Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Single-molecule observations of RNA–RNA kissing interactions in a DNA nanostructure†
Yosuke
Takeuchi
a,
Masayuki
Endo
*b,
Yuki
Suzuki
a,
Kumi
Hidaka
a,
Guillaume
Durand
cd,
Eric
Dausse
cd,
Jean-Jacques
Toulmé
*cd and
Hiroshi
Sugiyama
*ab
aDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawa-oiwakecho, Sakyo-ku, Kyoto 606-8502, Japan
bInstitute for Integrated Cell-Material Sciences (WPI-iCeMS), Kyoto University, Yoshida-ushinomiyacho, Sakyo-ku, Kyoto, 606-8501, Japan. E-mail: endo@kuchem.kyoto-u.ac.jp; hs@kuchem.kyoto-u.ac.jp
cARNA laboratory, University of Bordeaux, 146 rue Léo Saignat, 33076 Bordeaux, France. E-mail: jean-jacques.toulme@inserm.fr
dInserm U869, 146 rue Léo Saignat, 33076 Bordeaux, France
First published on 6th October 2015
Abstract
RNA molecules uniquely form a complex through specific hairpin loops, called a kissing complex. The kissing complex is widely investigated and used for the construction of RNA nanostructures. Molecular switches have also been created by combining a kissing loop and a ligand-binding aptamer to control the interactions of RNA molecules. In this study, we incorporated two kinds of RNA molecules into a DNA origami structure and used atomic force microscopy to observe their ligand-responsive interactions at the single-molecule level. We used a designed RNA aptamer called GTPswitch, which has a guanosine triphosphate (GTP) responsive domain and can bind to the target RNA hairpin named Aptakiss in the presence of GTP. We observed shape changes of the DNA/RNA strands in the DNA origami, which are induced by the GTPswitch, into two different shapes in the absence and presence of GTP, respectively. We also found that the switching function in the nanospace could be improved by using a cover strand over the kissing loop of the GTPswitch or by deleting one base from this kissing loop. These newly designed ligand-responsive aptamers can be used for the controlled assembly of the various DNA and RNA nanostructures.
Introduction
Structural diversity of RNA is one of the important properties of RNA molecules, which exhibit unique functions such as specific complex formation and catalysis. One of the variations of the complex formation between RNA molecules includes a “kissing complex,” which enables assembly of complementary RNA loops via Watson–Crick base pairing.1–8 This specific kissing complex formation with native RNA motifs has been used to create various RNA architectures such as polygonal structures and three-dimensional assemblies.9–11 Aptamers with a kissing RNA loop have been artificially developed by in vitro selection.12–15 A ligand-responsive kissing aptamer, called a guanosine triphosphate switch (GTPswitch), has been recently reported.16 This GTPswitch has a GTP-binding domain and a kissing domain that binds to a target RNA loop (Aptakiss) in the presence of GTP.16 Herein, we tried to visualize this unique ligand-responsive switching interaction between the GTPswitch and its counterpart Aptakiss at single-molecule resolution.Direct observation of interactions between biomolecules by using atomic force microscopy (AFM) is one of the practical methods for characterizing the properties of complex formation.17 Especially using a high-speed AFM and nano-sized DNA origami scaffold called DNA frame,17,18 dynamic formation of G-quadruplex structure and double-stranded DNA has been visualized.19–22 This single-molecule observation system should be used to observe the interactions of kissing aptamers for characterizing their properties.
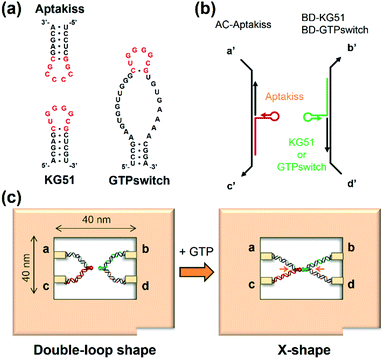
In this study, we placed the GTPswitch and the target kissing loop Aptakiss into the cavity of a DNA frame and directly observed their interaction (Fig. 1a).19–22 The GTPswitch was generated based on a KG51 RNA kissing hairpin, which can bind to the Aptakiss only in the presence of GTP.16 We elongated the 5′ end of each RNA molecule and hybridized these molecules to the supporting DNA strands. These constructs were incorporated into the DNA frame through four ssDNA linkers (Fig. 1b). When the GTPswitch binds to the Aptakiss upon the addition of GTP, the configuration change of the supporting DNA strands from the unbound “double-loop” to the “X-shape” should be observed in the DNA frame (Fig. 1c). We investigated the ligand-responsive binding of the GTPswitch to the Aptakiss by observing the structural changes in the supporting DNA strands in the DNA frame, and examined the experimental conditions to improve the switching function.
Experimental section
Preparation of RNA molecules
A template dsDNA containing T7 promoter was used to prepare RNA by in vitro transcription. The sequences are shown in ESI (Fig. S1 and Table S1†). Transcription was performed in a solution containing 0.5 μM template dsDNA, 40 mM Tris-HCl (pH 8.0), 10 mM DTT, 23 mM MgCl2, 2 mM spermidine, 4.0 mM NTPs, and 2.5 U μL−1 T7 RNA polymerase (Takara Bio, Kusatsu, Japan) at 37 °C for 20 h. The transcribed RNA was purified by polyacrylamide gel extraction. Gel piece containing target RNA was cut out from the gel and crushed. Then RNA was extracted from the crushed gel pieces using elution buffer (0.3 M NaOAc buffer pH 5.2, 10 mM EDTA). The eluted RNA was collected by ethanol precipitation. The products were confirmed by gel electrophoresis.Preparation of the DNA frame and incorporation of RNA molecules
The DNA frame and DNA strands containing RNA molecules were prepared separately. Then the DNA frame and two DNA strands were annealed together. The DNA frame was prepared as described previously. Briefly, for the preparation of the DNA frame,22b a sample solution containing 25 nM M13mp18, 125 nM staple strands (5 eq.), 10 mM Tris-HCl (pH 7.6), and 10 mM MgCl2 was annealed from 75 °C to 15 °C at a rate of −1.0 °C min−1.19 For the preparation of DNA strands containing aptamers, sample solutions containing 0.83 μM Aptakiss (or KG51, or GTPswitch), 0.17 μM supporting DNA strands (AC96 and AC32 or BD96 and BD32, see ESI Fig. S1†), 10 mM Tris-HCl (pH 7.6), and 10 mM MgCl2 were annealed from 75 °C to 15 °C at a rate of −1.0 °C min−1.After the first annealing, 8.0 μL of DNA frame solution, 6.0 μL of Aptakiss solution, and 6.0 μL of KG51 (or GTPswitch) solution were mixed and then annealed from 40 °C to 15 °C at a rate of −1.0 °C min−1. At this second annealing, the solution contains 10 nM DNA frame and 50 nM DNA strands containing aptamers. These DNA frames having target RNA/DNA hybrid strands were purified by a gel filtration column (Sephacryl 400, GE Helthcare, Uppsala, Sweden).
AFM imaging of the kissing interaction
AFM images were obtained using Dimension FastScan (Bruker AXS, Madison, WI) with cantilever, BL-AC40TS-CS (Olympas, Tokyo, Japan). Purified samples were diluted ten times using observation buffer. Observation buffer for Aptakiss-KG51 contained 10 mM Tris-HCl pH 7.6, and 10 mM MgCl2; for Aptakiss-GTPswitch (with cover strand), 10 mM Tris-HCl pH 7.0, and 10 mM MgCl2, (1 mM GTP or ATP); for Aptakiss-GTPswitch mutant, 10 mM MOPS-KOH pH 6.5, 10 mM MgCl2, and 50 mM KCl, (1 mM GTP or ATP). The diluted solution (10 μL) was adsorbed onto mica plate for 5 min at room temperature and then washed three times using the same observation buffer to remove unadsorbed DNA strands and DNA frames. Scanning was performed in the same buffer solution using tapping mode.Results and discussion
Assembly of the target RNA molecules in the DNA frame
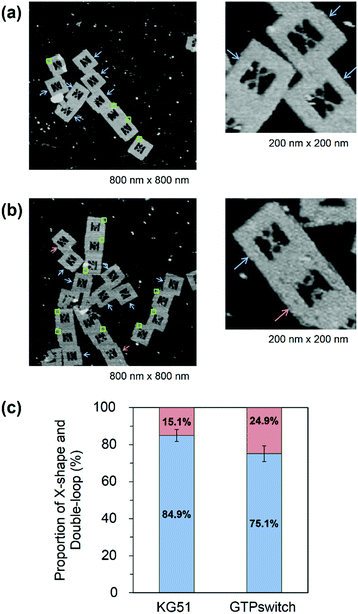
We used a DNA frame to evaluate the ligand-dependent activity of the GTPswitch at the single-molecule level. The DNA frame has a cavity (approximately 40 nm × 40 nm), in which four connectors are introduced to anchor the DNA strands. A pair of kissing RNA hairpins was placed in the cavity by incorporation into individual supporting DNA strands (DNA strands AC and BD), which were tethered between the specific connectors. Each strand comprised three parts: long ssDNA (AC96 or BD 96), short ssDNA (AC32 or BD32), and RNA that carried the designated sequence at its 3′ end (Fig. 1b). Here, we prepared three RNA/DNA hybrid strands: AC–Aptakiss, BD–KG51, and BD–GTPswitch. A pair of strands (AC–Aptakiss and BD–KG51 or BD–GTPswitch) was incorporated into the DNA frame through ssDNA linkers in the strands (Fig. 1b and S1†). Each linker a′, b′, c′, and d′ was connected to the corresponding connector a, b, c, and d, respectively. The binding of the RNA loops (kissing complex formation) was identified by configuration changes of the supporting DNA strands from the double-loop (unkissing) to the X-shape (kissing) (Fig. 1c). The difference in these structures was resolved in direct AFM imaging and quantified by statistical analysis of the AFM images (Fig. S2†).Observation of the interactions between KG51 and the GTPswitch in the DNA frame
First, we examined the interaction of a pair of kissing RNA hairpins, Aptakiss and KG51, in the DNA frame (Fig. 2a). KG51 is known to be capable of binding to the Aptakiss without the need for any additional cofactors and was used as an appropriate positive control for evaluating the data using this observation system. Quantitative analysis of micrographs of this sample revealed a high percentage of X-shaped structure (84.9%), suggesting that the KG51–Aptakiss system worked well in the DNA frame.We next substituted KG51 with the GTPswitch to examine the ligand-dependent binding property of the GTPswitch to the Aptakiss. Contrary to our expectation from the previous bulk experiment, 75.1% of DNA frame were observed to have the X-shaped structure even in the absence of GTP; this percentage was only ∼10% lower than that obtained for KG51 (Fig. 2c and Table 1). This unexpected high binding might have occurred because of an interaction between the bases in the kissing loop of the Aptakiss and the complementary bases of the unfolded free aptaswitch that was not previously detected in solution.16 We note that the two RNA sequences are located in relatively close positions in the DNA frame. The distance between the Aptakiss and the GTPswitch was estimated to be ∼10 nm. Assuming that the motion of each aptamer is limited to a sphere with a diameter of 10 nm, the hypothetical concentration can be estimated to be ∼1 mM. Although the molecular movement is constrained by fixation to the nanocavity, the molecules should behave as if they exist at a high concentration. This proximity effect may result in the ligand-independent binding.
| X-shape (%) | Double-loop shape (%) | ±S.D. | Counted numbers | |
|---|---|---|---|---|
| The data are represented as the mean ± S.D of triplicate experiments (n = 3). | ||||
| KG51 | 84.9 | 15.1 | 3.3 | 280 |
| GTPswitch | 75.1 | 24.9 | 4.3 | 285 |
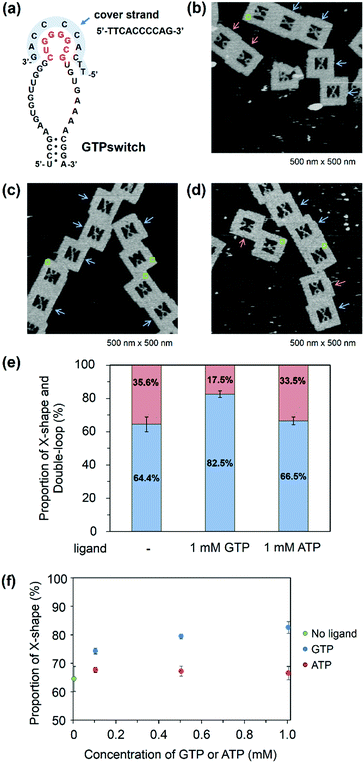
Observation of the switching function of the GTPswitch with a cover strand
To improve the ligand dependency of the GTPswitch in the DNA frame, we used a cover strand. Such a strategy was recently demonstrated to be successful and improved the specificity of an aptamer to adenosine.16 In our case, the chosen cover strand binds to the kissing loop of the GTPswitch and extends to part of the central loop that is the GTP binding site of the aptamer (Fig. 3a). This cover strand can be displaced by the addition of GTP, which allows the GTPswitch to bind to the Aptakiss in a ligand-dependent manner. The cover strand was hybridized to the GTPswitch by annealing, and the covered GTPswitch strand was then incorporated into the DNA frame together with the strand carrying the Aptakiss. In the absence of GTP, the percentage of X-shaped structure in the AFM images was calculated as 64.4% (Fig. 3b). This value reflected a ∼20% decrease in the percentage of the X-shape and is much lower than the values mentioned above for KG51 and the GTPswitch. The results indicate that the cover strand reduced the ligand-independent binding between the GTPswitch and the Aptakiss. Next, we examined the effect of introducing a cover strand-bound GTPswitch to the Aptakiss in the presence of GTP (Fig. 3c). Adenosine triphosphate (ATP) was also used to investigate the ligand specificity of the GTPswitch (Fig. 3d). From the AFM images, 82.5% of the cover strand-bound GTPswitch was in the X-shape in the presence of GTP; this value is similar to that for KG51 (84.9%). By contrast, in the presence of ATP, only 66.5% was in the X-shape; this value is close to that in the absence of GTP (64.4%). These results indicate that GTP could selectively induce binding between the GTPswitch and the Aptakiss, whereas ATP could not (Fig. 3e and Table 2).| GTPswitch/cover strand | X-shape (%) | Double-loop shape (%) | ±S.D. | Counted numbers |
|---|---|---|---|---|
| The data are represented as the mean ± S.D of triplicate experiments (n = 3). | ||||
| No ligand | 64.4 | 35.6 | 4.4 | 381 |
| 0.1 mM GTP | 74.3 | 25.7 | 1.0 | 238 |
| 0.5 mM GTP | 79.4 | 20.6 | 0.8 | 242 |
| 1.0 mM GTP | 82.5 | 17.5 | 2.1 | 324 |
| 0.1 mM ATP | 67.6 | 32.4 | 0.9 | 216 |
| 0.5 mM ATP | 67.1 | 32.9 | 1.7 | 229 |
| 1.0 mM ATP | 66.5 | 33.5 | 2.3 | 281 |
Moreover, we observed the binding in the presence of 0.1 and 0.5 mM of GTP and ATP. In the case of ATP, there is no change of the proportion. On the other hand, we found that at 0.1 mM of GTP the proportion of X-shape was decreased by ∼5% from the proportion at 0.5 and 1.0 mM of GTP (Fig. 3f).
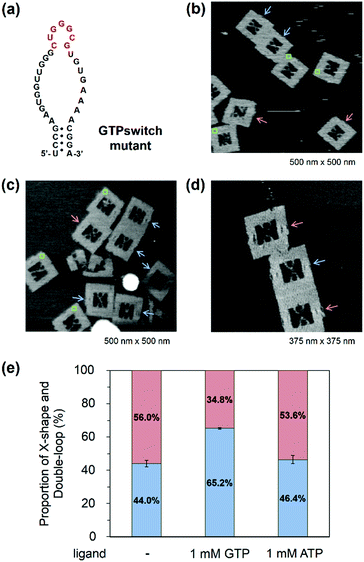
Observation of GTP switching in a mutant GTPswitch
When the Aptakiss and the GTPswitch were placed in the DNA frame, the two RNA hairpins bound easily because of the close packing in the nanospace. We tried to reduce the interaction of RNA hairpins by using a mutant GTPswitch, in which one G was deleted from the kissing loop of the GTPswitch (Fig. 4a). After assembling the Aptakiss and the mutant GTPswitch strands, we used AFM to observe the formation of the X-shape (Fig. 4b–d). In the absence of GTP, the X-shape formation between the Aptakiss and the mutant GTPswitch was 44.0%, which indicated a significant suppression of the interaction compared with the X-shape formed with the usual GTPswitch (75.1%). To examine the switching ability, formation of the X-shape was observed in the presence of GTP. The percentage in the X-shape was observed as 65.2%, which indicated a 21% increase in the binding of the GTPswitch and the Aptakiss when GTP was added. To confirm the ligand selectivity, we added ATP instead of GTP. In the presence of ATP, the percentage of the X-shape decreased by 19% to be 46.4% compared with that observed in the presence of GTP (Fig. 4e and Table 3) These results indicate that the mutant GTPswitch preserved the switching ability and ligand selectivity. These findings indicate that adjusting the association and dissociation of the kissing interaction of the RNA hairpins by deleting the nucleotide in the kissing domain was successful without losing the switching ability.| GTPswitch mutant | X-shape (%) | Double-loop shape (%) | ±S.D. | Counted numbers |
|---|---|---|---|---|
| The data are represented as the mean ± S.D of triplicate experiments (n = 3). | ||||
| No ligand | 44.0 | 56.0 | 2.0 | 202 |
| 1.0 mM GTP | 65.2 | 34.8 | 0.5 | 230 |
| 1.0 mM ATP | 46.4 | 53.6 | 2.5 | 214 |
Conclusions
We performed single-molecule observations of kissing complexes in the nanocavity of the DNA origami frame. Intriguingly, in the closely spaced condition, the GTPswitch could bind to the Aptakiss even in the absence of GTP in contrast to previous work.13,16 This GTP-independent binding could be suppressed by the addition of a cover strand against the kissing loop of the GTPswitch, and the GTP-dependent binding of the GTPswitch and Aptakiss was observed. The mutant GTPswitch also worked to control the kissing interaction and exhibited preserved switching ability and ligand selectivity. Although further optimization in the switching response is required, these findings support the potential applications of ligand-responsive kissing aptamers for dynamic systems that can be organized on DNA origami nanostructures. We believe that ligand-responsive kissing aptamers will enable us to regulate more global changes in nucleic acid nanostructures, such as programmed oligomerization into prescribed patterns.Acknowledgements
This work was supported by JSPS KAKENHI (grant numbers 15H03837, 24104002, 24225005, 26620133) to ME and HS. Financial supports from the Sekisui Chemical Research Grant and the Kurata Memorial Hitachi Science and Technology Foundation to ME are also acknowledged. We acknowledge the financial support of the Aquitaine Regional Government and ANR program VIBBnano (ANR-10-NANO-04) to JJT (Bordeaux).Notes and references
- F. Beaurain, C. Di Primo, J. J. Toulmé and M. Laguerre, Nucleic Acids Res., 2003, 31, 4275–4284 CrossRef CAS PubMed.
- I. Lebars, P. Legrand, A. Aime, N. Pinaud, S. Fribourg and C. Di Primo, Nucleic Acids Res., 2008, 36, 7146–7156 CrossRef CAS PubMed.
- J. C. Paillart, E. Skripkin, B. Ehresmann, C. Ehresmann and R. Marquet, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 5572–5577 CrossRef CAS.
- J. I. Tomizawa, Cell, 1986, 47, 89–97 CrossRef CAS PubMed.
- C. Persson, E. G. H. Wagner and K. Nordström, EMBO J., 1990, 9, 3761–3775 Search PubMed.
- L. Argaman and S. Altuvia, J. Mol. Biol., 2000, 300, 1101–1112 CrossRef CAS PubMed.
- C. Brunel, R. Marquet, P. Romby and C. Ehresmann, Biochimie, 2002, 84, 925–944 CrossRef CAS PubMed.
- E. Goux, S. Lisi, C. Ravelet, G. Durand, E. Fiore, E. Dausse, J. J. Toulmé and E. Peyrin, Anal. Bioanal. Chem., 2015, 407, 6515–6524 CrossRef CAS PubMed.
- H. Van Melckebeke, M. Devany, C. Di Primo, F. Beaurain, J. J. Toulmé, D. L. Bryce and J. Boisbouvier, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 9210–9215 CrossRef CAS PubMed.
- W. W. Grabow and L. Jaeger, Acc. Chem. Res., 2014, 47, 1871–1880 CrossRef CAS PubMed.
- A. Chworos, I. Severcan, A. Y. Koyfman, P. Weinkam, E. Oroudjev, H. G. Hansma and L. Jaeger, Science, 2004, 306, 2068–2072 CrossRef CAS PubMed.
- P. Guo, Nat. Nanotechnol., 2010, 5, 833–842 CrossRef CAS PubMed.
- F. Ducongé and J. J. Toulmé, RNA, 1999, 5, 1605–1614 CrossRef.
- K. Kikuchi, T. Umehara, K. Fukuda, J. Hwang, A. Kuno, T. Hasegawa and S. Nishikawa, J. Biochem., 2003, 133, 263–270 CrossRef CAS PubMed.
- S. Da Rocha Gomes, E. Dausse and J. J. Toulmé, Biochem. Biophys. Res. Commun., 2004, 322, 820–826 CrossRef CAS PubMed.
- G. Durand, S. Lisi, C. Ravelet, E. Dausse, E. Peyrin and J. J. Toulmé, Angew. Chem., Int. Ed., 2014, 53, 6942–6945 CrossRef CAS PubMed.
- M. Endo and H. Sugiyama, Acc. Chem. Res., 2014, 47, 1645–1653 CrossRef CAS PubMed.
- A. Rajendran, M. Endo and H. Sugiyama, Chem. Rev., 2014, 114, 1493–1520 CrossRef CAS PubMed.
- (a) Y. Sannohe, M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2010, 132, 16311–16313 CrossRef CAS PubMed; (b) A. Rajendran, M. Endo, K. Hidaka and H. Sugiyama, Angew. Chem., Int. Ed., 2014, 53, 4107–4112 CrossRef CAS PubMed.
- (a) A. Rajendran, M. Endo, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2013, 135, 1117–1123 CrossRef CAS PubMed; (b) M. Endo, M. Inoue, Y. Suzuki, C. Masui, H. Morinaga, K. Hidaka and H. Sugiyama, Chem. – Eur. J., 2013, 19, 16887–16890 CrossRef CAS PubMed.
- (a) M. Endo, Y. Yang, Y. Suzuki, K. Hidaka and H. Sugiyama, Angew. Chem., Int. Ed., 2012, 51, 10518–10522 CrossRef CAS PubMed; (b) Y. Yang, M. Endo, Y. Suzuki, K. Hidaka and H. Sugiyama, Chem. Commun., 2014, 50, 4211–4213 RSC.
- (a) M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2010, 132, 1592–1597 CrossRef CAS PubMed; (b) M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, Angew. Chem., Int. Ed., 2010, 49, 9412–9416 CrossRef CAS PubMed; (c) Y. Suzuki, M. Endo, Y. Katsuda, K. Ou, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2014, 136, 211–218 CrossRef CAS PubMed; (d) Y. Suzuki, M. Endo, C. Cañas, S. Ayora, J. C. Alonso, H. Sugiyama and K. Takeyasu, Nucleic Acids Res., 2014, 42, 7421–7428 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5bm00274e |
| This journal is © The Royal Society of Chemistry 2016 |