Open Access Article
Open Access ArticleBioorthogonal oxime ligation mediated in vivo cancer targeting†
Li
Tang‡
a,
Qian
Yin‡
a,
Yunxiang
Xu
a,
Qin
Zhou
b,
Kaimin
Cai
a,
Jonathan
Yen
a,
Lawrence W.
Dobrucki
c and
Jianjun
Cheng
*ac
aDepartment of Materials Science and Engineering, University of Illinois at Urbana–Champaign, 1304 West Green Street, Urbana, IL 61801, USA. E-mail: jianjunc@illinois.edu; Fax: +1-217-333-2736; Tel: +1-217-244-3924
bDepartment of Pharmaceutical Science, Guangdong Pharmaceutical University, Guangzhou, Guangdong 510006, China
cDepartment of Bioengineering, University of Illinois at Urbana–Champaign, Urbana, IL 61801, USA
First published on 2nd February 2015
Abstract
Current cancer targeting relying on specific biological interaction between the cell surface antigen and respective antibody or its analogue has proven to be effective in the treatment of different cancers; however, this strategy has its own limitations, such as the heterogeneity of cancer cells and immunogenicity of the biomacromolecule binding ligands. Bioorthogonal chemical conjugation has emerged as an attractive alternative to biological interaction for in vivo cancer targeting. Here, we report an in vivo cancer targeting strategy mediated by bioorthogonal oxime ligation. An oxyamine group, the artificial target, is introduced onto 4T1 murine breast cancer cells through liposome delivery and fusion. Poly(ethylene glycol)-polylactide (PEG-PLA) nanoparticles (NPs) are surface-functionalized with aldehyde groups as targeting ligands. The improved in vivo cancer targeting of PEG-PLA NPs is achieved through specific and efficient chemical reaction between the oxyamine and aldehyde groups.
Introduction
Current active cancer targeting mainly relies on specific and strong biological interaction between a natural target, such as a cancer cell surface antigen, and the corresponding biomacromolecule ligand, such as an antibody or its analogues.1–9 Although moderate cancer targeting efficiency can be achieved, this strategy has its intrinsic limitations. First, the biological interaction between antigen and antibody is generally weaker than covalent binding, which exhibits dissociation constants Kd below 10−8 M for many highly robust bioorthogonal reactions.10–12 Second, limited numbers of receptors per cell and heterogeneity of cancer cells could result in saturation and insufficient targeting or undesired off-targeting.13 Third, biomacromolecule binding ligands are generally more immunogenic and vulnerable than small molecule ligands and may be subjected to enzymatic degradation in vivo; the large size relative to small molecule ligands and the undesired physiochemical properties of the biomacromolecule binding ligands may hinder their access to the target.14 The incorporation of such ligands with anticancer drugs or nanomedicines also requires sophisticated techniques to maintain the binding site intact.15 Nanomedicine–ligand conjugates with a very high surface density of biomacromolecule binding ligands may have modified surface properties, resulting in reduced targeting effects and sometimes an undesired immune response.16Bioorthogonal chemical conjugation with high specificity, efficiency and biocompatibility has emerged as an attractive alternative to specific biological interaction in vivo.17–21 Rather than receptor–antibody interaction, such chemical conjugation is a covalent binding between two bioorthogonal functional groups. Compared with biological interactions, it has several advantages. First, an extremely high affinity between an unnatural target and its respective ligand, which bear a pair of bioorthogonal functional groups, could be achieved using highly specific and robust bioorthogonal chemistry. Second, an unnatural target containing one bioorthogonal functional group of the pair can be artificially introduced onto cancer cells in a controlled manner through various cell surface engineering methodologies.12,22 For example, metabolic glycoengineering has been widely used to express azide groups on the cell surface.22 A liposome fusion technique has successfully introduced bioorthogonal functional groups on the cell membrane.23,24 Third, small molecule ligands are generally easily synthesized on a large scale, stable under physiological conditions, easily conjugated with therapeutics including nanomedicines and have minimum impact on the nanoparticle (NP) properties due to their small size. Therefore, using bioorthogonal chemical conjugation could be a promising strategy for in vivo cancer targeting.25–32
