Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A labelled-ubiquicidin antimicrobial peptide for immediate in situ optical detection of live bacteria in human alveolar lung tissue†
Ahsan R.
Akram‡
a,
Nicolaos
Avlonitis‡
b,
Annamaria
Lilienkampf
b,
Ana M.
Perez-Lopez
b,
Neil
McDonald
a,
Sunay V.
Chankeshwara
b,
Emma
Scholefield
a,
Christopher
Haslett
a,
Mark
Bradley
*b and
Kevin
Dhaliwal
*a
aPulmonary Optical Molecular Imaging Group, MRC Centre for Inflammation Research, Queen's Medical Research Institute, 47 Little France Crescent, Edinburgh, EH16 4TJ, UK. E-mail: Kev.Dhaliwal@ed.ac.uk
bEaStCHEM, The University of Edinburgh School of Chemistry, Joseph Black Building, West Mains Road, Edinburgh EH9 3FJ, UK. E-mail: mark.bradley@ed.ac.uk
First published on 29th June 2015
Abstract
The in situ immediate detection of the presence of bacteria in the distal human lung is of significant clinical utility. Herein we describe the development and optimization of a bacterial binding fragment (UBI29–41) of the antimicrobial peptide, ubiquicidin (UBI), conjugated to an environmentally sensitive fluorophore to enable rapid live bacterial imaging within human lung tissue. UBI29–41 was modified for stability in the presence of human lung bronchoalveolar lavage fluid, for affinity to bacterial membranes and functionality in human lung tissue. The optimized cyclic structure yields an optical molecular Smartprobe for bacterial detection in human lung tissue.
Introduction
It is a global priority to reduce antibiotic resistance,1 and consequently novel methodologies to rapidly diagnose or exclude bacterial infection are urgently required to implement antibiotic stewardship. Current diagnostic approaches rely on detecting the non-specific clinical features of a developing infection alongside optimal sampling of an affected area and subsequent bacterial culture. However, these approaches are slow, prone to contamination and diagnose infection at a late stage. Hence, rapid, sensitive and in situ molecular imaging assays for bacterial infections address an unmet need for bacterial diagnostics2 and have the potential to lead to more effective antibiotic prescribing.Pneumonia is an important clinical disease in which to develop and apply bacterial diagnostics. It is the leading infectious cause of mortality in children,3 adults admitted to hospital with pneumonia have a mortality of up to 14%,4 and in critically ill individuals in the intensive care unit (ICU), those who develop ventilator associated pneumonia (VAP) can have an attributable mortality of over 70% with some etiologies.5 During the development of pneumonia, bacteria proliferate within the distal gas exchanging units of the human lung, an anatomical region in humans that is considered to be relatively sterile at basal microbiota levels. This is in contrast to other areas of the human body where bacterial burden from colonisation is high, such as the upper airway and gut, where distinguishing colonisation from pathogenic bacteria is a diagnostic challenge. Indeed current methodologies to diagnose pneumonia rely upon general sampling from upper airways and are prone to contamination and consequent ‘false-positive’ initiation of antibiotic therapy.
Optical molecular imaging (OMI), is a relatively novel diagnostic approach that has utility in preclinical and clinical applications.6 Advantages of OMI include: (i) real-time imaging, (ii) high resolution, (iii) absence of radiation-related risks and (iv) low costs. However, whilst the applications of molecular imaging are showing increasing utility in disease areas such as oncology, there are a limited number of functional effective molecular imaging approaches for bacterial detection.7
A rational and accepted approach for targeted bacterial molecular imaging has been to label antimicrobial peptides (AMPs), widespread components of the innate immune systems of virtually all multicellular organisms.8 AMPs possess a number of factors making them promising tools for bacterial OMI including their amino acid composition, amphipathicity and cationic charge that enable selective insertion and binding to the bacterial membrane.9 The most studied AMP for infection imaging is the 59 amino acid cationic AMP, ubiquicidin (UBI) and in particular a UBI fragment containing thirteen amino acid residues (UBI29–41) [Thr-Gly-Arg-Ala-Lys-Arg-Arg-Met-Gln-Tyr-Asn-Arg-Arg] which has been utilised in radionucleotide based imaging.10 However its use involves unavoidable radiation exposure and is not suitable where portable solutions are required, such as critically ill ventilated patients with suspected pneumonia. More recently a near-infrared labelled version of UBI29–41 was applied in a murine model of bacterial infection.11 Unfortunately, the synthetic route led to a multitude of products with the dye dominating the distribution of the construct, resulting in a similar distribution for the dye alone compared to the fluorescently labelled UBI29–41. In addition there are obvious “Achilles heels” in a labelled UBI29–41 peptide with respect to potential degradation in vivo, exacerbated even more so in sites of active inflammation by proteolysis and oxidation. Building on these developments and also in recognition of the feasibility of developing an OMI agent (Smartprobe) based upon a UBI scaffold, we embarked upon a lead optimisation approach to yield a Smartprobe suitable for application in diagnosing pneumonia (bacterial infection in the distal lung) when partnered with an optical imaging device. For this Smartprobe to be valuable bio-medically we optimised a series of prerequisites including;
(i) Specificity and selectivity to label clinically relevant bacteria,
(ii) High signal-to-noise contrast,
(iii) Resistance to degradation and stability in the presence of human lung bronchoalveolar lavage fluid (BALF) from patients with acute respiratory distress syndrome (ARDS),
(iv) High affinity and bacterial detection by a clinically approved optical imaging device in human lung alveolar tissue.
We demonstrate the development and optimisation of fluorescently labelled UBI29–41, through sequential modification of the fluorophore label, peptide sequence and secondary structure, and demonstrate the ability of the lead compound to detect live bacteria in human lung tissue.
Results and discussion
We synthesised a series of UBI peptides to assess structure and functional consequences for bacterial labelling (Table 1) and to remove proteolytic susceptibilities.| Compound code | Modification | Peptide sequence | Effect on stability and efficacy |
|---|---|---|---|
| a Ahx: 6-aminohexanoic acid, PEG: 8-amino-3,6-dioxaoctanoic acid, MeOPEG: 8-methoxy-3,6-dioxaoctanoic acid, FAM: 5(6)-carboxyfluorescein amide, NBD: 7-nitrobenz-2-oxa-1,3-diazole, MeArg: N-methyl-arginine, ARDS: acute respiratory distress syndrome. All D-amino acid denoted by prefix D. *A lysine reside was added to the amino terminus of UBI to enable cyclisation. | |||
| UBI-1 | UBI29–41 sequence labelled with carboxyfluorescein at the amino-terminus | FAM-Ahx-Thr-Gly-Arg-Ala-Lys-Arg-Arg-Met-Gln-Tyr-Asn-Arg-Arg-NH2 | Insufficient signal-to-noise |
| UBI-2 | Fluorophore change to NBD | NBD-Ahx-Thr-Gly-Arg-Ala-Lys-Arg-Arg-Met-Gln-Tyr-Asn-Arg-Arg-NH2 | Unstable at 37 °C in PBS |
| UBI-3 | Norleucine substitution of methionine to improve chemical stability | NBD-Ahx-Thr-Gly-Arg-Ala-Lys-Arg-Arg-Nle-Gln-Tyr-Asn-Arg-Arg-NH2 | Unstable in ARDS BALF |
| UBI-4 | N-Methylarginine (MeArg) insertion based on MALDI-TOF analysis of cleavage peptides | NBD-Ahx-Thr-Gly-Arg-Ala-Lys-MeArg-MeArg-Nle-Gln-Tyr-Asn-MeArg-MeArg-NH2 | Stable in ARDS BALF, poor bacterial affinity |
| UBI-5 | D-Amino acids (D-Val and D-Phe) incorporated at the C-terminus to limit proteolytic degradation | NBD-Ahx-Thr-Gly-Arg-Ala-Lys-Arg-MeArg-Nle-Gln-Tyr-Asn-Arg-Arg-D-Phe-D-Val-NH2 | Stable in ARDS BALF, poor bacterial affinity |
| UBI-6 | PEG insertion at the N and C-termini | MeO-PEG-Lys(NBD)-Thr-Gly-Arg-Ala-Lys-Arg-Arg-Nle-Gln-Tyr-Asn-Arg-Arg-PEG-PEG-NH2 | Unstable in ARDS BALF, loss of labelling efficacy |
| UBI-7 | PEG introduction at the C-terminus | NBD-Ahx-Thr-Gly-Arg-Ala-Lys-Arg-Arg-Nle-Gln-Tyr-Asn-Arg-Arg-PEG-PEG-NH2 | Unstable in ARDS BALF at 30 minutes, loss of labelling efficacy |
| UBI-8 | The all D-amino acid variant of UBI with the incorporation of D-Nle and a PEG spacer at the amino terminus | NBD-PEG-D-Thr-Gly-D-Arg-D-Ala-D-Lys-D-Arg-D-Arg-D-Nle-D-Gln-D-Tyr-D-Asn-D-Arg-D-Arg-OH | Stable in ARDS BALF, loss of labelling efficacy |
| UBI-9 | Retroinverse sequence of UBI (all D-amino acids) and NBD attached to the C-terminal amino acid. Peptide capped at the amino terminus with MeO-PEG | MeO-PEG- D- Arg- D- Arg- D- Asn- D- Tyr- D- Gln- D- Nle- D- Arg- D- Arg- D- Lys- D- Ala- D- Arg-Gly- D- Thr-Lys(NBD)-OH | Stable in ARDS BALF, loss of labelling efficacy |
| UBI-10* | Side-chain to tail cyclisation of UBI with Met to Nle substitution | NBD-Ahx-cyclo[Lys-Thr-Gly-Arg-Ala-Lys-Arg-Arg-Nle-Gln-Tyr-Asn-Arg-Arg-Gly] | Improved stability in ARDS BALF, improved bacterial labelling |
| UBI-11* | As in UBI-10 – a cyclic UBI with Met to Nle substitution and insertion of N-MeArg at the major cleavage site | NBD-Ahx-cyclo[Lys-Thr-Gly-Arg-Ala-Lys-Arg-Arg-Nle-Gln-Tyr-Asn-Arg-MeArg-Gly] | Stable in ARDS BALF, reduction of labelling efficacy |
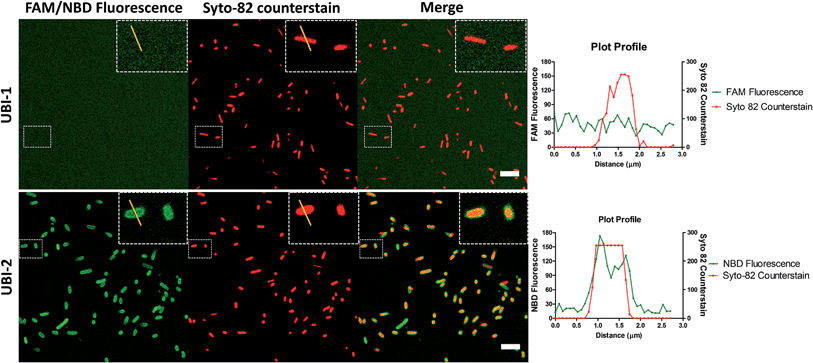
7-Nitrobenz-2-oxa-1,3-diazole (NBD) demonstrates increased signal to noise over carboxyfluorescein (FAM) UBI conjugates
UBI29–41 was firstly conjugated to carboxyfluorescein (UBI-1) and used to image bacteria with analysis via confocal microscopy. This yielded insufficient signal to noise to distinguish bacteria from background fluorescence (Fig. 1). We therefore took advantage of the environmental reporting properties of the fluorophore 7-nitrobenz-2-oxa-1,3-diazole (NBD)12 and hypothesized there would be fluorescence amplification of NBD in the apolar hydrophobic bacterial membrane. UBI-2 demonstrated good signal to noise and was able to detect bacteria (Fig. 1).Removing and replacing the methionine residue improves stability
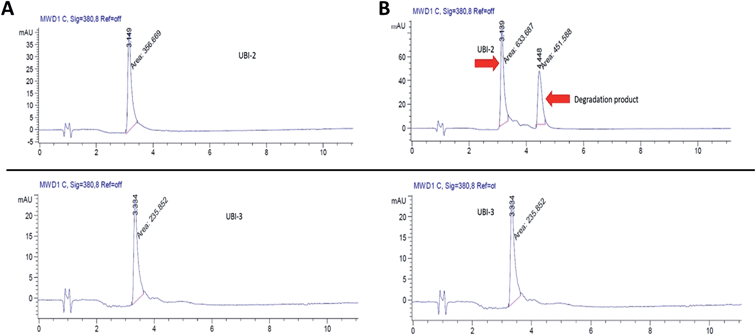
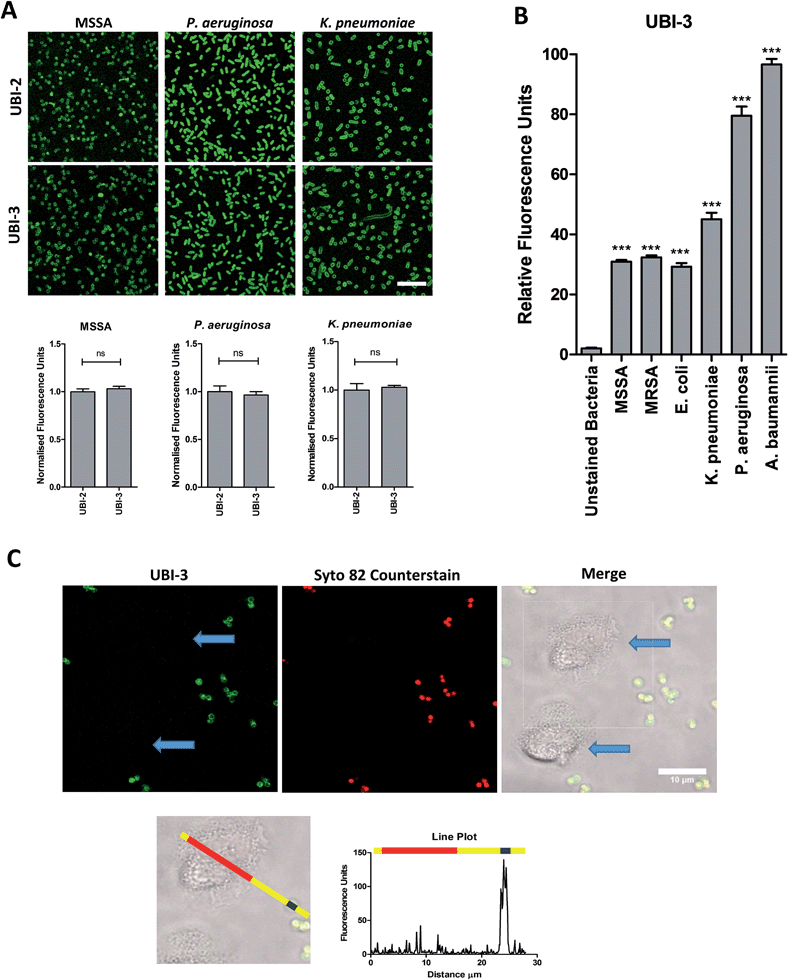
UBI-2 underwent chemical degradation upon storage in phosphate buffered saline at 37 °C, which was prevented by substitution of the Met to Nle, giving rise to UBI-3 (Fig. 2). This modification is particularly of relevance due to the expected in vivo inflammatory oxidative environment within an infected pneumonic lung due to infiltrating host inflammatory cells such as neutrophils,13 and therefore all subsequent modifications incorporated Nle in place of Met.To assess the bacterial labelling consequences of this single amino acid substitution, UBI-2 and UBI-3 were screened against a panel of Gram positive (methicillin-sensitive Staphylococcus aureus (MSSA)) and Gram negative pathogenic bacteria (Pseudomonas aeruginosa and Klebsiella pneumoniae) demonstrating retained bacterial labelling with specificity over eukaryotic cells (Fig. 3).
Controlling degradation in human bronchoalveolar lavage fluid (BALF)
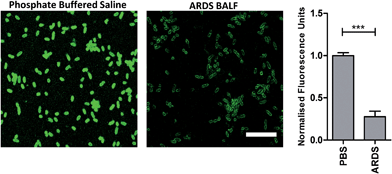
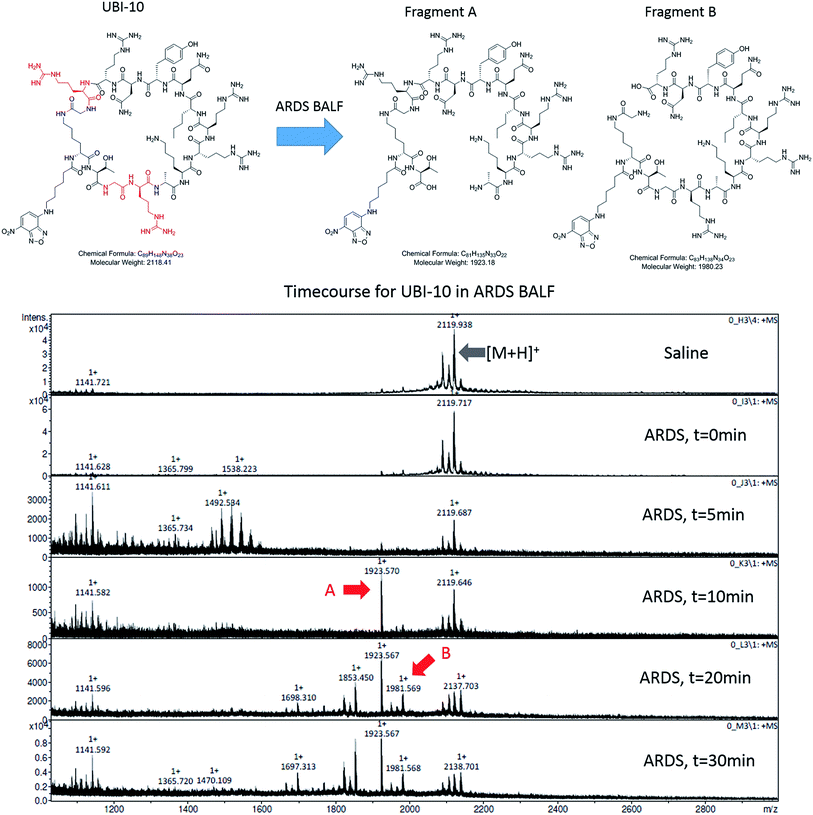
To determine the ability of UBI-3 to image bacteria in the continued presence of a complex inflammatory environment, bacteria were labelled with UBI-3 in the presence of broncho-alveolar lavage fluid (BALF) retrieved from ventilated ICU patients with Acute Respiratory Distress Syndrome (ARDS). ARDS BALF was utilised as ARDS is a severe life threatening pulmonary syndrome, characterized by a massive proteolytic and oxidative inflammatory environment within the distal alveolar region.14 Under these harsh experimental conditions, there was a significant reduction in the fluorescent labelling intensity of bacteria with UBI-3 (Fig. 4). This was due to the rapid (<5 min) degradation of the peptide as demonstrated by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) analysis (ESI Fig. S1†). To design out susceptible amino acid cleavage points, a number of modifications were made (Table 1) and the functional consequences assessed. The chemical approaches pursued to improve resistance to degradation included replacing amino acids at the cleavage positions with non-natural amino acids such as N-methyl or D-amino acids,15 and cyclisation of the peptide,16 which could have the added benefit of enhancing affinity towards bacterial membrane.17Modification of mass spectrometry identified degradation sites
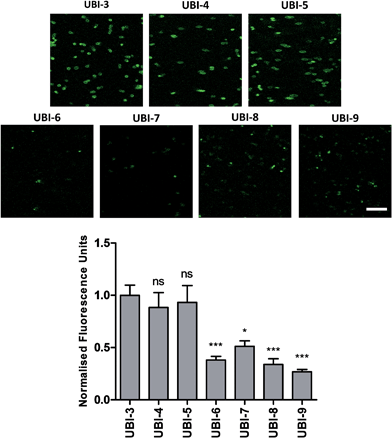
The Arg–Arg bonds were identified as the prominent position of degradation (Fig. 5) and Compounds UBI-4–9 (Table 1) were synthesised with a variety of substitutions around the degradation sites. The susceptible peptide bonds at P1–P2 and P7–P8 were blocked using N-MeArg (UBI-4), and to further mask these positions, a long polyethylene glycol (PEG) chain and/or a D-amino acid were positioned at the carboxylic and/or amino termini (UBI-5–7). N-MeArg insertion (UBI-4) or N-MeArg with two D-amino acid residues at the C-terminal (UBI-5) improved stability in ARDS BALF (ESI Fig. S2†) and allowed bacterial labelling at the same intensity as UBI-3 (Fig. 6). However, insertion of a PEG chain at both termini (UBI-6) and constructs with entirely D-amino acids (UBI-8 and UBI-9), whilst improving stability, resulted in loss of labelling on bacteria (ESI Fig. S2 and 6†). A single PEG at the C-terminus alone (UBI-7) did not improve stability in ARDS BALF, and resulted in loss of bacterial labelling.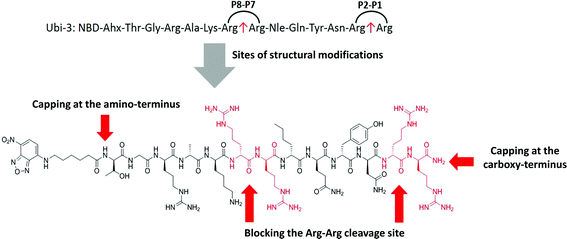 | ||
| Fig. 5 Sites of structural modifications undertaken on UBI-3 based on MALDI-TOF analysis of proteolytic degradation. | ||
Cyclisation of the peptide sequence
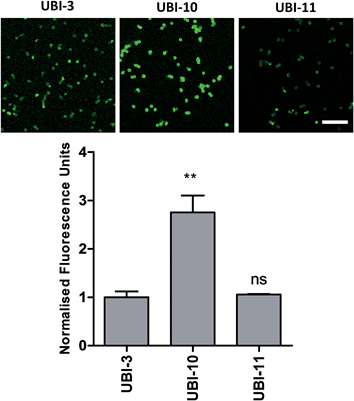
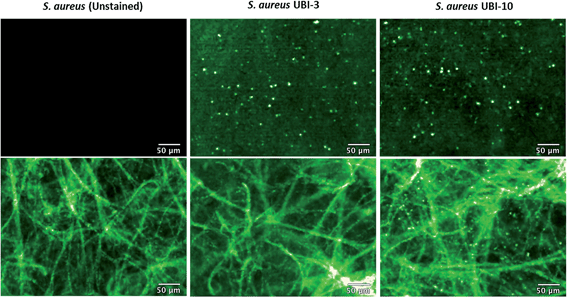
We hypothesized that cyclisation of the sequence would allow for greater stability and also improve bacterial affinity and therefore cyclic variants of UBI-3 were synthesized (UBI-10 and UBI-11), which were both found to be stable in BALF for 5 minutes (Fig. 7 and S3 ESI†). Compared to the UBI-3 there was a significantly higher signal for equivalent concentrations of the cyclic construct (UBI-10) (Fig. 8). However, these studies also highlighted the degradation of UBI-10 at 10 minutes, guiding the temporal constraints of Smartprobe delivery and optical imaging. UBI-11 demonstrated that a single amino acid change imparted stability in BALF for over 30 minutes and the compound showed comparable bacterial labelling to UBI-3, (ESI Fig. S3†) however the enhanced bacterial labelling seen with UBI-10 was lost (Fig. 8).Bacterial affinity evaluation
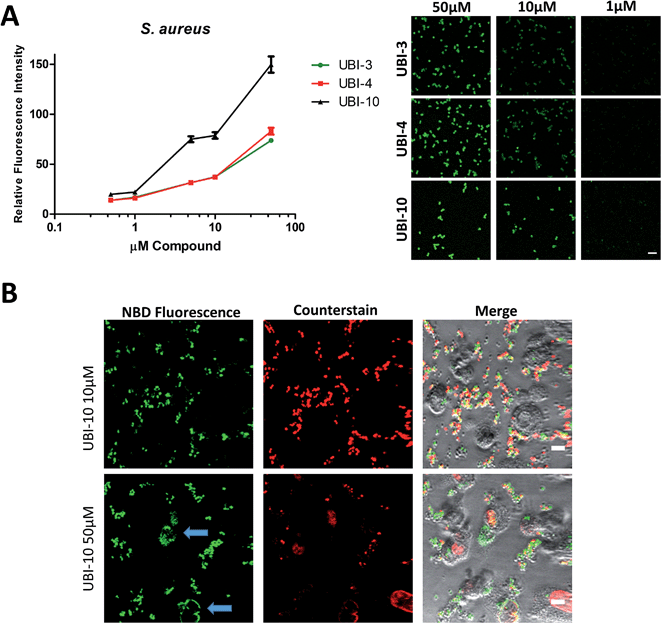
Three compounds (UBI-3, UBI-5 and UBI-10) were assessed for further evaluation. The rationale for assessing these variants was that UBI-3 and UBI-10 were native UBI sequences in linear or cyclic secondary structures respectively and UBI-5 was stable and demonstrated a similar bacterial labelling to UBI-3. Bacterial labelling was demonstrated in a concentration dependent manner for all three compounds. The relative fluorescence of UBI-10 labelled bacteria was higher than UBI-3 or UBI-5 labelled bacteria (Fig. 9). At 10 μM, UBI-10 retained bacterial specificity over neutrophils. However at 50 μM, some neutrophil labelling was observed (Fig. 9). In vitro affinity of bacterial binding was assessed by incorporating a wash step in the live confocal imaging protocol with a panel of bacteria with the ‘wash-off’ representing a surrogate indicator of Smartprobe-bacterial labelling affinity. This demonstrated that following labelling and a wash, there was significantly lower fluorescence for the linear compounds (UBI-3 and UBI-5) than the cyclic compound without modification (UBI-10) (ESI Fig. S4†) against three different bacterial species. UBI-10, therefore, was assessed further in a clinically relevant biological system.Bacterial detection by a clinical fibre-based optical imaging in human lung tissue
Fibered confocal fluorescence microscopy (FCFM) with a 488 nm laser system enables in vivo cellular and subcellular resolution imaging deep in human lung.18 This system allows imaging of the intrinsic autofluorescence of elastin thereby enabling alveolar imaging. Therefore, to assess whether this platform has the resolution to detect fluorescent bacteria, we pre-labeled bacteria (with calcein AM) and then demonstrated that we were able to detect fluorescently labeled bacteria on the background of human lung autofluorescence (ESI Fig. S5†).Since NBD is also excited at 488 nm, the utility and detection of the Smartprobes with FCFM was next evaluated. Bacteria were labelled with UBI-3 and UBI-10 and imaged in suspension demonstrating the punctate fluorescence pattern of labelled bacteria. These were then delivered onto ex vivo human lung tissue fragments. Bacteria labelled with UBI-3 immediately lost their fluorescent signal whereas UBI-10 labelled bacteria, were readily identified above elastin autofluorescence (Fig. 10).
Summary
We synthesised a library of fluorescently labelled UBI29–41 mimetic peptides for rapid, live, bacterial imaging; with the ambition to develop Smartprobes that demonstrate potential clinical utility in conditions of high unmet need; such as the rapid diagnosis of suspected pneumonia in critically ill individuals. We discovered key determinants of peptide stability and bacterial binding affinity as potential obstacles for clinical utility. A number of structural modifications were subsequently engineered to drive improved stability, resistance to degradation and improve function. A cyclic variant of NBD–UBI (UBI-10) proved to be the most functional and specific Smartprobe for bacterial detection and showed proof of concept for detection of bacteria over intrinsic lung elastin autofluorescence in human lung tissue.These findings provide pivotal insights into future strategies to develop chemical Smartprobes capable to detect bacteria within the distal human lung.
Acknowledgements
We thank Professor John Govan from the University of Edinburgh who provided the clinical isolates of A. baumannii and P. aeruginosa. We also thank Dr Andrew Conway Morris from the University of Edinburgh for the use of BALF from ICU cohorts. This work was funded in part by the Department of Health and Wellcome Trust through the Health Innovation Challenge Fund (HICF). Funding Reference Number: 0510-069 – Immediate Point of Care Molecular Diagnostics for Lung Inflammation/Infection in Critical Care. A. L. was supported by an EU Marie Curie Intra-European Fellowship for career development (Project No. PIEF-GA-2010-276116).References
- A. Huttner, S. Harbarth, J. Carlet, S. Cosgrove, H. Goossens, A. Holmes, V. Jarlier, A. Voss and D. Pittet, Antimicrob. Resist. Infect. Contr., 2013, 2, 31 CrossRef PubMed.
- X. Ning, S. Lee, Z. Wang, D. Kim, B. Stubblefield, E. Gilbert and N. Murthy, Nat. Mater., 2011, 10, 602–607 CrossRef CAS PubMed; P. Panizzi, M. Nahrendorf, J. L. Figueiredo, J. Panizzi, B. Marinelli, Y. Iwamoto, E. Keliher, A. A. Maddur, P. Waterman, H. K. Kroh, F. Leuschner, E. Aikawa, F. K. Swirski, M. J. Pittet, T. M. Hackeng, P. Fuentes-Prior, O. Schneewind, P. E. Bock and R. Weissleder, Nat. Med., 2011, 17, 1142–1146 CrossRef PubMed.
- C. L. Fischer Walker, I. Rudan, L. Liu, H. Nair, E. Theodoratou, Z. A. Bhutta, K. L. O'Brien, H. Campbell and R. E. Black, Lancet, 2013, 381, 1405–1416 CrossRef.
- W. S. Lim, J. T. Macfarlane, T. C. Boswell, T. G. Harrison, D. Rose, M. Leinonen and P. Saikku, Thorax, 2001, 56, 296–301 CrossRef CAS PubMed.
- J. Chastre and J. Y. Fagon, Am. J. Respir. Crit. Care Med., 2002, 165, 867–903 CrossRef PubMed; E. Tejerina, F. Frutos-Vivar, M. I. Restrepo, A. Anzueto, F. Abroug, F. Palizas, M. Gonzalez, G. D'Empaire, C. Apezteguia and A. Esteban, J. Crit. Care, 2006, 21, 56–65 CrossRef PubMed.
- D. A. Dorward, C. D. Lucas, A. G. Rossi, C. Haslett and K. Dhaliwal, Pharmacol. Ther., 2012, 135, 182–199 CrossRef CAS PubMed.
- A. Bunschoten, M. M. Welling, M. F. Termaat, M. Sathekge and F. W. van Leeuwen, Bioconjugate Chem., 2013, 24, 1971–1989 CrossRef CAS PubMed.
- M. Welling, A. Paulusma-Annema, H. S. Balter, E. K. J. Pauwels and P. H. Nibbering, Eur. J. Nucl. Med., 2000, 27, 291–301 CrossRef CAS; M. M. Welling, S. Mongera, A. Lupetti, H. S. Balter, V. Bonetto, U. Mazzi, E. K. Pauwels and P. H. Nibbering, Nucl. Med. Biol., 2002, 29, 413–422 CrossRef PubMed; R. E. W. Hancock and H.-G. Sahl, Nat. Biotechnol., 2006, 24, 1551–1557 CrossRef PubMed.
- K. A. Brogden, Nat. Rev. Microbiol., 2005, 3, 238–250 CrossRef CAS PubMed.
- A. Ostovar, M. Assadi, K. Vahdat, I. Nabipour, H. Javadi and M. Eftekhari, Clin. Nucl. Med., 2013, 38, 413–416 CrossRef PubMed; J. Sepulveda-Mendez, C. A. de Murphy, J. C. Rojas-Bautista and M. Pedraza-Lopez, Nucl. Med. Commun., 2010, 31, 889–895 Search PubMed.
- C. Liu and Y. Gu, J. Innovative Opt. Health Sci., 2013, 6, 1350026 CrossRef.
- S. Mukherjee, A. Chattopadhyay, A. Samanta and T. Soujanya, J. Phys. Chem., 1994, 98, 2809–2812 CrossRef CAS.
- H. Rosen, S. J. Klebanoff, Y. Wang, N. Brot, J. W. Heinecke and X. Fu, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 18686–18691 CrossRef CAS PubMed.
- R. B. Goodman, J. Pugin, J. S. Lee and M. A. Matthay, Cytokine Growth Factor Rev., 2003, 14, 523–535 CrossRef CAS PubMed.
- H. Lam, D. C. Oh, F. Cava, C. N. Takacs, J. Clardy, M. A. de Pedro and M. K. Waldor, Science, 2009, 325, 1552–1555 CrossRef CAS PubMed.
- H. Kessler, Angew. Chem., Int. Ed., 1982, 21, 512–523 CrossRef CAS; J. Chatterjee, B. Laufer and H. Kessler, Nat. Protoc., 2012, 7, 432–444 CrossRef PubMed.
- Z. Oren and Y. Shai, Biochemistry, 2000, 39, 6103 CrossRef CAS PubMed; M. Dathe, H. Nikolenko, J. Klose and M. Bienert, Biochemistry, 2004, 43, 9140 CrossRef PubMed.
- L. Thiberville, M. Salaun, S. Lachkar, S. Dominique, S. Moreno-Swirc, C. Vever-Bizet and G. Bourg-Heckly, Eur. Respir. J., 2009, 33, 974–985 CrossRef CAS PubMed; M. Salaun, F. Roussel, G. Bourg-Heckly, C. Vever-Bizet, S. Dominique, A. Genevois, V. Jounieaux, G. Zalcman, E. Bergot, J. M. Vergnon and L. Thiberville, Eur. Respir. J., 2013, 42, 1646–1658 CrossRef PubMed; R. C. Newton, S. V. Kemp, G. Z. Yang, D. S. Elson, A. Darzi and P. L. Shah, Respir. Med., 2012, 106, 127–137 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details and Fig. S1–S5. See DOI: 10.1039/c5sc00960j |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |