Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Potent inhibition of cyclic diadenylate monophosphate cyclase by the antiparasitic drug, suramin†
Clement
Opoku-Temeng
ab and
Herman O.
Sintim
*bc
aGraduate Program in Biochemistry, University of Maryland, College Park, Maryland 20742, USA
bDepartment of Chemistry, Purdue University, West Lafayette, IN 47907, USA
cCenter for Drug Discovery, Purdue University, West Lafayette, IN 47907, USA. E-mail: hsintim@purdue.edu; Tel: +1 (765) 496-6078
First published on 22nd January 2016
Abstract
C-di-AMP synthases are essential in several bacteria, including human pathogens; hence these enzymes are potential antibiotic targets. However, there is a dearth of small molecule inhibitors of c-di-AMP metabolism enzymes. Screening of 2000 known drugs against DisA has led to the identification of suramin, an antiparasitic drug as potent inhibitor of c-di-AMP synthase.
Cyclic diadenylate monophosphate (c-di-AMP) is an important second messenger found in Gram-positive Firmicutes and Actinobacteria.1–3 It is synthesized from two molecules of adenosine triphosphate (ATP) by diadenylate cyclases, DAC (see Fig. 1), and degraded by c-di-AMP-specific phosphodiesterases (PDE).4–7 C-di-AMP has been shown to regulate important processes in bacteria, such as virulence,8 cell wall formation,8,9 cell size,10 ion transport11 among others. For some Gram-positive bacteria, such as Bacillus subtilis and Listeria monocytogenes, it has been demonstrated that low intracellular levels of c-di-AMP increased the sensitivity of the cells to β-lactam antibiotic treatment; an effect attributed to defective peptidoglycan synthesis.12,13 Attempts to delete DAC from human pathogens L. monocytogenes13 and Streptococcus pneumoniae,14 failed and this was probably due to the essentiality of DAC for maintaining the integrity of the peptidoglycan. Considering that the majority of antibiotics target peptidoglycan synthesis, there is an obvious interest in discovering inhibitors of DAC enzymes.15–17
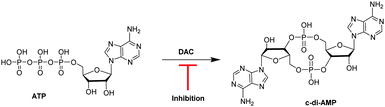 | ||
| Fig. 1 Schematic of the synthesis of c-di-AMP by DisA from ATP. A DAC inhibitor will hinder the synthesis of c-di-AMP.15 | ||
Motivated by the potential of DAC inhibitors as antibacterial targets we developed an interesting fluorescent assay for monitoring c-di-AMP synthesis, using readily available coralyne fluorophore.18 Using this assay, we discovered the first small molecule inhibitor of a DAC (DisA), bromophenol thiohydantoin (or bromophenol-TH).15 Bromophenol-TH is however a weak inhibitor of DisA (IC50 of 56 μM at 5 μM DisA)15 and attempts to improve its potency via modifications were unfruitful.17 Recently, Müller et al. showed that cordycepin triphosphate (3′-deoxyATP) was an inhibitor of Thermotoga maritima DisA (TmaDisA) with an IC50 of 3 μM at 26 nM TmaDisA.16
In an effort to identify more potent and non-nucleotide-based inhibitors of c-di-AMP synthesis, we employed the coralyne assay to screen a library of 2000 known drugs against DisA. This effort led to the identification of suramin (see Fig. 2), a drug used to treat parasitic infections, as a potent inhibitor of DisA.
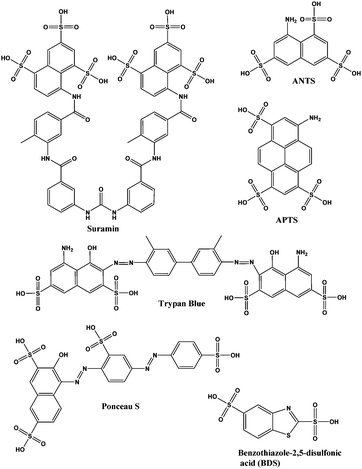 | ||
| Fig. 2 Structures of suramin and other sulfonated molecules tested against DisA. Abbreviations: ANTS is 8-aminonaphthalene-1,3,6-trisulfonic acid; APTS is 8-aminopyrene-1,3,6-trisulfonic acid. | ||
Suramin is a symmetrical polysulfonated urea derivative that has long been used as an anti-parasitic drug for the treatment of African trypanosomiasis as well as onchocerciasis.19,20 To delineate which moieties on suramin are responsible for DisA inhibition, we tested the enzyme against five structurally related compounds (see Fig. 2) using the coralyne assay. To eliminate the possibility that a positive hit from the coralyne assay was not due to the quenching of coralyne's fluorescence by the tested compounds, we also used HPLC to monitor the DisA reaction in presence of the compounds. 8-Aminopyrene-1,3,6-trisulfonic acid (APTS), 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS), Ponceau S and benzothiazole-2,5-disulfonic (compounds that contain two or more sulfonic acid groups) did not inhibit DisA (see Fig. 3 and Fig. S1, ESI†), indicating that the presence of sulfonic acid alone was not sufficient to cause DisA inhibition. Both trypan blue and suramin inhibited DisA (see Fig. 3) but suramin was a better inhibitor. Using HPLC to monitor the DisA reaction (see Fig. S1, ESI†), we noted that that at 20 μM, suramin inhibited DisA activity by 90% after 30 min while trypan blue only showed 20% inhibition. The coralyne assay indicated about 80% inhibition with trypan blue; the discrepancy between the coralyne and HPLC results for trypan blue inhibition is probably due to partial fluorescence quenching by trypan blue, which contains an azo moiety. Therefore, although the coralyne assay is more convenient than HPLC analysis for high throughput screening to discover c-di-AMP synthase inhibitors, it is crucial to use counter screens, such as HPLC analysis, to confirm “hits”.
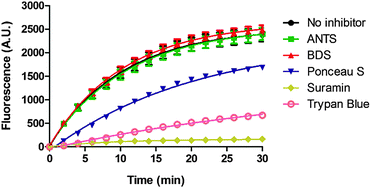 | ||
| Fig. 3 Coralyne assay of suramin and suramin-related compounds. For the coralyne assay, emission fluorescence intensity of coralyne (at 475 nm) increases as the concentration of c-di-AMP increases.18 BDS stands for benzothiazole-2,5-disulfonic acid. 8-Aminopyrene-1,3,6-trisulfonic acid (APTS) is highly fluorescent so could not be tested using the coralyne assay. Each experiment was done in triplicate. | ||
We wondered if suramin was inhibiting DisA via ATP-competitive mechanism. Previous report by Avliyakulov et al. indicated that binding of suramin to L3, a ribosomal protein from Trypanoplasma borreli, completely abolished ATP binding.21 Also, Morgan et al. crystallized suramin bound to the ATP binding site of Leishmania mexicana pyruvate kinase.22 These precedents provide evidence of suramin binding to the nucleotide binding pockets of proteins.
Docking of suramin with a monomer of TmaDisA(PDB:3C1Z)3 revealed that suramin bound to the ATP binding site of TmaDisA (Fig. 4) with binding affinity of −9.4 kcal mol−1. Future crystallography studies, beyond the scope of this work, should reveal if the docked structure is accurate and could provide more insight into how to design potent DisA inhibitors. B. subtilis DisA is similar to TmaDisA; computationally modelled 3D structure23 of B. subtilis DisA aligned well with TmaDisA (see Fig. S2, ESI†).
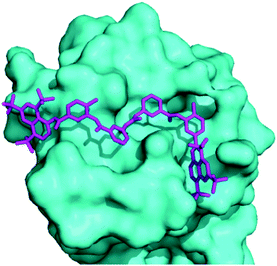 | ||
| Fig. 4 Molecular docking used to obtain a suramin/TmaDisA complex. Suramin (magenta) binds in the nucleotide-binding pocket of TmaDisA. Image was generated with PyMOL. | ||
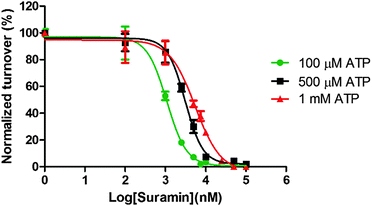
To provide some experimental confirmation that suramin and ATP compete for the same binding site in DisA, we determined the IC50 values at various ATP concentrations (100 μM, 500 μM and 1 mM, see Fig. 5). The formation of α-32P-c-di-AMP/c-di-AMP from α-32P-ATP/ATP by 1 μM DisA in the presence of increasing concentrations of suramin at the various ATP concentrations was monitored via TLC. IC50 values of 1.1 μM, 3.1 μM and 5.4 μM were obtained at the ATP concentrations of 100 μM, 500 μM and 1 mM respectively. The observed increase in IC50 values upon increasing ATP concentration is consistent with both molecules competing for similar binding site.
We also compared the potency of suramin's inhibition with that of 3′-deoxyATP and bromophenol-TH. Here, IC50 values of 2.3 μM, 3.8 μM and 67.2 μM (DisA concentration was 1 μM) were obtained for suramin, 3′-deoxyATP and bromophenol-TH respectively. This indicates that suramin is more potent than either of the two previously identified inhibitors of DisA (see Fig. S3, ESI†).
The addition of suramin to DisA resulted in a decrease in the protein intrinsic fluorescence (see Fig. S4, ESI†), probably due to changes in the microenvironment(s) of the tyrosine residues in the protein. Therefore we were able to determine the apparent Kd of suramin binding to DisA via fluorescence titration of the inhibitor with 5 μM DisA. The apparent Kd was determined to be 5.4 μM (Fig. 6A), assuming a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding. Bromophenol-TH bound to DisA with an apparent Kd of 21 μM,15 so it appears that suramin binds tighter to DisA, compared to bromophenol-TH. We also analysed the fluorescence quenching data using the Stern–Volmer equation24,25 (see Fig. S4, ESI†) and by the modified form of the Stern–Volmer equation (eqn (1) and Fig. 6B),26
1 binding. Bromophenol-TH bound to DisA with an apparent Kd of 21 μM,15 so it appears that suramin binds tighter to DisA, compared to bromophenol-TH. We also analysed the fluorescence quenching data using the Stern–Volmer equation24,25 (see Fig. S4, ESI†) and by the modified form of the Stern–Volmer equation (eqn (1) and Fig. 6B),26
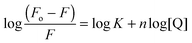 | (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding between DisA and suramin.
1 binding between DisA and suramin.
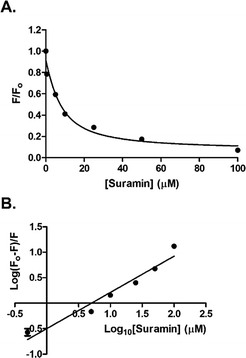 | ||
| Fig. 6 (A) Plot of relative fluorescence of DisA (5 μM) at 340 nm as a function of suramin concentration. The data was fitted to the non-linear regression, eqn (S3) (see ESI,† Experimental section). (B) Stern–Volmer plot generated by fitting the fluorescence emission at 340 nm to eqn (1). GraphPad Prism was used to obtain both plots from triplicate measurements. | ||
Suramin did not inhibit the activity of B. subtilis YybT, a c-di-AMP specific phosphodiesterase (see Fig. S5, ESI†), indicating that the observed DisA inhibition is somehow specific.
In conclusion, we have identified suramin as a potent inhibitor of DisA. Suramin is already used in the clinic to treat parasitic infection and has also been shown to have anti-cancer27 and anti-viral28 properties. Recently Nautiyal et al. showed that suramin inhibited Mycobacterium tuberculosis RecA protein with submicromolar IC50.29 They also showed that suramin potentiated the activity of ciprofloxacillin against M. smegmatis.29 Here, we show that suramin also inhibits cyclic diadenylate cyclase enzyme and represents an interesting scaffold, which could be used to develop cyclic dinucleotide signalling inhibitors. The advances made in the identification of c-di-AMP related enzymes, receptor proteins and RNA far outpace the number of small molecule inhibitors of these proteins. This paper sets the tone for the discovery of such small molecules, which could find use in unravelling the intricacies of cyclic dinucleotide signalling as well as being used as antibacterial agents.
This work was supported by funding from National Science Foundation (CHE1307218) and Camille Dreyfus Foundation. We thank Dr Karl-Peter Hopfner and Dr Angelika Gründling for plasmids encoding DisA and also Dr Zhao-Xun Liang for YybT plasmid.
Notes and references
- R. M. Corrigan and A. Gründling, Nat. Rev. Microbiol., 2013, 11, 513–524 CrossRef CAS PubMed.
- (a) U. Römling, Sci. Signaling, 2008, 1, pe39 CrossRef PubMed; (b) D. Kalia, G. Merey, S. Nakayama, Y. Zheng, J. Zhou, Y. Luo, M. Guo, B. T. Roembke and H. O. Sintim, Chem. Soc. Rev., 2013, 42, 305–341 RSC.
- G. Witte, S. Hartung, K. Buettner and K.-P. Hopfner, Mol. Cell, 2008, 30, 167–178 CrossRef CAS PubMed.
- Y. Bai, J. Yang, X. Zhou, X. Ding, L. E. Eisele and G. Bai, PLoS One, 2012, 7, e35206 Search PubMed.
- T. Kamegaya, K. Kuroda and Y. Hayakawa, Nagoya J. Med. Sci., 2011, 73, 49–57 CAS.
- T. N. Huynh, S. K. Luo, D. Pensinger, J. D. Sauer, L. Tong and J. J. Woodward, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E747–E756 CrossRef CAS PubMed.
- Y. Bai, J. Yang, L. E. Eisele, A. J. Underwood, B. J. Koestler, C. M. Waters, D. W. Metzger and G. Bai, J. Bacteriol., 2013, 195, 5123–5132 CrossRef CAS PubMed.
- C. E. Witte, A. T. Whiteley, T. P. Burke, J.-D. Sauer, D. A. Portnoy and J. J. Woodward, mBio, 2013, 4, e00282-13 CrossRef PubMed.
- M. Kaplan Zeevi, N. S. Shafir, S. Shaham, S. Friedman, N. Sigal, R. Nir Paz, I. G. Boneca and A. A. Herskovits, J. Bacteriol., 2013, 195, 5250–5261 CrossRef PubMed.
- R. M. Corrigan, J. C. Abbott, H. Burhenne, V. Kaever and A. Gründling, PLoS Pathog., 2011, 7, e1002217 CAS.
- Y. Bai, J. Yang, T. M. Zarrella, Y. Zhang, D. W. Metzger and G. Bai, J. Bacteriol., 2014, 196, 614–623 CrossRef PubMed.
- Y. Luo and J. D. Helmann, Mol. Microbiol., 2012, 83, 623–639 CrossRef CAS PubMed.
- J. J. Woodward, A. T. Iavarone and D. A. Portnoy, Science, 2010, 328, 1703–1705 CrossRef CAS PubMed.
- J. H. Song, K. S. Ko, J. Y. Lee, J. Y. Baek, W. S. Oh, H. S. Yoon, J. Y. Jeong and J. Chun, Mol. Cells, 2005, 19, 365–374 CAS.
- Y. Zheng, J. Zhou, D. A. Sayre and H. O. Sintim, Chem. Commun., 2014, 50, 11234–11237 RSC.
- M. Müller, T. Deimling, K. P. Hopfner and G. Witte, Biochem. J., 2015, 469, 367–374 CrossRef PubMed.
- Y. Zheng, J. Zhou, S. M. Cooper Jr, C. Opoku-Temeng, A. M. De Brito and H. O. Sintim, Tetrahedron, 2016 DOI:10.1016/j.tet.2015.10.073.
- J. Zhou, D. A. Sayre, Y. Zheng, H. Szmacinski and H. O. Sintim, Anal. Chem., 2014, 86, 2412–2420 CrossRef CAS PubMed.
- F. Hawking, Adv. Pharmacol. Chemother., 1978, 15, 289–322 CAS.
- C. C. Wang, Annu. Rev. Pharmacol. Toxicol., 1995, 35, 93–127 CrossRef CAS PubMed.
- N. K. Avliyakulov, J. Lukes, A. V. Kajava, B. Liedberg, I. Lundstrom and S. P. S. Svensson, Eur. J. Biochem., 2000, 267, 1723–1731 CrossRef CAS PubMed.
- H. P. Morgan, I. W. McNae, M. W. Nowicki, W. Zhong, P. A. M. Michels, D. S. Auld, L. A. Fothergill-Gilmore and M. D. Walkinshaw, J. Biol. Chem., 2011, 286, 31232–31240 CrossRef CAS PubMed.
- L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass and M. J. E. Sternberg, Nat. Protoc., 2015, 10, 845–858 CrossRef CAS PubMed.
- J. R. Lakowicz and G. Weber, Biochemistry, 1973, 12, 4161–4170 CrossRef CAS PubMed.
- J. R. Lakowicz and G. Weber, Biochemistry, 1973, 12, 4171–4179 CrossRef CAS PubMed.
- O. K. Abou-Zied and O. I. K. Al-Shihi, J. Am. Chem. Soc., 2008, 130, 10793–10801 CrossRef PubMed.
- R. V. La Rocca, C. A. Stein, R. Danesi and C. E. Myers, J. Steroid Biochem. Mol. Biol., 1990, 37, 893–898 CrossRef CAS PubMed.
- I. C. Albulescu, M. van Hoolwerff, L. A. Wolters, E. Bottaro, C. Nastruzzi, S. C. Yang, S.-C. Tsay, J. R. Hwu, E. J. Snijder and M. J. van Hemert, Antiviral Res., 2015, 121, 39–46 CrossRef CAS PubMed.
- A. Nautiyal, K. N. Patil and K. Muniyappa, J. Antimicrob. Chemother., 2014, 69, 1834–1843 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: HPLC analysis of DisA and YybT with select compounds, Experimental section. See DOI: 10.1039/c5cc10446g |
| This journal is © The Royal Society of Chemistry 2016 |