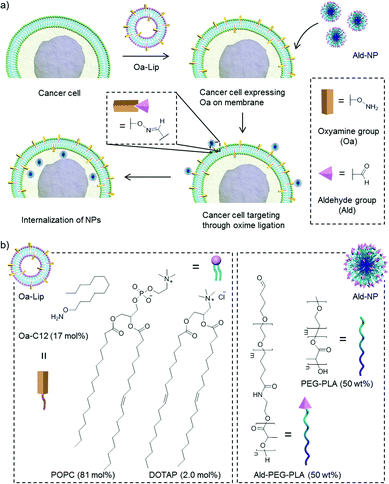
Oxime ligation is a highly efficient click-type bioorthogonal condensation reaction between an oxyamine and an aldehyde or a ketone.11,18,33 The formed oxime is highly stable toward hydrolysis (Kd ≤ 10−8 M) compared with the corresponding imines or hydrazones.34 Thus, oxime ligation is compatible with most biomolecule functionalities, including amines, and ideally suited for application in living systems.23,35,36 However, in vivo cancer targeting using an oxime ligation reaction has not been reported. Herein, we report a novel chemical conjugation mediated in vivo cancer targeting of poly(ethylene glycol)-b-polylactide (PEG-PLA) NPs through bioorthogonal oxime ligation (Scheme 1a). To prove the concept, the artificial target, an oxyamine group, was labelled on 4T1 murine breast cancer cells through liposome delivery and fusion. The cancer targeting efficiency was evaluated in vitro and in vivo using PEG-PLA NPs functionalized with aldehyde groups on the surface (Ald-NP, Scheme 1b).
Results and discussion
Effective surface engineering of target cancer cells to present bioorthogonal function groups is the prerequisite for cancer targeting via chemical conjugation. Metabolic glycoengineering has proven to be a powerful methodology to present glycan with a specific chemical group on the plasma membrane. However, this method perturbs cellular physiology and may have the risk of interfering with significant biochemical pathways or cellular functions of normal cells when these glycans are off target.37 Inspired by the recent work of Yousaf and others,24,38,39 we explored the use of tailored liposomes to introduce an oxyamine group on cancer cells though liposome delivery and fusion. We synthesized O-dodecyloxyamine (Oa-C12) and incorporated it into a liposome composed of a neutral lipid, egg palmitoyl-oleoyl phosphatidylcholine (POPC) and a cationic lipid, 1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP) at a mole ratio of Oa-C12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPC
POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOTAP = 17
DOTAP = 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 81
81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (Oa-Lip, Scheme 1b). The hydrophobic tail facilitated the spontaneous insertion of Oa-C12 into the liposome membrane.24 The cationic lipid, DOTAP, was added to enhance the interaction between the synthesized Oa-Lip and negatively charged cell membrane through electrostatic destabilization and thus increase the liposome fusion and presentation of Oa groups on cell surface. The formulated Oa-Lip had a hydrodynamic size of 136 nm determined by dynamic light scattering (DLS) and transmission electronic microscopy (TEM) (Fig. S1†). They also exhibited a positive ζ-potential on the surface (12.3 mV, Table S1†) at neutral pH owing to Oa-C12 and DOTAP.
2 (Oa-Lip, Scheme 1b). The hydrophobic tail facilitated the spontaneous insertion of Oa-C12 into the liposome membrane.24 The cationic lipid, DOTAP, was added to enhance the interaction between the synthesized Oa-Lip and negatively charged cell membrane through electrostatic destabilization and thus increase the liposome fusion and presentation of Oa groups on cell surface. The formulated Oa-Lip had a hydrodynamic size of 136 nm determined by dynamic light scattering (DLS) and transmission electronic microscopy (TEM) (Fig. S1†). They also exhibited a positive ζ-potential on the surface (12.3 mV, Table S1†) at neutral pH owing to Oa-C12 and DOTAP.
We next used the Oa-Lip to engineer the 4T1 murine breast cancer cell surface. In order to verify the successful cell surface labelling, we first conjugated a fluorescent dye, rhodamine (Rhd), to Oa-C12 to prepare Rhd-C12 and formulated the fluorescent liposome with POPC/DOTAP at the same mole ratio (Rhd-Lip, Fig. S3a†). Rhd-Lips were then added to the 4T1 cells and incubated in cell culture for 4 h (Fig. S3b†). The high fluorescence signal of Rhd shown in the confocal microscopy image indicates the successful Rhd-Lip fusion and display of Rhd-C12 on the 4T1 cells (Fig. S3c†). Direct addition of Rhd-C12 to the cell culture only resulted in minimum cell surface labelling of fluorescence suggesting the importance of the liposome binding and fusion process.23,24 Furthermore, we investigated whether the Oa groups displayed on the cell surface were still active for oxime ligation. The 4T1 cell surface was engineered with Oa-Lip similarly to present Oa groups on the 4T1 cell membrane (denoted as o4T1). Rhd dye bearing an aldehyde group (Ald-Rhd) was subsequently added after washing off the excess Oa-Lip (Fig. S4a†). A markedly increased fluorescence signal was observed on the 4T1 cells with surface-expressed Oa groups (o4T1) through the specific oxime ligation with Ald-Rhd. In contrast, the o4T1 cells treated with non-functional Rhd or native 4T1 cells treated with Ald-Rhd resulted in a limited level of fluorescence signal likely due to the nonspecific and poorly efficient conjugation or adsorption (Fig. S4b†). Therefore, these results demonstrated that the cancer cell surface could be effectively engineered using a liposome fusion strategy to express chemically active Oa groups to further facilitate specific oxime ligation on the cell membrane.
Intracellular fluorescence was observed during the liposome fusion process suggesting endocytosis of these liposomes or their components may occur after membrane fusion (Fig. S3c†). This could provide a possible method for internal membrane engineering for intracellular targeting.24 However, it was noticed that sufficient functional groups were present on cell surface to facilitate the oxime ligation with the corresponding ligands (Fig. S4b†). Such intracellular membrane labelling may be beneficial in the scenario of targeted intracellular drug delivery.
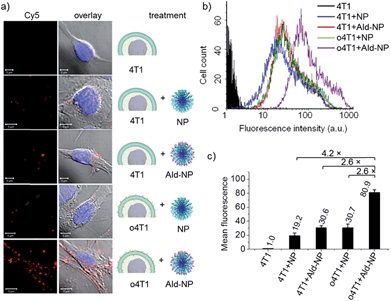
Next, we studied the in vitro cancer cell targeting of the nanomedicine toward 4T1 cells using the oxime ligation reaction. PEG and PLA are highly biocompatible polymers widely used in the formulation of polymeric nanomedicines for cancer therapy. Small molecule anticancer drugs could be easily incorporated into the PEG-PLA NPs prepared with the block copolymer via physical encapsulation or chemical conjugation.40,41 We used a fluorescent dye, Cy5, as a model drug to fluorescently label the NPs for the cancer cell binding and internalization study.42 To introduce Ald groups to the NP surface, a block polymer of Ald functionalized PEG-PLA was co-precipitated with PEG-PLA at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio to form Ald-NPs (Scheme 1b). The resulting Ald-NPs were about 119 nm in size with a narrow size distribution (Fig. S2†). Native 4T1 or o4T1 cells were then treated with Cy5 labeled NPs or Ald-NPs and analyzed by confocal fluorescence microscopy and flow cytometry (Fig. 1). Native 4T1 cells incubated with PEG-PLA NPs showed minimum cell binding and/or internalization (Fig. 1a). Native 4T1 cells treated with Ald-NPs or o4T1 cells treated with NPs without Ald on the surface showed a slightly higher level of NP binding or uptake. The former may be due to some non-specific reactions between the cell surface groups and Ald; the latter could be the result of increased electrostatic interaction between the surface Oa groups on o4T1 and the negatively charged NPs (−10.0 mV ζ-potential, Table S1†). When the o4T1 cells were treated with Ald-NPs, the cellular binding and/or internalization was remarkably enhanced, likely because of the specific and efficient oxime ligation between the cell surface Oa groups and Ald groups on Ald-NPs. Further quantification with flow cytometry indicated a 2.6 fold increase in cell uptake of o4T1 + Ald-NP compared with 4T1 + Ald-NP or o4T1 + NP, and a 4.2 fold increase compared with 4T1 + NP (Fig. 1b and c). The results demonstrated that Oa–Ald recognition largely improves cancer targeting via oxime ligation. It is also noticeable that a significant amount of the Ald-NPs were internalized into the o4T1 cells. These observations indicate that the bioorthogonal oxime ligation mediated not only the enhanced cancer cell–NP binding but also the subsequent endocytosis of the NPs, which is important for intracellular delivery of anticancer drugs to achieve improved efficacy against cancer cells.
1 weight ratio to form Ald-NPs (Scheme 1b). The resulting Ald-NPs were about 119 nm in size with a narrow size distribution (Fig. S2†). Native 4T1 or o4T1 cells were then treated with Cy5 labeled NPs or Ald-NPs and analyzed by confocal fluorescence microscopy and flow cytometry (Fig. 1). Native 4T1 cells incubated with PEG-PLA NPs showed minimum cell binding and/or internalization (Fig. 1a). Native 4T1 cells treated with Ald-NPs or o4T1 cells treated with NPs without Ald on the surface showed a slightly higher level of NP binding or uptake. The former may be due to some non-specific reactions between the cell surface groups and Ald; the latter could be the result of increased electrostatic interaction between the surface Oa groups on o4T1 and the negatively charged NPs (−10.0 mV ζ-potential, Table S1†). When the o4T1 cells were treated with Ald-NPs, the cellular binding and/or internalization was remarkably enhanced, likely because of the specific and efficient oxime ligation between the cell surface Oa groups and Ald groups on Ald-NPs. Further quantification with flow cytometry indicated a 2.6 fold increase in cell uptake of o4T1 + Ald-NP compared with 4T1 + Ald-NP or o4T1 + NP, and a 4.2 fold increase compared with 4T1 + NP (Fig. 1b and c). The results demonstrated that Oa–Ald recognition largely improves cancer targeting via oxime ligation. It is also noticeable that a significant amount of the Ald-NPs were internalized into the o4T1 cells. These observations indicate that the bioorthogonal oxime ligation mediated not only the enhanced cancer cell–NP binding but also the subsequent endocytosis of the NPs, which is important for intracellular delivery of anticancer drugs to achieve improved efficacy against cancer cells.
We next evaluated the potential of oxime ligation for in vivo cancer targeting. A subcutaneous 4T1 tumor model was established on both flanks of BALB/c mice. To prove the concept of cancer cell surface engineering in vivo using the liposome fusion technique and subsequent oxime ligation mediated cancer targeting, we injected Oa-Lip intratumorally to label the cancer cells in vivo. We first verified the cancer cell surface presentation of Oa groups inside the tumor tissues by injecting fluorescent Rhd-Lip intratumorally (Fig. S5a†). Fluorescently labeled Oa-C12 was expected to be present on the cell membrane inside the tumors through a similar liposome fusion process observed in vitro. The tumors were collected 4 h or 24 h post injection (p.i.), sectioned and analyzed by confocal microscopy. As expected, a strong florescence signal from Rhd-C12 was observed on the tumor cells at 4 h p.i. indicating the successful introduction of functional groups on plasma membrane in vivo through liposome delivery and membrane fusion (Fig. S5b†). The fluorescence decreased at 24 h p.i. likely due to the clearance or metabolizing of the Rhd-C12 (Fig. S5b†).
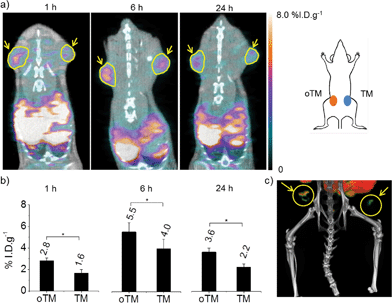
We then injected the Oa-Lip intratumorally to the left tumor (oTM) and PBS to the right one (TM) as a control (Fig. 2a) to study the cancer targeting in vivo. The mice received intravenous injection of Ald-NPs through tail veins 4 h after the introduction of Oa groups to the cells in the left tumors. In order to monitor the in vivo tumor targeting kinetics, the injected Ald-NPs were radio labeled with the isotope 64Cu using a chelating ligand, DOTA, conjugated to the polymer chain for dynamic positron emission tomography (PET) imaging over time without sacrificing the animals.43 All of the PET images were overlaid with micro X-ray computed tomography (CT) images to identify the position of the tumors (yellow circles and arrows in Fig. 2a). Ald-NPs started to accumulate in oTM (2.8% I.D. g−1) as early as 1 h p.i. with 75.0% higher amount than that in TM (1.6% I.D. g−1). At 6 h p.i., the accumulation of Ald-NPs in oTM increased to 5.5% I.D. g−1, which was 37.5% higher than that in TM (4.0% I.D. g−1) with statistical significance (Fig. 2b). The three-dimensional reconstructed image (Fig. 2c) also provides evidence for the significantly enhanced accumulation of Ald-NPs in oTM at 6 h p.i. At 24 h p.i., the tumor accumulation decreased slightly on both sides compared with the amount at 6 h. However, Ald-NPs persisted in oTM (3.6% I.D. g−1) with an amount 63.6% higher than that in TM (2.2% I.D. g−1). These results were confirmed by ex vivo measurement of the accumulation of Ald-NPs in the excised tumors using a γ-counter (Fig. S6†). A similar experiment was performed whereby PEG-PLA NPs without Ald groups on the surface were intravenously injected into the mice bearing oTM and TM on each side. No significant difference in tumor accumulation of the NPs on the two sides at 6 h or 24 h p.i. was observed (Fig. S7†) and the accumulation of NPs in oTM was lower than that of the Ald-NPs in oTM (Fig. S7†vs.Fig. 2b) indicating that the specific oxime ligation was responsible for the increased tumor accumulation and retention of Ald-NPs in oTM. These observations suggest that in vivo cancer targeting could indeed be achieved using bioorthogonal oxime ligation.
Cancer pretargeting strategies, which use antibodies to target tumors for the first step, followed by the second step of using an imaging or therapeutic agent to bind to the tumor-localized antibody, have proven to be superior to direct targeting by the conventional antibody conjugates in clinical trials.44–46 Here, we described a similar two-step pretargeting strategy, using liposome delivery and fusion for the first-step passive cancer targeting, and then guiding the nanomedicine to the tumor through an oxime ligation reaction with the bioorthogonal groups introduced by the liposome. Compared with the antibody-based pretargeting method, our strategy does not involve any potentially immunogenic biomacromolecules or depend on the heterogeneous cell surface antigens.14 As a proof of concept, we used intratumoral injection of Oa-Lip to label the cancer cells in vivo. However, liposomes bearing Oa groups could also be dosed systemically in the pretargeting step to achieve sufficient presentation of the target functional groups on the plasma membrane inside tumors via a passive tumor targeting mechanism, i.e. an enhanced permeation and retention (EPR) effect of the nanomedicine, followed by the administration of the second component, the drug-containing nanomedicine modified with the ligand functional groups.47
Conclusions
In conclusion, we demonstrated a novel cancer targeting strategy in vitro and in vivo using a bioorthogonal oxime ligation reaction. One bioorthogonal functional group (i.e. Oa) as an unnatural target was efficiently introduced to cancer cells using a liposomal delivery and fusion technique. Ald groups, as the targeting ligand for Oa, could be easily synthesized and incorporated into nanomedicines. Specific and efficient oxime ligation led to markedly improved cancer targeting in vivo through chemical Oa–Ald recognition. Given the versatility of the liposome fusion method for different bioorthogonal groups and the large variety of bioorthogonal chemistry available, we anticipate that many bioorthogonal reaction-mediated cancer targeting strategies can be easily developed, optimized and compared. Future efficacy studies using targeted, drug-loaded biocompatible nanomedicine combined with the cancer pretargeting method through systemic delivery will be important to further evaluate the potential of such strategies for targeted cancer therapy.Acknowledgements
This work is supported by the National Institute of Health (Director's New Innovator Award 1DP2OD007246-01), and National Science Foundation (DMR 13-09525). L.T., Q.Y. and K.C. were funded at UIUC from the NIH National Cancer Institute Alliance for Nanotechnology in Cancer “Midwest Cancer Nanotechnology Training Center” Grant R25 CA154015A. We thank Ms Catherine Yao for drawing the 3D pictures.Notes and references
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nat. Nanotechnol., 2007, 2, 751–760 CrossRef CAS PubMed.
- M. E. Davis, Z. Chen and D. M. Shin, Nat. Rev. Drug Discovery, 2008, 7, 771–782 CrossRef CAS PubMed.
- R. A. Petros and J. M. DeSimone, Nat. Rev. Drug Discovery, 2010, 9, 615–627 CrossRef CAS PubMed.
- H. Wei, J. G. Schellinger, D. S. H. Chu and S. H. Pun, J. Am. Chem. Soc., 2012, 134, 16554–16557 CrossRef CAS PubMed.
- A. F. Adler and K. W. Leong, Nano Today, 2010, 5, 553–569 CrossRef CAS PubMed.
- D. Kim, Y. Y. Jeong and S. Jon, ACS Nano, 2010, 4, 3689–3696 CrossRef CAS PubMed.
- X. A. Jiang, Y. R. Zheng, H. H. Chen, K. W. Leong, T. H. Wang and H. Q. Mao, Adv. Mater., 2010, 22, 2556–2560 CrossRef CAS PubMed.
- J. H. Myung, K. A. Gajjar, J. Saric, D. T. Eddington and S. Hong, Angew. Chem., Int. Ed., 2011, 50, 11769–11772 CrossRef CAS PubMed.
- L. Tang, X. Yang, L. W. Dobrucki, I. Chaudhury, Q. Yin, C. Yao, S. Lezmi, W. G. Helferich, T. M. Fan and J. Cheng, Angew. Chem., Int. Ed., 2012, 51, 12721–12726 CrossRef CAS PubMed.
- J. P. Landry, Y. Y. Fei and X. D. Zhu, Assay Drug Dev. Technol., 2012, 10, 250–259 CrossRef CAS PubMed.
- W. R. Algar, D. E. Prasuhn, M. H. Stewart, T. L. Jennings, J. B. Blanco-Canosa, P. E. Dawson and I. L. Medintz, Bioconjugate Chem., 2011, 22, 825–858 CrossRef CAS PubMed.
- J. A. Prescher and C. R. Bertozzi, Nat. Chem. Biol., 2005, 1, 13–21 CrossRef CAS PubMed.
- D. Hanahan and R. A. Weinberg, Cell, 2011, 144, 646–674 CrossRef CAS PubMed.
- D. Schrama, R. A. Reisfeld and J. C. Becker, Nat. Rev. Drug Discovery, 2006, 5, 147–159 CrossRef CAS PubMed.
- A. M. Wu and P. D. Senter, Nat. Biotechnol., 2005, 23, 1137–1146 CrossRef CAS PubMed.
- D. R. Elias, A. Poloukhtine, V. Popik and A. Tsourkas, Nanomedicine: Nanotechnology, Biology and Medicine, 2013, 9, 194–201 CrossRef CAS PubMed.
- R. K. Iha, K. L. Wooley, A. M. Nystrom, D. J. Burke, M. J. Kade and C. J. Hawker, Chem. Rev., 2009, 109, 5620–5686 CrossRef CAS PubMed.
- R. K. V. Lim and Q. Lin, Chem. Commun., 2010, 46, 1589–1600 RSC.
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974–6998 CrossRef CAS PubMed.
- J. M. Baskin and C. R. Bertozzi, QSAR Comb. Sci., 2007, 26, 1211–1219 CAS.
- J. M. Baskin, J. A. Prescher, S. T. Laughlin, N. J. Agard, P. V. Chang, I. A. Miller, A. Lo, J. A. Codelli and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 16793–16797 CrossRef CAS PubMed.
- J. A. Prescher, D. H. Dube and C. R. Bertozzi, Nature, 2004, 430, 873–877 CrossRef CAS PubMed.
- D. Dutta, A. Pulsipher, W. Luo and M. N. Yousaf, J. Am. Chem. Soc., 2011, 133, 8704–8713 CrossRef CAS PubMed.
- D. Dutta, A. Pulsipher, W. Luo, H. Mak and M. N. Yousaf, Bioconjugate Chem., 2011, 22, 2423–2433 CrossRef CAS PubMed.
- C. L. Washington-Hughes, Y. Cheng, X. Duan, L. Cai, L. A. Lee and Q. Wang, Mol. Pharm., 2012, 10, 43–50 CrossRef PubMed.
- S. Hapuarachchige, W. Zhu, Y. Kato and D. Artemov, Biomaterials, 2014, 35, 2346–2354 CrossRef CAS PubMed.
- J. B. Haun, N. K. Devaraj, S. A. Hilderbrand, H. Lee and R. Weissleder, Nat. Nanotechnol., 2010, 5, 660–665 CrossRef CAS PubMed.
- R. Rossin, P. R. Verkerk, S. M. van den Bosch, R. C. M. Vulders, I. Verel, J. Lub and M. S. Robillard, Angew. Chem., Int. Ed., 2010, 49, 3375–3378 CrossRef CAS PubMed.
- D. J. Vugts, A. Vervoort, M. S.-v. Walsum, G. W. M. Visser, M. S. Robillard, R. M. Versteegen, R. C. M. Vulders, J. D. M. Herscheid and G. A. M. S. van Dongen, Bioconjugate Chem., 2011, 22, 2072–2081 CrossRef CAS PubMed.
- H. Koo, S. Lee, J. H. Na, S. H. Kim, S. K. Hahn, K. Choi, I. C. Kwon, S. Y. Jeong and K. Kim, Angew. Chem., Int. Ed., 2012, 51, 11836–11840 CrossRef CAS PubMed.
- F. Emmetiere, C. Irwin, N. T. Viola-Villegas, V. Longo, S. M. Cheal, P. Zanzonico, N. Pillarsetty, W. A. Weber, J. S. Lewis and T. Reiner, Bioconjugate Chem., 2013, 24, 1784–1789 CrossRef CAS PubMed.
- B. M. Zeglis, K. K. Sevak, T. Reiner, P. Mohindra, S. D. Carlin, P. Zanzonico, R. Weissleder and J. S. Lewis, J. Nucl. Med., 2013, 54, 1389–1396 CrossRef CAS PubMed.
- S. Ulrich, D. Boturyn, A. Marra, O. Renaudet and P. Dumy, Chem.–Eur. J., 2014, 20, 34–41 CrossRef CAS PubMed.
- J. Kalia and R. T. Raines, Angew. Chem., Int. Ed., 2008, 47, 7523–7526 CrossRef CAS PubMed.
- D. A. Nauman and C. R. Bertozzi, Biochim. Biophys. Acta, 2001, 1568, 147–154 CrossRef.
- K. J. Yarema, L. K. Mahal, R. E. Bruehl, E. C. Rodriguez and C. R. Bertozzi, J. Biol. Chem., 1998, 273, 31168–31179 CrossRef CAS PubMed.
- D. Rabuka, M. B. Forstner, J. T. Groves and C. R. Bertozzi, J. Am. Chem. Soc., 2008, 130, 5947–5953 CrossRef CAS PubMed.
- A. Csiszar, N. Hersch, S. Dieluweit, R. Biehl, R. Merkel and B. Hoffmann, Bioconjugate Chem., 2010, 21, 537–543 CrossRef CAS PubMed.
- D. Sarkar, P. K. Vemula, W. A. Zhao, A. Gupta, R. Karnik and J. M. Karp, Biomaterials, 2010, 31, 5266–5274 CrossRef CAS PubMed.
- R. Tong and J. Cheng, Polym. Rev., 2007, 47, 345–381 CrossRef CAS.
- R. Tong and J. Cheng, Angew. Chem., Int. Ed., 2008, 47, 4830–4834 CrossRef CAS PubMed.
- E. J. Chaney, L. Tang, R. Tong, J. Cheng and S. A. Boppart, Mol. Imaging, 2010, 9, 153–162 CAS.
- Q. Yin, R. Tong, Y. Xu, K. Baek, L. W. Dobrucki, T. M. Fan and J. Cheng, Biomacromolecules, 2013, 14, 920–929 CrossRef CAS PubMed.
- D. M. Goldenberg, R. M. Sharkey, G. Paganelli, J. Barbet and J. F. Chatal, J. Clin. Oncol., 2006, 24, 823–834 CrossRef CAS PubMed.
- O. C. Boerman, F. G. van Schaijk, W. J. G. Oyen and F. H. M. Corstens, J. Nucl. Med., 2003, 44, 400–411 Search PubMed.
- C.-H. Chang, R. M. Sharkey, E. A. Rossi, H. Karacay, W. McBride, H. J. Hansen, J.-F. Chatal, J. Barbet and D. M. Goldenberg, Mol. Cancer Ther., 2002, 1, 553–563 CAS.
- S. K. Hobbs, W. L. Monsky, F. Yuan, W. G. Roberts, L. Griffith, V. P. Torchilin and R. K. Jain, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 4607–4612 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis and characterization of compounds and nanoparticles, in vitro and in vivo tumor cell labelling with liposomes, PET/CT imaging and biodistribution. See DOI: 10.1039/c5sc00063g |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |