA m7GpppA 1 yielded 47% of N2-benzyl-m7GpppA 13 and was confirmed by HPLC and MS (ESI Fig. 1†).Encouraged by these results, we supposed that enzymatic introduction of a 4-vinylbenzyl residue might also be feasible. This modification is an interesting platform for further chemical reactions, especially PC and IEDDA (Fig. 1). We therefore synthesized 5′-[(R/S)-[(3S)-3-amino-3-carboxypropyl]4-vinylbenzylsulfonio]-5′-deoxyadenosine (AdoViBe, 3), a novel AdoMet-analog carrying 4-vinylbenzyl instead of the methyl group, from commercially available 4-vinylbenzylchloride and S-adenosyl-L-homocysteine (SAH, 4). Formation of 3 was confirmed by mass spectrometry and NMR (ESI Fig. 2†).
AdoViBe 3 was also accepted by our RNA methyltransferase variant. Bioconversions of m7GpppA 1 with 3 catalyzed by GlaTgs2-Var1 yielded a new peak in HPLC analysis (tR = 14.6 min, Fig. 2). This peak was absent in control reactions that were performed without 1 or 3, and if denatured enzyme was used. The expected product P1-adenosine(5′)-P3-[N2-(4-vinylbenzyl),7-methylguanosine(5′)]triphosphate (N2-(4-vinylbenzyl)-m7GpppA) 6 was confirmed by mass spectrometry (ESI Fig. 3†). Based on HPLC analysis, up to 30% (300 μM) of 6 were formed in a 3 hour reaction at a catalyst loading of 10 mol%.
In a similar way, N2-allyl-m7GpppA 5 can be produced in 90% yield, as reported previously.12 Therefore, we now have access to 5′-cap structures modified with different types of terminal alkenes, namely N2-allyl-m7GpppA 5 and N2-(4-vinylbenzyl)-m7GpppA 6, by a highly site-specific enzymatic conversion.
Photoclick reactions with alkene-modified 5′-caps
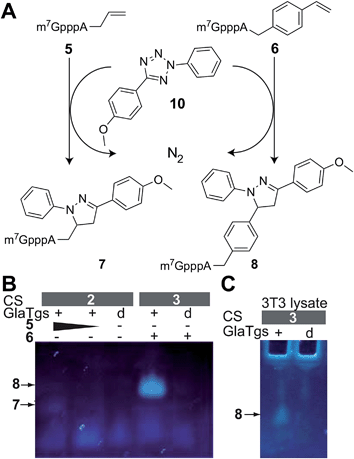
We wanted to test these modified 5′-cap structures in cell-compatible bioorthogonal click reactions. Photoclick reaction of an alkene with a tetrazole is particularly attractive because the reaction is intrinsically fluorogenic—a desirable feature for intracellular applications, where excess reagent cannot be washed away and would make it hard to distinguish the labeled biomolecule from the excess reagent.1,48 The choice of tetrazole is known to affect the kinetics of the reaction with a given dipolarophile51 and the fluorescence properties of the resulting pyrazoline adduct.52–54 The effect of the dipolarophile attached to the biomolecule however, has not been used to tune the reaction or the product.We tested the two differently N2-modified caps 5 and 6, produced in the bioconversion for reactions with 2-(4-methoxyphenyl)-5-phenyl-2H-tetrazole 10 (Fig. 3).
Photoclick reactions were initiated by irradiation with UV-light (λ = 254 nm, 5 min) and reacted for 16 hours at 4 °C. PC reactions were then analyzed for product formation by imaging in-gel fluorescence after electrophoretic separation and by ESI-MS (Fig. 3B, ESI Fig. 4 and 5†).
Both 5 and 6 reacted with 10, yielding the pyrazolines 7 and 8, respectively. As expected, both of these products were fluorescent, but 8 showed a much stronger fluorescent band compared to 7 (Fig. 3). By carrying out control and reference reactions we could rule out that a different excess of reagent 10, a UV-induced side reaction, or the aromatic 4-vinylbenzyl residue itself were responsible for this difference in fluorescence (Fig. 3B and ESI Fig. 6†). A quantum yield of 0.03 ± 0.02 could be determined for 8, whereas the signal of 7 was too weak to determine a reliable value. Taken together, our data suggest that the photoclick reaction with the vinylbenzyl-modified cap leads to a stronger fluorescing product (8) than with the allyl-modified cap, which is a desirable feature for eventual intracellular applications.
Notably, formation of modified cap 6 and subsequent labeling by PC worked also in the lysate of eukaryotic 3T3 cells, a fibroblast cell line (Fig. 3C). A fluorescent product band could only be detected by in-gel fluorescence in samples containing 6 and not in negative controls that were performed with denatured enzyme.
IEDDA works with 4-vinylbenzyl but not with allyl-modified 5′-caps
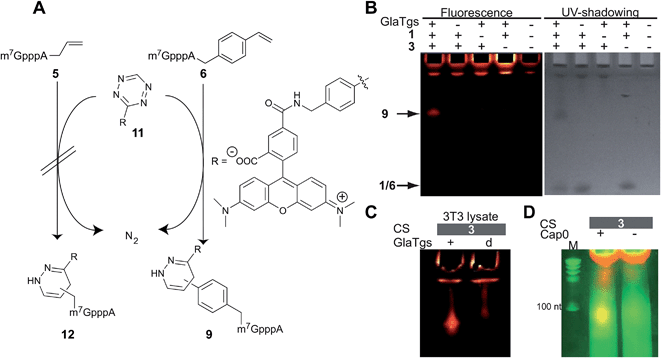
Alkene-functionalized biomolecules can also be selectively converted in aqueous solution by reactions other than the PC.9,12,62 Especially tetrazine ligation based on the IEDDA reaction has recently become popular, because it is fast and bioorthogonal without the need for catalyst.43,63 Mostly strained alkenes and alkynes (especially norbornene and bicyclononyne) have been introduced as modifications into biomolecules, although vinyl-modified nucleobases were recently reported to be suitable too.34,45,50Since IEDDA is also controlled by frontier orbitals, we figured that using different types of alkenes should influence the reactivity with a given tetrazine and thus allow tuning of the reaction. We therefore reacted enzymatically produced 5 and 6, respectively, directly as dienophiles in IEDDA using tetrazine-5-TAMRA™545/575 14 as diene (Fig. 4). In a chemo-enzymatic one-pot reaction, enzymatically produced N2-4-vinylbenzyl-m7GpppA 6 reacted with 11 to form the fluorescently labeled cap 9 that can be imaged upon irradiation at 520 nm (Fig. 4B). Controls show that in the absence of either AdoViBe 3, cap 1, or enzyme, no fluorescent band is formed and that excess of 11 can be found in the gel pockets. Formation of 9 was also confirmed by mass spectrometry (ESI Fig. 7†).
UV-shadowing of the same PAA gel shows an upshifted band for the newly formed 9 bearing the TAMRA label relative to the unmodified or vinylbenzyl-modified cap (1 and 6, lower band). Measurements of substrate decrease by reversed-phase HPLC suggests that IEDDA reaction leads to quantitative conversion of 6 to 9 (ESI Fig. 8†). IEDDA reaction with the N2-allyl-modified cap 5, however, was not successful (ESI Fig. 9†). This underscores the importance of being able to introduce different types of alkene modifications to tune the reactivity for certain reactions.
To test whether chemo-enzymatic labeling of the 5′-cap with PC and IEDDA is bioorthogonal, we also performed both reaction steps in the lysate of eukaryotic NIH 3T3 cells. Fig. 3C and 4C show the result of enzymatic labeling with 3 followed by PC or IEDDA, respectively. After PAGE, fluorescently labeled 5′-caps were only found in samples containing the modified cap 6 and not in negative controls that were performed with heat-inactivated enzyme. These data show that labeling was also specific in the presence of other proteins, like cap-binding proteins, and small molecules.
Finally, we tested our approach for longer capped RNAs. We therefore produced a 106 nt long capped RNA by in vitro transcription followed by capping with the vaccinia capping enzyme. A control RNA was produced in the same way but without methylation of the cap at position N7. Both RNAs were subjected to the chemo-enzymatic modification protocol using AdoViBe 3 and IEDDA to achieve labeling with tetrazine-5-TAMRA™545/575 11. As expected, imaging these samples after reaction and separation on a PAA gel showed that only correctly capped RNA was fluorescently labeled (Fig. 4D) confirming the specificity of this chemo-enzymatic strategy.
Conclusions
In conclusion, we demonstrate for the first time the enzymatic transfer of a 4-vinylbenzyl moiety that gives access to efficient PC and IEDDA reactions. Labeling of the 5′-cap was achieved in a chemo-enzymatic one-pot reaction starting with GlaTgs2-Var1-catalyzed bioconversions followed by the respective click chemistry. Both reactions are bioorthogonal, do not require a catalyst, and do not produce toxic side products, which makes them suitable for cellular applications. The increased fluorogenic properties of the 4-vinylbenzyl moiety in the PC reaction and its high reactivity in the IEDDA reaction, provide significant advantages over previously reported modifications.11,12The PC reaction has, so far, not been used to label RNA. Here we used it to label both 4-vinylbenzyl- and allyl-modified 5′-caps. Our data indicate that the 1,3,5-triarylpyrazoline 8 resulting from reaction with the 4-vinylbenzyl-modification shows a significantly higher quantum yield than the 1,3-diarylpyrazoline 7. Our results show how enzymatic transfer of different dipolarophiles (i) provides a means to tune the PC reaction and (ii) gives access to products with different fluorescence properties. We anticipate that the light-triggered PC reaction in combination with the bright fluorescence of 8 will prove valuable for applications in mRNA imaging.
The IEDDA reaction has not been used for enzymatically modified RNA (only for chemically and enzymatically synthesized RNA)—presumably because an appropriate transferable modification was not available. Varying the alkene modification represents a useful alternative strategy to influence the reaction itself as well as the properties of the product and thus complements the more common strategy to alter the reagent.
Since a lot of RNA-,6,7,12 but also DNA-4,5 and protein-methyltransferases3,10,59 show considerable promiscuous activities even as wildtype enzymes, we anticipate that our new AdoMet-analog 3 will also be accepted by many other methyltransferases. In combination with PC and IEDDA reaction our approach should thus prove useful for labeling different classes of biomolecules. Moreover, a 4-vinylbenzyl residue might also be incorporated by different approaches (e.g. as non-natural amino acid into proteins). Since PC is and IEDDA can also be made fluorogenic64,65 (which was not observable in our case), we anticipate that the approach will be fruitful for biomolecular labeling, and provide advantages when it comes to intracellular applications.
Acknowledgements
A.R. gratefully acknowledges financial support from the Emmy Noether-Programme of the Deutsche Forschungsgemeinschaft (RE 2796/2-1) and the Fonds der Chemischen Industrie. This work was partly supported by the Deutsche Forschungsgemeinschaft, DFG EXC 1003 Cells in Motion – Cluster of Excellence, Münster, Germany. J. M. H. thanks the Fonds der Chemischen Industrie for a doctoral fellowship. We acknowledge the MS- and NMR-facility of the Department of Organic Chemistry, University of Münster, as well as the MS facility of the University of Hamburg for assistance with analytical measurements. We thank Prof. Hahn (University of Hamburg) for providing the DNA template for transcription. We would like to thank Prof. Zhaohui “Sunny” Zhou (Northeastern University, Boston, MA USA) as well as Prof. Birgit Dräger (University of Halle, Germany) for plasmids encoding LuxS and MTAN.Notes and references
- W. Song, Y. Wang, J. Qu and Q. Lin, J. Am. Chem. Soc., 2008, 130, 9654–9655 CrossRef CAS PubMed.
- K. E. Beatty, J. D. Fisk, B. P. Smart, Y. Y. Lu, J. Szychowski, M. J. Hangauer, J. M. Baskin, C. R. Bertozzi and D. A. Tirrell, ChemBioChem, 2010, 11, 2092–2095 CrossRef CAS PubMed.
- W. Peters, S. Willnow, M. Duisken, H. Kleine, T. Macherey, K. E. Duncan, D. W. Litchfield, B. Luscher and E. Weinhold, Angew. Chem., Int. Ed., 2010, 49, 5170–5173 CrossRef CAS PubMed.
- C. Dalhoff, G. Lukinavicius, S. Klimasauskas and E. Weinhold, Nat. Chem. Biol., 2006, 2, 31–32 CrossRef CAS PubMed.
- C. Dalhoff, G. Lukinavicius, S. Klimasauakas and E. Weinhold, Nat. Protoc., 2006, 1, 1879–1886 CrossRef CAS PubMed.
- Y. Motorin, J. Burhenne, R. Teimer, K. Koynov, S. Willnow, E. Weinhold and M. Helm, Nucleic Acids Res., 2011, 39, 1943–1952 CrossRef CAS PubMed.
- M. Tomkuviene, B. Clouet-d'Orval, I. Cerniauskas, E. Weinhold and S. Klimasauskas, Nucleic Acids Res., 2012, 40, 6765–6773 CrossRef CAS PubMed.
- N. J. Agard, J. A. Prescher and C. R. Bertozzi, J. Am. Chem. Soc., 2004, 126, 15046–15047 CrossRef CAS PubMed.
- A. Niederwieser, A. K. Spate, L. D. Nguyen, C. Jungst, W. Reutter and V. Wittmann, Angew. Chem., Int. Ed., 2013, 52, 4265–4268 CrossRef CAS PubMed.
- O. Binda, M. Boyce, J. S. Rush, K. K. Palaniappan, C. R. Bertozzi and O. Gozani, ChemBioChem, 2011, 12, 330–334 CrossRef CAS PubMed.
- J. M. Holstein, D. Schulz and A. Rentmeister, Chem. Commun., 2014, 50, 4478–4481 RSC.
- D. Schulz, J. M. Holstein and A. Rentmeister, Angew. Chem., 2013, 52, 7874–7878 CrossRef CAS PubMed.
- E. Lecuyer, H. Yoshida, N. Parthasarathy, C. Alm, T. Babak, T. Cerovina, T. R. Hughes, P. Tomancak and H. M. Krause, Cell, 2007, 131, 174–187 CrossRef CAS PubMed.
- E. Bertrand, P. Chartrand, M. Schaefer, S. M. Shenoy, R. H. Singer and R. M. Long, Mol. Cell, 1998, 2, 437–445 CrossRef CAS.
- S. Baumann, T. Pohlmann, M. Jungbluth, A. Brachmann and M. Feldbrugge, J. Cell Sci., 2012, 125, 2740–2752 CrossRef CAS PubMed.
- J. Konig, S. Baumann, J. Koepke, T. Pohlmann, K. Zarnack and M. Feldbrugge, EMBO J., 2009, 28, 1855–1866 CrossRef PubMed.
- M. S. Rook, M. Lu and K. S. Kosik, J. Neurosci., 2000, 20, 6385–6393 CAS.
- I. J. Cajigas, G. Tushev, T. J. Will, S. Tom Dieck, N. Fuerst and E. M. Schuman, Neuron, 2012, 74, 453–466 CrossRef CAS PubMed.
- K. C. Martin and A. Ephrussi, Cell, 2009, 136, 719–730 CrossRef CAS PubMed.
- G. J. Bassell and S. T. Warren, Neuron, 2008, 60, 201–214 CrossRef CAS PubMed.
- D. A. Lyons, S. G. Naylor, A. Scholze and W. S. Talbot, Nat. Genet., 2009, 41, 854–858 CrossRef CAS PubMed.
- Y. S. Aulchenko, I. A. Hoppenbrouwers, S. V. Ramagopalan, L. Broer, N. Jafari, J. Hillert, J. Link, W. Lundstrom, E. Greiner, A. Dessa Sadovnick, D. Goossens, C. Van Broeckhoven, J. Del-Favero, G. C. Ebers, B. A. Oostra, C. M. van Duijn and R. Q. Hintzen, Nat. Genet., 2008, 40, 1402–1403 CrossRef CAS PubMed.
- S. Kummer, A. Knoll, E. Socher, L. Bethge, A. Herrmann and O. Seitz, Bioconjugate Chem., 2012, 23, 2051–2060 CrossRef CAS PubMed.
- J. S. Rinne, T. P. Kaminski, U. Kubitscheck and A. Heckel, Chem. Commun., 2013, 49, 5375–5377 RSC.
- J. C. Politz, K. L. Taneja and R. H. Singer, Nucleic Acids Res., 1995, 23, 4946–4953 CrossRef CAS PubMed.
- J. C. Politz, E. S. Browne, D. E. Wolf and T. Pederson, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6043–6048 CrossRef CAS.
- R. L. Strack, M. D. Disney and S. R. Jaffrey, Nat. Methods, 2013, 10, 1219–1224 CrossRef CAS PubMed.
- T. Ozawa, Y. Natori, M. Sato and Y. Umezawa, Nat. Methods, 2007, 4, 413–419 CAS.
- H. Yoshimura, A. Inaguma, T. Yamada and T. Ozawa, ACS Chem. Biol., 2012, 7, 999–1005 CrossRef CAS PubMed.
- J. S. Paige, K. Y. Wu and S. R. Jaffrey, Science, 2011, 333, 642–646 CrossRef CAS PubMed.
- S. J. Kellermann, A. K. Rath and A. Rentmeister, ChemBioChem, 2013, 14, 200–204 CrossRef CAS PubMed.
- H. Y. Park, H. Lim, Y. J. Yoon, A. Follenzi, C. Nwokafor, M. Lopez-Jones, X. Meng and R. H. Singer, Science, 2014, 343, 422–424 CrossRef CAS PubMed.
- T. Lionnet, K. Czaplinski, X. Darzacq, Y. Shav-Tal, A. L. Wells, J. A. Chao, H. Y. Park, V. de Turris, M. Lopez-Jones and R. H. Singer, Nat. Methods, 2011, 8, 165–170 CrossRef CAS PubMed.
- J. Schoch, S. Ameta and A. Jaschke, Chem. Commun., 2011, 47, 12536–12537 RSC.
- M.-L. Winz, A. Samanta, D. Benzinger and A. Jäschke, Nucleic Acids Res., 2012, 40, e78 CrossRef CAS PubMed.
- C. Y. Jao and A. Salic, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15779–15784 CrossRef CAS PubMed.
- E. Paredes and S. R. Das, ChemBioChem, 2011, 12, 125–131 CrossRef CAS PubMed.
- M. Grammel, H. Hang and N. K. Conrad, ChemBioChem, 2012, 13, 1112–1115 CrossRef CAS PubMed.
- E. J. Kim, D. W. Kang, H. F. Leucke, M. R. Bond, S. Ghosh, D. C. Love, J. S. Ahn, D. O. Kang and J. A. Hanover, Carbohydr. Res., 2013, 377, 18–27 CrossRef CAS PubMed.
- C. S. McKay, J. A. Blake, J. Cheng, D. C. Danielson and J. P. Pezacki, Chem. Commun., 2011, 47, 10040–10042 RSC.
- S. B. Engelsma, L. I. Willems, C. E. van Paaschen, S. I. van Kasteren, G. A. van der Marel, H. S. Overkleeft and D. V. Filippov, Org. Lett., 2014, 16, 2744–2747 CrossRef CAS PubMed.
- R. K. V. Lim and Q. Lin, Acc. Chem. Res., 2011, 44, 828–839 CrossRef CAS PubMed.
- M. L. Blackman, M. Royzen and J. M. Fox, J. Am. Chem. Soc., 2008, 130, 13518–13519 CrossRef CAS PubMed.
- C. P. Ramil and Q. Lin, Chem. Commun., 2013, 49, 11007–11022 RSC.
- D. S. Liu, A. Tangpeerachaikul, R. Selvaraj, M. T. Taylor, J. M. Fox and A. Y. Ting, J. Am. Chem. Soc., 2012, 134, 792–795 CrossRef CAS PubMed.
- A. M. Pyka, C. Domnick, F. Braun and S. Kath-Schorr, Bioconjugate Chem., 2014, 25, 1438–1443 CrossRef CAS PubMed.
- M. A. Tasdelen and Y. Yagci, Angew. Chem., Int. Ed., 2013, 52, 5930–5938 CrossRef CAS PubMed.
- W. J. Song, Y. Z. Wang, Z. P. Yu, C. I. R. Vera, J. Qu and Q. Lin, ACS Chem. Biol., 2010, 5, 875–885 CrossRef CAS PubMed.
- Z. Yu, T. Y. Ohulchanskyy, P. An, P. N. Prasad and Q. Lin, J. Am. Chem. Soc., 2013, 135, 16766–16769 CrossRef CAS PubMed.
- H. Busskamp, E. Batroff, A. Niederwieser, O. S. Abdel-Rahman, R. F. Winter, V. Wittmann and A. Marx, Chem. Commun., 2014, 50, 10827–10829 RSC.
- Y. Wang, W. Song, W. J. Hu and Q. Lin, Angew. Chem., Int. Ed., 2009, 48, 5330–5333 CrossRef CAS PubMed.
- K. Rurack and U. Resch-Genger, Chem. Soc. Rev., 2002, 31, 116–127 RSC.
- Z. Yu, L. Y. Ho, Z. Wang and Q. Lin, Bioorg. Med. Chem. Lett., 2011, 21, 5033–5036 CrossRef CAS PubMed.
- C. J. Fahrni, L. C. Yang and D. G. VanDerveer, J. Am. Chem. Soc., 2003, 125, 3799–3812 CrossRef CAS PubMed.
- S. Ito, Y. Tanaka, A. Kakehi and K.-I. Kondo, Bull. Chem. Soc. Jpn., 1976, 49, 1920–1923 CrossRef CAS.
- D. Schulz and A. Rentmeister, RNA Biol., 2012, 9, 577–586 CrossRef CAS PubMed.
- T. Monecke, A. Dickmanns and R. Ficner, Nucleic Acids Res., 2009, 37, 3865–3877 CrossRef CAS PubMed.
- S. Fery-Forgues and D. Lavabre, J. Chem. Educ., 1999, 76, 1260–1264 CrossRef CAS.
- K. Islam, I. Bothwell, Y. Chen, C. Sengelaub, R. Wang, H. Deng and M. Luo, J. Am. Chem. Soc., 2012, 134, 5909–5915 CrossRef CAS PubMed.
- J. Li, H. Wei and M.-M. Zhou, J. Med. Chem., 2011, 54, 7734–7738 CrossRef CAS PubMed.
- Q. Lin, F. Y. Jiang, P. G. Schultz and N. S. Gray, J. Am. Chem. Soc., 2001, 123, 11608–11613 CrossRef CAS PubMed.
- S. Wittrock, T. Becker and H. Kunz, Angew. Chem., Int. Ed., 2007, 46, 5226–5230 CrossRef CAS PubMed.
- N. K. Devaraj, R. Weissleder and S. A. Hilderbrand, Bioconjugate Chem., 2008, 19, 2297–2299 CrossRef CAS PubMed.
- N. K. Devaraj, S. Hilderbrand, R. Upadhyay, R. Mazitschek and R. Weissleder, Angew. Chem., Int. Ed., 2010, 49, 2869–2872 CrossRef CAS PubMed.
- L. G. Meimetis, J. C. Carlson, R. J. Giedt, R. H. Kohler and R. Weissleder, Angew. Chem., 2014, 53, 7531–7534 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: See DOI: 10.1039/c4sc03182b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |