Archaeal ammonium oxidation coupled with bacterial nitrite oxidation in a simulated drinking water premise plumbing system†
Gem E.
Santillana‡
ab,
Heidi J.
Smith
ac,
Mark
Burr
a and
Anne K.
Camper
*ad
aCenter for Biofilm Engineering, Montana State University, Bozeman, MT 59717, USA. E-mail: anne_c@erc.montana.edu; Fax: 406 994 6098; Tel: 406 994 4906
bDepartment of Microbiology and Immunology, Montana State University, Bozeman, MT 59717, USA
cDepartment of Land Resources and Environmental Science, Montana State University, Bozeman, MT 59717, USA
dDepartment of Civil Engineering, Montana State University, Bozeman, MT 59717, USA
First published on 19th January 2016
Abstract
Simulated copper and PVC premise plumbing reactors modeling chloramine decay were monitored for complete nitrification of 0.71 mg NH4-N L−1 ammonium to nitrate with no nitrite detected. PCR, qPCR, fluorescent in situ hybridization (FISH) and DNA sequencing were used to investigate the microbial community responsible for nitrification in the reactors' influent and biofilm on copper and PVC surfaces. No bacterial ammonium oxidizers were detected by directly targeting the bacterial amoA gene or 16S rRNA gene amplicons. FISH images indicated an archaeal population on both surfaces. Archaeal 16S rRNA and amoA gene sequences showed 98.6% and 87.6% similarity to the known archaeal ammonium oxidizer, Candidatus Nitrosotenuis uzonenis. Copy numbers of the archaeal 16S rRNA gene and archaeal amoA approximated a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, suggesting that any archaea in the systems are likely to be ammonium oxidizers. Further, there was evidence for the presence of bacterial nitrite oxidizers. Copper surfaces supported fewer archaea as detected using the archaeal 16S rRNA and amoA genes. The results provide strong evidence for biofilms in a drinking water premise plumbing system composed of archaeal ammonium oxidizers and bacterial nitrite oxidizers, capable of complete oxidation of ammonium to nitrate. Since no bacterial ammonium oxidizers were found, this study adds to the growing body of research indicating an important role for archaeal ammonium oxidizers in freshwater/drinking water environments in the conversion of ammonium to nitrite.
1 ratio, suggesting that any archaea in the systems are likely to be ammonium oxidizers. Further, there was evidence for the presence of bacterial nitrite oxidizers. Copper surfaces supported fewer archaea as detected using the archaeal 16S rRNA and amoA genes. The results provide strong evidence for biofilms in a drinking water premise plumbing system composed of archaeal ammonium oxidizers and bacterial nitrite oxidizers, capable of complete oxidation of ammonium to nitrate. Since no bacterial ammonium oxidizers were found, this study adds to the growing body of research indicating an important role for archaeal ammonium oxidizers in freshwater/drinking water environments in the conversion of ammonium to nitrite.
Water impactChloramines added for disinfection of drinking water can decay to release ammonium, which then acts as a substrate for nitrification. Nitrification causes undesired changes in water quality and can lead to corrosion, particularly in premise plumbing. This study demonstrates the possible role of archaea in drinking water distribution system nitrification, prompting the consideration of the importance of this group of organisms in water distribution systems. |
Introduction
Driven by the need to comply with the Stage 2 Disinfectant/Disinfection By-Product (DDBP) Rule,1 there has been an increasing trend towards the use of monochloramine as a secondary disinfectant in drinking water in the U.S. An undesirable outcome is nitrification, a microbial oxidation process that converts ammonium to nitrite and/or nitrate. Nitrification leads to a loss of disinfectant residual, increased nitrate and nitrite concentrations, elevated microbial counts, and decreased pH in poorly buffered waters.2,3 Utilities struggle to control nitrification because of the limitations placed on operational conditions and concentrations of disinfectants. In a case study from a chloraminated drinking water treatment plant, significant nitrification episodes persisted despite an annual mitigation regime. This illustrates the complexity of drinking water disinfection and how utilities can struggle with balancing disinfectant choice, regulations, and water quality.4In premise plumbing, extended water age and reaction of chloramine with plumbing materials may lead to chloramine decay and the release of ammonia.5,6 Subsequent nitrification and reactions with plumbing materials, solder and fixtures can impact the concentrations of lead and copper in the water,7–9 the levels of which are regulated by the US EPA via the Lead and Copper Rule. Additionally, pipe material is expected to affect nitrification3 since compounds such as copper can be toxic or inhibitory to microorganisms.10–12
Traditionally, nitrification in drinking water has been attributed to ammonium oxidizing bacteria (AOB) and nitrite oxidizing bacteria (NOB). The first step of ammonium oxidization is often considered rate-limiting due to the slow growth of AOB and their sensitivity to environmental conditions.13–15 AOB have long been thought to be responsible for ammonium oxidation in mesophilic environments, and pure cultures of these organisms have been routinely used in disinfection studies.16–18 The past decade has seen this “bacteriocentric” view challenged,19 especially in environments where AOB were detected in low abundances and ammonium concentrations were below limits of AOB affinity.20 In particular, molecular methods have enabled the detection of difficult-to-culture archaeal ammonia oxidizers (AOA) (or more conservatively, putative AOA where the amoA gene encodes the enzyme ammonia mono-oxygenase subunit A), as potential candidates for nitrification.19
AOA have been overlooked because these archaea have long been considered strict extremophiles, with roles in the global nitrogen cycle largely limited to marine or thermal environments.14,19,21 However, mesophilic AOA and putative AOA with the amoA gene have been increasingly detected by molecular methods in quantities rivaling or exceeding AOB, for example, in soil,22 freshwater,23,24 the rhizosphere,25 wastewater treatment plants (reviewed by Limpiyakorn),13 and drinking water treatment and distribution systems.4,26–34 Nonetheless, environmental factors affecting niche differentiation and selection processes for AOA or AOB in a specific environment are poorly understood.19,35 More importantly, the role of AOA is unresolved, as there is little evidence for their contribution to nitrification in disinfected drinking water. Additionally, it was recently demonstrated by both van Kessel et al.36 and Daims et al.37 that complete nitrification, or the oxidation of ammonia to nitrate, can be achieved by a single Nitrospira spp. This single step nitrification process is referred to as comammox, and is utilized by organisms as an energy conservation mechanism. These recent studies report that there is the genetic potential for this process from a variety of environments,37 and the role of these organisms in microbial nitrogen cycling is only now being recognized.
The current study used well-established (nitrification at pseudo-steady state for over six years; full conversion of ammonium to nitrate) laboratory reactors simulating premise plumbing with periods of flow and stagnation. Research with these reactors had shown differences in nitrification depending on the surfaces (copper or polyvinyl chloride (PVC)) in response to potential control mechanisms (addition of copper to PVC, dosing with chlorite, increasing chlorine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ammonia ratios).9 To further elucidate potential reasons for these differences in nitrification, a more thorough understanding of the microbial ecology governing nitrification was sought. Specific objectives were to 1) determine differences in populations and diversity between the two types of reactors (copper and PVC) at two time points, 2) identify the organisms responsible for nitrification, 3) assess the relative abundance of these organisms in the biofilms on two surfaces (copper and PVC), and 4) evaluate the diversity and abundance of bacterial and archaeal 16S rRNA genes and genes for nitrification. A culture-independent approach was coupled with the use of propidium monoazide (PMA) to analyze both total cells and those with intact membranes.36
ammonia ratios).9 To further elucidate potential reasons for these differences in nitrification, a more thorough understanding of the microbial ecology governing nitrification was sought. Specific objectives were to 1) determine differences in populations and diversity between the two types of reactors (copper and PVC) at two time points, 2) identify the organisms responsible for nitrification, 3) assess the relative abundance of these organisms in the biofilms on two surfaces (copper and PVC), and 4) evaluate the diversity and abundance of bacterial and archaeal 16S rRNA genes and genes for nitrification. A culture-independent approach was coupled with the use of propidium monoazide (PMA) to analyze both total cells and those with intact membranes.36
Because complex factors control nitrification and due to the general inability to mitigate the process once it begins, there is a need to better understand the microbial ecology of nitrification in drinking water systems. This understanding would lead to improved predictive capabilities by identifying appropriate targets for detection which will enable the elucidation of reasonable control strategies.
Methods
Reactor set-up
Reactors simulating premise plumbing that had been actively nitrifying (complete conversion of ammonium to nitrate) for six years9 were used for this study. Baseline conditions (feed of 0.71 mg L−1 NH4-N) from the previous work were continuously maintained. There were four replicate reactors containing copper or PVC coupons that had been in place for a minimum of six years. Reactors were kept in the dark and at room temperature. To simulate premise plumbing, periods of flushing and stagnation were included in the operational conditions of the reactors (one reactor volume, three times daily).Reactors were fed a combination of mineral amended reverse osmosis (RO) water, Bozeman tap water treated by passing through a biologically active carbon (BAC) column (BAC water; surface water source, no background ammonium, chlorinated), and a humic substances organic feed extracted from Elliot silt loam soil (International Humic Substances Society). Constituents of the feedwater have been reported previously.9 Ammonium sulfate was added to a final concentration of 0.71 mg L−1 as N, equivalent to the ammonia concentration from the complete decay of 4 mg L−1 chloramine. BAC water provided a continuous inoculum of indigenous organisms (104 CFU mL−1 of heterotrophic plate count (HPC)). No other inoculum of nitrifiers was added. All reactors showed signs of stable, complete nitrification as measured by conversion of ammonia to nitrate.
Sample preparation
BAC water was sampled for DNA extraction/denaturing gradient gel electrophoresis (DGGE) and qPCR. Biofilm was sampled for DNA extraction/DGGE and subsequent cloning, qPCR and for fluorescent in situ hybridization (FISH) analysis. All sampling was done at the end of an 8 hour stagnation period over a period of four years. Biofilms were scraped from coupons (1.5 × 1.7 cm) using a rubber policeman and homogenized. For BAC sampling, 500 mL were collected at the same time biofilm was collected and filtered through a 47 mm diameter 0.2 micron pore size polycarbonate membrane (http://www.millipore.com). Membranes were shredded and placed into individual 1.5 mL clear centrifuge tubes with 500 μL filtered BAC water. Biofilm samples were re-suspended in 1 mL filtered BAC water, vortexed, and split (500 μL aliquots) into clear 1.5 mL microcentrifuge tubes. PMA treatment on one of the two parallel samples followed the method of Nocker et al.38 DNA was extracted from PMA treated and untreated tubes using the FastDNA® SPIN Kit for Soil (MP Biomedicals, OH) modified to include a step to remove humics with guanidine thiocyanate.39 For PMA controls, killed samples were prepared by autoclaving samples at 121 °C for 20 minutes.Endpoint polymerase chain reaction (PCR)
End point PCR amplifications were conducted in an Eppendorf Mastercycler® ep (Eppendorf North America, http://www.eppendorfna.com). Reactions used 25 μL volumes (0.2 μM primers, 1X Go Taq® Green Master Mix (http://www.promega.com), DEPC treated water, 1 μg μL−1 ultrapure BSA (Ambion) and approximately 5 ng of template DNA). Oligonucleotide primers were synthesized by IDT (Integrated DNA Technologies, http://www.idtdna.com). Primer sequences and PCR thermal cycler conditions are reported in ESI† Table S1. PCR products were run on a 0.8% agarose gel in 1X TBE buffer for 45 minutes at 40 V then stained with ethidium bromide and visualized using a FluorChem™ 8800 fluorescence imager (http://www.alphainnotech.com).The functional gene encoding for bacterial amoA was targeted using primers amoA1-F and amoA2-R40 most widely used in environmental studies.41 This primer pair targets a region conserved in all betaproteobacterial AOB but does not cover gammaproteobacterial AOB. Preliminary sequencing of DGGE bands from 16S rRNA gene amplification showed no evidence of gammaproteobacteria (see Results and discussion). DNA from Nitrosomonas europaea Winogradsky (ATCC strain 25978) was used as a positive control. To detect NOB, the nxrA primer pair for Nitrobacter spp.42 and the nxrB primer pair for most Nitrospira spp. were used.43 For AOA, the functional archaeal amoA gene was amplified using arch-amoAF and arch-amoAR.44
Denaturing gradient gel electrophoresis (DGGE)
DGGE was used to separate PCR products based on sequence differences.45 The 16S rRNA gene was targeted using the primers 1055F and 1392R46 (with GC clamp) for bacteria and ARC344F-GC and ARC915R47 for archaea. For the archaea, products from an initial PCR reaction were used as a template for a nested PCR with the second set of primers.48 This step was necessary because dilution to remove PCR inhibitors also diluted the target DNA. For DGGE analysis of archaeal amoA, the primer pair Arch-amoA-for and Arch-amoA-rev (with GC clamp) was used.49 DGGE was performed using a DCode™ system (http://www.biorad.com). Denaturing gels with denaturant concentration of 40–60% from top to bottom were used for separating the PCR amplicons, where 100% denaturant is defined as 7 M urea and 40% formamide. Gels also contained an 8 to 12% polyacrylamide gradient from top to bottom. Ten microliters of each sample was loaded per well. Electrophoresis was done at 60 V for 16 hours. Gels were stained with Sybr®Gold (http://www.invitrogen.com) and visualized using a FluorChem™ 8800 fluorescence imager (http://www.alphainnotech.com). Marker lanes using amplicons from five unidentified 16S rRNA gene clones targeting the vector insert and selected for their different migration distances spanning the entire gel were included in each DGGE gel to allow for comparison between gels.DGGE bands in the images were identified using GelCompar II software (Version 6.1, Applied Maths, Inc.) and confirmed visually. Pairwise correlation analysis of bands was done to determine if the occurrence of one band was correlated to another/other bands which may suggest that a single operational taxonomic unit (OTU) was represented by more than one band. Phylotypes of each sample were determined by counting the total number of distinct bands of each sample's DGGE profile. A binary matrix (band presence–absence data) was created from the normalized DGGE gels and saved as a Comma Delimited Format (CDL) file and used in the subsequent analysis in R v.2.11.1 (R Development Core Team, 2010).
The Dice coefficient of similarity/Sorensen's coefficient of similarity was computed as previously reported.50,51 The computed Dice coefficient was then used for cluster analysis using flexible beta in the package cluster.52
Bands were excised from the DGGE gels and re-suspended in DEPC treated water. DNA was extracted using a freeze thaw cycle (3 cycles of 1 hour freezing and 1 hour at room temperature). One microliter was used as the PCR template (1055F/1392R + GC). Resulting amplicons were run on DGGE to verify the position of the bands and to make sure that each amplicon produced only one band.
DNA sequencing
PCR products from excised DGGE bands were gel purified using QIAquick® Gel Extraction Kit (QIAGEN) and cloned into plasmid vector pCR™4-TOPO® using the TOPO® TA Cloning kit (Invitrogen, http://www.invitrogen.com). Transformants were inoculated into 10 mL of Luria-Bertani (LB) broth plus 50 mg mL−1 ampicillin and incubated overnight at 37 °C in a shaking incubator. One clone was chosen for every excised band and plasmid DNA was purified from each individual clone using the Wizard Plus SV Minipreps DNA Purification System (http://www.promega.com) and quantified using the NanoDrop ND-1000 spectrophotometer (Nano Drop, Wilmington USA). Clones were Sanger sequenced by the Research Technology Support Facility (RTSF) at Michigan State University using the M13F primer. Sequences were checked for chimeras using Chimera Slayer53 or Bellerophon54 and were compared with known sequences in the GenBank database using the Basic Local Alignment Sequence Tool (BLAST; Altschul et al.55http://blast.ncbi.nlm.nih.gov/Blast.cgi). All sequences were deposited in the GenBank NCBI database (ESI† Table S2).Quantitative PCR
Quantitative PCR (qPCR) was used to determine the relative abundance of bacterial and archaeal amoA and 16S rRNA genes in PMA treated and untreated samples of BAC and biofilm from one copper and one PVC reactor at one time point. Primers pairs 338F/518R and 931F/m1100R were used for bacterial and archaeal 16S rRNA genes, respectively.56 Primer pairs amoA-1F/amoA-2R40 and Arch-amoA-for/Arch-amoA-rev49 were used for bacterial and archaeal amoA genes, respectively. Primer sequences and thermal cycling conditions are reported in ESI† Table S1. The qPCR reaction mixture consisted of 1X Power SYBR® Green PCR Master Mix (http://www.appliedbiosystems.com), 0.2 μM each of the forward and reverse primers, 1 μg μL−1 ultrapure BSA (50 mg mL−1, Ambion, http://www.ambion.com) and DEPC water. For each 25 μL reaction, 8 μL of template DNA was used.qPCR was performed in a Rotor-Gene 3000 real time PCR cycler (QIAGEN, http://www.qiagen.com) in a 72-well rotor. Data were acquired using the FAM/Sybr detection channel during the extension step. Standards and samples were prepared in duplicate and negative controls containing no template DNA were included. Melt curve analysis was performed from 60–95 °C in 0.1 °C increments held for 5 s with an initial pre-melt hold for 90 s at the first step to verify amplification of correct PCR products. Univariate analysis of variance (ANOVA) was performed on the gene copy abundance data with a general linear model using Minitab 16 (Minitab).
16S rRNA gene amplicons were cloned into pCR™4-TOPO® using the TOPO® TA Cloning Kit (Invitrogen, http://www.invitrogen.com). A purified plasmid was randomly chosen and was sequenced as described previously to verify a 16S rRNA gene insert. Plasmid concentration was determined using a ND-100 spectrophotometer (Nano Drop, Wilmington USA) and a Sybr®Gold (http://www.invitrogen.com) assay standardized with concentrations of double stranded Lambda DNA (Promega) measured with a ND-3300 fluorospectrometer (Nano Drop, Wilmington USA). Copy number was calculated by considering the size of the plasmid (3890 bp) plus insert length (180 bp) and assuming a molecular mass of 660 Da for each base pair. qPCR standards were prepared by diluting the plasmid suspension (8.8 × 103 to 8.8 × 107 target gene copies per μL, tenfold dilutions). The same approach was used for the bacterial amoA qPCR standards but with the 491 bp long PCR amplicon amplified from N. europaea Winogradsky (ATCC strain 25978). The bacterial 16S rRNA gene standard sequence was deposited in GenBank with the accession number JQ406518.
PCR inhibition was investigated by using dilutions of sample DNA as templates for the qPCR reaction along with standards. Ct values were compared to that of the standards. The lowest dilution falling within the linear range that was parallel to that of the standards was chosen for analysis. For validation, a spiked sample dilution was prepared by adding a known amount of standard template to the sample dilution. The Ct value of the spiked dilution was compared to that of a standard with an equal concentration of DNA.
Phylogenetic analyses of 16S rRNA and amoA genes
Amplified 16S rRNA and amoA sequences from the reactors were screened for the presence of chimeric sequences using Bellerophon.54 Respectively, reference sequences of either 16S rRNA or amoA gene sequences were compiled from publically available sources: NCBI BLASTN function (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi, Altschul et al.55) using the nucleotide collection and the whole-genome shotgun contigs databases. The amplified 16S rRNA and amoA sequences were aligned to respective reference sequences with Clustal W.57 Prior to constructing phylogenetic tress all alignments were manually refined. Phylogenetic trees were constructed in Geneious using the neighbor-joining method58 and the Jukes–Cantor distance model59 with bootstrap values of 1000 replicates.Fluorescence in situ hybridization (FISH)
FISH analysis was performed on biofilm that was not treated with PMA. Biofilm was scraped from coupons and re-suspended in 1 mL phosphate-buffered saline (PBS) (0.8% NaCl in 10 mM phosphate, pH 7.2). Re-suspended biofilm (500 μL) was mixed with ice cold 4% paraformaldehyde (PFA) solution in PBS. Samples were incubated at 4 °C for 4 hours. A centrifugation (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 5 min), supernatant removal, and PBS washing step was repeated three times. Samples were then re-suspended in one volume of ice cold PBS and one volume of ice cold 96% (v/v) ethanol (500 μL total volume).
000 × g, 5 min), supernatant removal, and PBS washing step was repeated three times. Samples were then re-suspended in one volume of ice cold PBS and one volume of ice cold 96% (v/v) ethanol (500 μL total volume).
Five microliters of fixed sample was deposited on a Teflon coated slide and air dried at 46 °C for 10 min. Samples were dehydrated by dipping sequentially in 50, 80 and 100% ethanol baths. Ten microliters of hybridization buffer (0.9 M NaCl, 20 mM Tris HCl, 35% formamide, 0.01% (w/v) SDS and 0.3 ng of archaeal and bacterial specific probes (ESI† Table S1) were added and incubated at 46 °C for 3 hours. The slide was then transferred to a 50 ml tube containing the washing buffer (0.07 M NaCl, 0.02 mM Tris HCl, 5 mM EDTA) at 46 °C for 10 minutes. The slide was then dipped in ice cold water and dried. Samples were mounted with Citifluor AFI antifadent (Citifluor Ltd, Leicester, UK).
A Leica TCS-SPZ AOBS laser scanning confocal microscope was used for imaging. A 561 nm laser was used to excite Cy3 and a 633 nm laser to excite Cy5. Fluorescence was collected from 568–618 for Cy3 and from 660–800 nm for Cy5. Cy3 fluorescence was false colored red and Cy5 green. Samples were imaged using a HCX PL APO CS100×1.4NA oil objective.
Results and discussion
The purpose of this research was to characterize the microbial community in simulated premise plumbing laboratory reactors that had been nitrifying for six years as determined by ammonium loss and nitrate production. One of the reasons for doing so was the difference shown previously60 in nitrification in the same copper vs. PVC reactors sampled in the current study; copper systems began nitrifying several months after the PVC systems. Once nitrification was established, ammonium was depleted within three hours in the PVC system while copper systems required four hours for ammonium removal during the eight hour stagnation periods.60 Other notable characteristics were (i) copper added to PVC reactors at levels up to 1.3 mg L−1 (regulatory limit of copper as per the EPA's Lead and Copper Rule) did not impact nitrification, (ii) the copper reactor was more sensitive to the addition of 20 mg L−1 chlorite, (iii) chlorite at concentrations below 20 mg L−1 and at levels determined to control nitrification in some field studies61,62 did not affect nitrification in either reactor type, and (iv) nitrification in copper reactors recovered more slowly after termination of chloramination at a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 chorine/ammonium ratio.9 In these experiments Rahman et al.9 reported most probable number (MPN) values for ammonium and nitrite oxidizing organisms in the bulk phase. Results were greatly dependent upon treatment, the abundance of ammonium oxidizing organisms ranged from 7 to 50 MPN mL−1 while the abundance of nitrite oxidizing organisms was from below detection limit to approximately 300 MPN mL−1. It is important to note that the MPN method is based on culturing and microbial identities inferred by substrate conversion. Consequently, phylogenetic analysis was required to give greater insight into the organisms responsible for ammonium and nitrite oxidation.
1 chorine/ammonium ratio.9 In these experiments Rahman et al.9 reported most probable number (MPN) values for ammonium and nitrite oxidizing organisms in the bulk phase. Results were greatly dependent upon treatment, the abundance of ammonium oxidizing organisms ranged from 7 to 50 MPN mL−1 while the abundance of nitrite oxidizing organisms was from below detection limit to approximately 300 MPN mL−1. It is important to note that the MPN method is based on culturing and microbial identities inferred by substrate conversion. Consequently, phylogenetic analysis was required to give greater insight into the organisms responsible for ammonium and nitrite oxidation.
An initial study was done to determine if there was a potential inhibitory effect of copper on the community that would lead to damaged cells, for which PMA use would be beneficial as a diagnostic tool. Two representative bacterial 16S rRNA gene DGGE profiles (PMA treated and untreated, not shown) were produced from the influent BAC water and biofilm from one copper and one PVC reactor. Dice coefficient/Sorensen's pairwise similarity coefficients of the DGGE profiles were 84% for BAC, 98% for PVC biofilm, and 96% for copper biofilm. No profile for the same sample (PMA treated vs. untreated) matched 100%, indicating the removal of some members from the community, presumably because they were dead and/or had compromised cell membranes. Only faint bands were removed by PMA treatment from profiles and no new bands appeared. Therefore, it did not appear that there was inactivation of organisms on the copper surfaces but that there could be selection based on surface material.
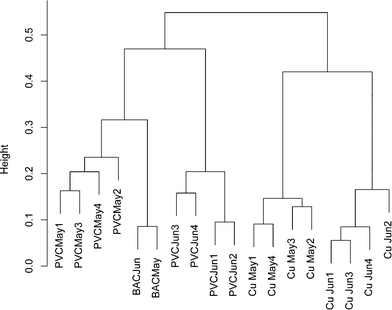
To ensure that any comparisons between copper and PVC were not the result of time dependent variability, a bacterial 16S rRNA gene DGGE banding pattern was obtained from replicate influent BAC water and copper/PVC biofilm samples collected from two sequential months (ESI† Fig. S1). All samples were PMA treated to restrict the analysis to DNA from intact cells. A total of 37 distinct DGGE bands/OTUs were detected. Pairwise correlations no greater than 0.75 suggest each band is representative of a unique OTU. The average number of DGGE profile bands for BAC (n = 2) was 17.5, for copper biofilms (n = 8) 21.4, and for PVC biofilms (n = 8) 19.4. Band richness of BAC, copper and PVC samples were not significantly different from each other (p ≥ 0.58) and were in the same range as those in ESI† Tables S3 (copper) and S4 (PVC). Only one band was present across all samples, and three bands were found only in the PVC biofilm. Based on the cluster analysis of DGGE banding patterns a community profile, over time and sample type, was detected (Fig. 1). The influent BAC and PVC samples clustered more closely together compared to the copper. This suggests that the PVC biofilm organisms most closely resembled those of the reactor influent compared to the copper. However, BAC samples clustered more strongly compared to the PVC samples, which is indicative of greater temporal variability in the PVC samples compared to the influent BAC water. Additionally, copper samples clustered separately from the PVC and influent BAC samples, indicating some level of selection on the copper. There was also similarity between months indicating that the population was stable over this time period. These data also supported the concept that each surface selected for a different community.
Once reproducibility of sampling times and communities were established, the research focused on identifying the organism(s) responsible for ammonium oxidation. Since there were no reports of the presence of AOA in premise plumbing systems, it was assumed that AOB were responsible for the first step in nitrification. PCR targeting the functional amoA bacterial gene was performed. However, even with repeated attempts over the course of the research to optimize conditions, no amplicon indicative AOB was ever obtained from any sample. To confirm that the selected primers were accurately targeting and amplifying the bacterial amoA gene, a positive control was included with every amplification that was preformed, and an amplicon was consistently obtained. Simultaneously, bacterial studies were extended to investigate the second step in nitrification; the conversion of nitrite to nitrate. Using nxrA primers designed for Nitrobacter, an amplicon was obtained from biofilm from the PVC reactor, but not from the copper reactor. When nxrB primers designed to amplify nxrB from most Nitrospira were used, amplicons were obtained from both PVC and copper. The presence of amplification products from the two sets of primers suggests the probable presence of more than one nitrite oxidizing genus as well as organisms capable of comammox. Recently identified organisms that are capable of comammox belong to Nitrospira lineage II. Upon the comparison of the 16S rRNA sequences from the PVC reactors, to the 16S ribosomal database, sequences were most closely related to a Nitrospria japonica sp. (ranging from 93–98% similarity). Similarly, the only 16S rRNA sequence retrieved from the copper biofilm identified as a Nitrospira sp., had the closest similarity (96%) to the same organism, Nitrospria japonica sp. when compared to the 16S ribosomal database. While Nitrospria japonica sp. is a member of Nitrospira lineage II based off from findings from Daims et al.37 it phylogentically belongs to a different cluster than does the identified comammox organisms.37 While this does not provide definitive evidence in support or against comammox in these systems, based on the current understanding of comammox the organisms in our system are not most closely related to organisms possessing the ability for complete nitrification. Since the primary focus was on ammonium oxidation, amplicons obtained using nxr primer sets were not sequenced and presumed positive PCR results were based on obtaining an nxrB amplicon of the expected length.
To further investigate the prominent members of the communities of the copper and PVC biofilms as detected by DGGE, an initial study of bacterial diversity was conducted by targeting the bacterial 16S rRNA gene because it had the potential to identify sequences similar to those of known bacterial ammonium and nitrite oxidizers. Total DNA from a copper and PVC reactor was used instead of DNA from PMA treated samples to assess the microbial diversity of the entire microbial community. ESI† Tables S3 (copper) and S4 (PVC) show the BLAST taxonomic identities of the bands obtained from the DGGE profiles. Twelve of 15 bands from the biofilm from the copper coupon and 14 of 16 bands from the biofilm from the PVC coupon were successfully re-amplified to yield DNA sequences considered sufficiently long for phylogenetic analysis (≥200 bp). No sequence was common to both reactor types. Additional diversity may have been present but below some threshold such that no visible band was produced from these templates as band intensity was not considered; this is a potential limitation for DGGE. However, this method did confirm the initial inability to detect bacterial ammonium oxidizers using amoA genes as targets, and no sequences from either biofilm sample obtained using the 16S rRNA gene as a target had significant percent identity to known bacterial ammonium oxidizers. Several sequences were highly similar to sequences from NOB belonging to the genus Nitrospira (ESI† Table S3, band 8; Table S4, bands 3, 5, 11, 12). The detection of Nitrospira-like 16S rDNA sequences with high percent identity scores (e.g., ESI† Table S4, band 8), supports the detection of nxr genes by PCR, reported above.
The information gained from the 16S rRNA sequences was used to evaluate the potential diversity of the most common members of the biofilm populations. Obtained sequences (ESI† Table S2) had closest GenBank relatives from a variety of environments, many completely unrelated to drinking water systems. However, an interesting exception is band 2 from the copper reactor (ESI† Table S3) that shares 100% sequence identity with (a) 11 clones from a laboratory study of the relationship between drinking water distribution system bacteria and chloramine decay,63 (b) seven clones from a study of the effects of plumbing materials (but not including copper) on microbial community composition,64 and (c) a single clone from two other studies of drinking water microbial communities.65,66 These reported clones are not listed because they were not taxonomically identified in GenBank. The richness in the PVC biofilm compared favorably with the 17 OTUs reported by Lin et al.64 for biofilm grown on PVC in a similar laboratory study. Jang et al.67 found distinct profiles for replicate samples of copper, stainless steel, cast iron, and PVC surfaces. No sequences associated with bacterial ammonium oxidizers were detected.
Because there was no evidence of bacterial ammonium oxidizers from bacterial amoA PCR or bacterial 16S rRNA derived sequences based on the primers utilized in this study, it appeared possible that archaea were responsible for ammonium oxidation. This conclusion is also supported by the fact that the archaea population is dominated by only one species where amoA gene copy numbers are in agreement with the abundance of archaea. Lebedeva et al.68 also reported failure to detect AOB by amoA and 16S rRNA gene PCR in a nitrifying environment where they did detect AOA. Likewise, Park et al.69 failed to detect AOB in an AOA enrichment culture from marine sediment. FISH was performed with domain specific probes and both archaea and bacteria were detected in PVC and copper biofilms (Fig. 2), while control probes confirmed the absence of nonspecific binding. Archaea appeared to be relatively more abundant in the PVC biofilm compared to the copper.
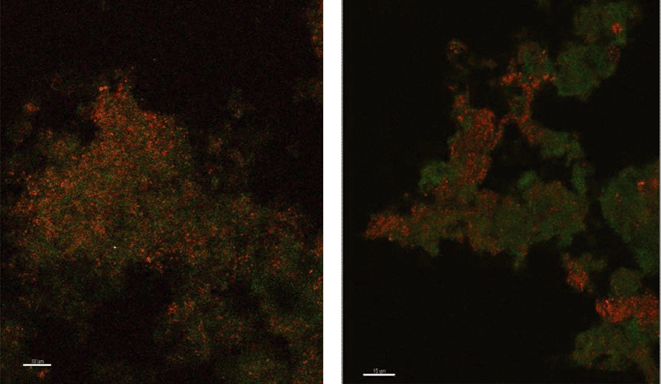 | ||
| Fig. 2 Fluorescence in situ hybridization microscopy images of biofilms from copper (left) and PVC (right) coupons showing bacteria (red) and archaea (green). | ||
Based on evidence obtained from the FISH results and the lack of evidence for bacterial ammonium oxidizing organisms, the research then focused on the potential presence of ammonium oxidizing archaea. A single, near full length, archaeal 16S rRNA gene fragment from DNA collected from biofilms growing on both the PVC and the copper and the BAC water was amplified using archaeal primers regardless of sample date or PMA treatment. Supporting the earlier noted differences between copper and PVC, a two-step PCR was needed to amplify the fragment from the copper sample. Even though there was apparently lower abundance of the archaea on copper, screening of clone libraries generated from these amplicons indicated identical sequences amplified from both PVC and copper. The 710 bp sequence was submitted to GenBank as accession JQ717299. In addition, four shorter archaeal amoA fragments were found over the course of the research: 617 bp, (JQ406520) from one PVC reactor, 254 bp (JQ717297), and 253 bp (JQ717298) in the BAC influent and coupon biofilms and 256 bp (JQ406519), from PVC biofilm. The latter was used as a qPCR standard. All four of the amoA gene sequences were highly similar to each other (>99%), and for the purpose of this study the longest amplified sequence (617 bp, JQ406520) was used as a representative amoA gene sequence.
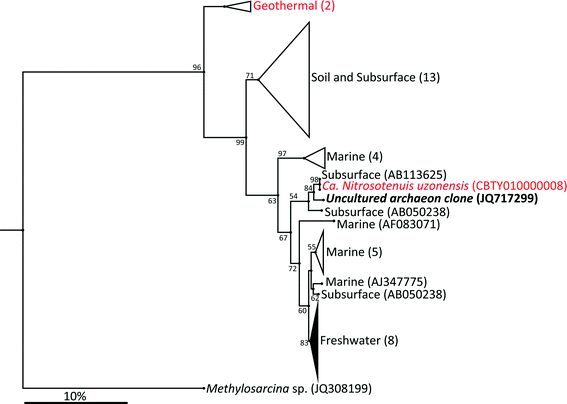
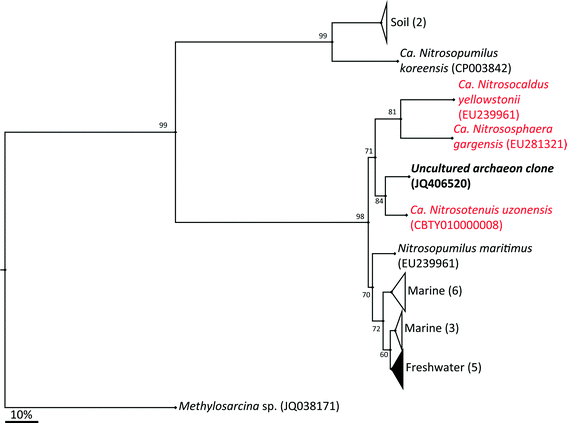
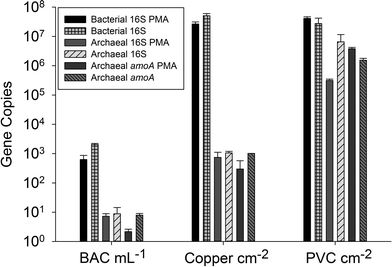
The archaeal 16S rRNA gene sequence was most closely related to the ammonium oxidizing organism Ca. Nitrosotenuis uzonensis (98.6% identity, Fig. 3) isolated from a geothermal feature.68 When compared to the identity obtained using the amoA gene sequence, the closest relative is also Ca. Nitrosotenuis uzonensis68 (87.6% identity, Fig. 4). Additionally, the amoA gene amplified from the reactor influent and biofilms is more closely related to other archaea isolated from geothermal environments than it is to other freshwater or marine habitats. Both the amplified 16S rRNA and amoA gene had the same closest relative, suggesting that the amplified 16S rRNA and amoA gene sequences are from the same organism. Since these were the only sequences retrieved after exhaustive sequencing efforts, it is speculated that there is only one AOA present in these reactors. Other evidence supporting the hypothesis of the presence of one type of AOA and the absence of bacterial ammonia oxidizers in the reactors are the qPCR results that determined the relative copy numbers of the archaeal and bacterial 16S rRNA and amoA genes in PMA treated and untreated samples from copper and PVC reactors. As with endpoint PCR, no bacterial amoA was amplified during qPCR from any sample. Fig. 5 reports gene copy numbers in the BAC influent water (copies per mL) and in copper and PVC biofilms (copies per cm2). In general, there were no significant differences between PMA treated and untreated samples, and PMA treatment indicates that the detected genes came predominately from intact bacterial and archaeal cells.
There were almost two logs more bacterial than archaeal 16S rRNA gene copies in the reactor BAC influent (p = 0.006). Bacterial 16S rRNA gene abundance in the biofilm from the copper and PVC coupons were not significantly different (p = 0.927), while there were significantly (three logs, p = 0.004) more archaeal 16S rRNA gene copies in the biofilm from the PVC coupon compared to the copper coupon. This agrees with the visual observation in the FISH images of more archaea in PVC biofilm compared to copper. There were significantly more bacterial than archaeal 16S rRNA gene copies in the biofilm from the copper coupon (four log difference, p = 0.001. Bacterial and archaeal 16S rRNA gene copies were not significantly different for non PMA treated biofilm from the PVC coupons (p = 0.487). However, the PMA treated PVC biofilm sample had approximately 1.5 logs fewer archaeal than bacterial 16S rRNA gene copies (p = 0.012). There were almost four logs fewer archaeal amoA copies in the copper biofilm compared to the PVC (p = 0.008). This is another indication of the potential selective effect of the copper surface on archaea compared to the more inert PVC surface.
There is need for caution in interpreting relative copy numbers of amoA and 16S rRNA genes from archaea and bacteria obtained by qPCR. Herrmann et al.25 cited Kapplenbach et al.70 and Leininger et al.22 to justify a correction based on the assumption that AOB have an average of 2.5 copies of amoA and 3.6 copies of the 16S rRNA gene, while to date AOA have been shown to have only single copies of these two genes. Gene abundance per se may have little correlation with ammonium oxidizer activity.20 Mußmann et al.71 reported cases where the number of AOA estimated by qPCR of amoA could not have been supported autotrophically by the amount of ammonium measured in the system.
The most important result from the qPCR data is the remarkable agreement in copy numbers between archaeal 16S rRNA and amoA genes, which holds true for all three pairings of these genes (BAC, copper biofilm, and PVC biofilm). This is strong evidence that the archaeal population, which appears from DNA sequencing data to be limited to a single phylotype, is an AOA. The archaeal cells detected in the system probably carry a single copy of both genes, which is consistent with what is currently known about the archaeal ammonium oxidizers.72 Kasuga et al.32 also reported archaeal 16S rRNA gene and amoA copy numbers that were consistently within the same order of magnitude during a four month period on filters used in drinking water treatment. In an enrichment of an archaeal ammonium oxidizer, Hatzenpichler et al.19 also suggested that 16S rRNA gene and amoA sequences came from the same organism, but were only able to confirm that by subsequent near-complete genome sequencing.68 The same process would be required to confirm that our archaeal amoA and 16S rRNA gene sequences came from the same organism.
The question then arises as to why AOA and not AOB were responsible for nitrification in the model premise plumbing reactors. Prosser and Nicol35 did not believe that any single factor explained the selection of AOA vs. AOB, but thought that the three most important were ammonium concentration, mixotrophy, and pH. Work by Martens-Habbena et al.73 showed that Candidatus Nitrosopumulus maritimus in pure culture had a low Km of 133 nM total ammonium and a low substrate threshold of ≤10 nM and could therefore effectively compete with other organisms in the ocean environment. A tempting hypothesis is that the presence of only AOA was determined primarily by the ammonium feed concentration (0.71 mg NH4-N L−1). Several studies in water systems suggest that AOA are present at lower ammonium concentrations while AOB are more likely to be found at levels over 1 mg NH4-N L−1.23,26,27,31,34 However, the importance of ammonium concentration is not clear cut, and there are studies where ammonium concentration did not appear to explain selection of AOB vs. AOA, or where the results were inconclusive.28,29,33
In summary, we have described the microbial community in simulating premise plumbing systems that have been nitrifying for six years. Ammonium oxidation appears to be performed by archaea with diversity limited to a single organism. Nitrite oxidation is most likely performed by bacteria. Therefore, complete nitrification is accomplished by an archaeal-bacterial consortium. From previous work9 it was shown that the addition of copper to a PVC system at the regulated limit of 1.3 mg L−1 did not inhibit nitrification, but copper reactors were less robust to potential nitrification mitigation strategies (addition of chlorite, changes in chlorine:ammonium ratios to form chloramine). The current work suggests that there is a different community of organisms on copper vs. PVC. It was more difficult to detect AOA on copper vs. PVC by molecular methods (including FISH). Copper had a lower abundance of archaea as detected using the 16S rRNA and amoA genes. PMA treatment results suggested that the two surfaces selected for different populations in addition to copper having a greater effect on cell viability as detected through cell wall integrity; this effect was most pronounced on the archaea. However, both the PVC and copper reactors were capable of fully oxidizing 0.71 mg L−1 added ammonium to nitrate in the 8 hour stagnation period.
An understanding of the diversity of organisms that impact nitrification in freshwater systems, including drinking water, requires further investigation. An awareness of the types of organisms and their responses to environmental and operational conditions will lead to better mechanisms for predicting, identifying, and controlling nitrification events in water systems that have naturally occurring ammonium or use chloramine for secondary disinfection. An extension of this information to premise plumbing will be beneficial in determining what systems are at risk for the detrimental aspects of nitrification. With the recognition that AOA may be the prevalent group of organisms responsible for nitrification in some systems, it is now important to determine the environmental factors that favor AOB over AOA and the implications for the dominance of one organism over the other in system performance and operations.
Acknowledgements
The authors wish to acknowledge the Fulbright Philippine Agriculture Scholarship Program and the Center for Biofilm Engineering for their financial support. Special thanks are extended to Dr. Al Parker for assistance with statistics and Andy Olson for his help in the laboratory. Kristen Brileya and Betsey Pitts are acknowledged for their assistance with FISH and microscopy.References
- USEPA.2000. M/DBP FACA Support Document Search PubMed.
- J. Skadsen, Nitrification in a distribution system, J. - Am. Water Works Assoc., 1993, 85(7), 95–103 CAS.
- Y. Zhang, N. Love and M. Edwards, Nitrification in drinking water systems, Crit. Rev. Environ. Sci. Technol., 2009, 39, 153–208 CrossRef CAS.
- H. Wang, C. R. Proctor, M. A. Edwards, M. Pryor, J. W. Santo Domingo, H. Ryu, A. K. Camper, A. Olson and A. Pruden, Microbial community response to chlorine conversion in a chloraminated drinking water distribution system, Environ. Sci. Technol., 2014, 48(18), 10624–10633 CrossRef CAS PubMed.
- C. Nguyen, C. Elfland and M. Edwards, Impact of advanced water conservation features and new copper pipe on rapid chloramine decay and microbial regrowth, Water Res., 2012, 46, 611–621 CrossRef CAS PubMed.
- Y. Zhang and M. Edwards, Accelerated chloramine decay and microbial growth by nitrification in premise plumbing, J. - Am. Water Works Assoc., 2009, 101(11), 51–62 CAS.
- Y. Zhang, A. Griffin, M. Rahman, A. K. Camper, H. Baribeau and M. Edwards, Lead contamination of potable water due to nitrification, Environ. Sci. Technol., 2009, 43(6), 1890–1895 CrossRef CAS PubMed.
- Y. Zhang, A. Griffin and M. Edwards, Effect of nitrification on corrosion of galvanized iron, copper and concrete, J. - Am. Water Works Assoc., 2010, 102(4), 83–93 CAS.
- M. S. Rahman, G. Encarnacion and A. K. Camper, Nitrification and potential control mechanisms in simulated premises plumbing, Water Res., 2011, 45, 5511–5522 CrossRef CAS PubMed.
- J. T. Trevors and C. M. Cotter, Copper toxicity and uptake in microorganisms, J. Ind. Microbiol., 1990, 6, 77–84 CrossRef CAS.
- M. J. Lehtola, I. T. Miettinen, T. Lampola, A. Hirvonen, T. Vartiainen and P. J. Martikainen, Pipeline materials modify the effectiveness of disinfectants in drinking water distribution systems, Water Res., 2005, 39, 1962–1971 CrossRef CAS PubMed.
- D. van der Kooij, H. R. Veenendaal and W. J. H. Scheffer, Biofilm formation and multiplication of Legionella in a model warm water system with pipes of copper, stainless steel and cross-linked polyethylene, Water Res., 2005, 39, 2789–2798 CrossRef CAS PubMed.
- T. Limpiyakorn, M. Furhacker, R. Haberl, T. Chodanon, P. Srithep and P. Sonthiphand, amoA-encoding archaea in wastewater treatment plants: a review, Appl. Microbiol. Biotechnol., 2013, 97, 1425–1439 CrossRef CAS PubMed.
- H. Urakawa, W. Martens-Habbena and D. A. Stahl, Physiology and genomics of ammonia-oxidizing Archaea, in Nitrification, ed. B. B. Ward, D. J. Arp and M. G. Kotz, ASM Press, Washington DC, 2011 Search PubMed.
- J. You, A. Das, E. M. Dolan and Z. Hu, Ammonia-oxidizing archaea involved in nitrogen removal, Water Res., 2009, 43, 1801–1809 CrossRef CAS PubMed.
- C. Chauret, C. Volk, L. Stover, T. S. Dykstra, R. C. Andrews and G. A. Gagnon, Effect of disinfectants on microbial ecology in model distribution systems, J. Water Health, 2005, 3, 359–369 CrossRef CAS PubMed.
- D. G. Wahman, K. A. Wulfeck-Kleier and J. G. Pressman, Monochloramine disinfection kinetics of Nitrosomonas europea by propidium monoazide quantitative PCR and live/dead BacLight methods, Appl. Environ. Microbiol., 2009, 75(17), 5555–5562 CrossRef CAS PubMed.
- J. M. Regan, G. W. Harrington, H. Baribeau, R. De Leon and D. R. Noguera, Diversity of nitrifying bacteria in full-scale chloraminated distribution systems, Water Res., 2003, 37(1), 197–205 CrossRef CAS PubMed.
- R. Hatzenpichler, E. V. Lebedeva, E. Spieck, K. Stoecker, A. Richter, H. Daims and M. Wagner, A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2134–2139 CrossRef CAS PubMed.
- M. Monteiro, J. Seneca and C. Magalhaes, The history of aerobic ammonia oxidizers: from the first discoveries to today, J. Microbiol. Methods, 2014, 52(7), 537–547 CAS.
- G. W. Nicol, S. Leininger and C. Schleper, Distribution and activity of ammonia-oxidizing archaea in natural environments. In Nitrification, ed. B. B. Ward, D. J. Arp and M. G. Kotz, ASM Press, Washington DC, 2011 Search PubMed.
- S. Leininger, T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster and C. Schleper, Archaea predominate among ammonia-oxidizing prokaryotes in soils, Nature, 2006, 442, 806–809 CrossRef CAS PubMed.
- J.-C. Auguet, X. Triado-Margarit, N. Nomokonova, L. Camarero and E. O. Casamayor, Vertical segregation and phylogenetic characterization of ammonia-oxidizing Archaea in a deep oligotrophic lake, ISME J., 2012, 6, 1786–1797 CrossRef CAS PubMed.
- E. French, J. A. Kozlowski, M. Mukherjee, G. Bullerjahn and A. Bollmann, Ecophysiological characterization of ammonia-oxidizing archaea and bacteria from freshwater, Appl. Environ. Microbiol., 2012, 78(16), 5773–5780 CrossRef CAS PubMed.
- M. Herrmann, A. M. Saunders and A. Schramm, Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora, Appl. Environ. Microbiol., 2008, 74, 3279–3283 CrossRef CAS PubMed.
- V. Gomez-Alvarez, K. A. Schrantz, J. G. Pressman and D. G. Wahman, Biofilm community dynamics in bench-scale annular reactors simulating arrestment of chloraminated drinking water nitrification, Environ. Sci. Technol., 2014, 48, 5448–5457 CrossRef CAS PubMed.
- C. O. Lee, R. Boe-Hansen, S. Musovic, B. Smets, H.-J. Albrechtsen and P. Binning, Effects of dynamic operating conditions on nitrification in biological rapid sand filters for drinking water treatment, Water Res., 2014, 64, 226–236 CrossRef CAS PubMed.
- J. Niu, I. Kasuga, F. Kurisu, H. Furumai and T. Shigeeda, Evaluation of autotrophic growth of ammonia-oxidizers associated with granular activated carbon used for drinking water purification by DNA-stable isotope probing, Water Res., 2013, 47, 7053–7065 CrossRef CAS PubMed.
- S. Feng, X. Zhang, Q. Wang, R. Wan, C. Chen and S. Xie, Heterogeneity of ammonia-oxidizing community structures in a pilot-scale drinking water biofilter, Int. Biodeterior. Biodegrad., 2012, 70, 148–152 CrossRef CAS.
- W. W. J. M. de Vet, I. J. T. Dinkla, G. Muyzer, L. C. Rietveld and M. C. M.van Loosdrecht, Molecular characterization of microbial populations in groundwater sources and sand filters for drinking water production, Water Res., 2009, 43, 182–194 CrossRef CAS PubMed.
- W. W. J. M. de Vet, R. Kleeresbezem, P. W. J. J. van der Wielen, L. C. Rietveld and M. C. M. van Loosdrecht, Assessment of nitrification in groundwater filters for drinking water production by qPCR and activity measurement, Water Res., 2011, 45, 4008–4018 CrossRef CAS PubMed.
- I. Kasuga, H. Nakagaki, F. Kurisu and H. Furumai, Predominance of ammonia-oxidizing archaea on granular activated carbon used in a full-scale advanced drinking water treatment plant, Water Res., 2010, 44, 5039–5049 CrossRef CAS PubMed.
- I. Kasuga, H. Nakagaki, F. Kurisu and H. Furumai, Abundance and diversity of ammonia-oxidizing archaea and bacteria on biological activated carbon in a pilot-scale drinking water treatment plant with different treatment processes, Water Sci. Technol., 2010, 61(12), 3070–3077 CrossRef CAS PubMed.
- P. W. van der Wielen, S. Voost and D. van der Kooij, Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems, Appl. Environ. Microbiol., 2009, 75, 4687–4695 CrossRef CAS PubMed.
- J. I. Prosser and G. W. Nicol, Archeal and bacterial ammonia-oxidisers in soil: the quest for niche specialization and differentiation, Trends Microbiol., 2012, 20(11), 523–531 CrossRef CAS PubMed.
- M. A. H. J. van Kessel, D. R. Speth, M. Albertsen, P. H. Nielsen, H. J. M. Op den Camp, B. Kartal, M. S. M. Jetten and S. Lucker, Complete nitrification by a single microorganism, Nature, 2015, 528, 555–559 CrossRef PubMed.
- H. Daims, E. V. Lebedeva, P. Jjevac, P. Han, C. Herbold, M. Albertsen, N. Jehmlich, M. Palatinszky, J. Vierheilig, A. Bulaev, R. H. Kirkegaard, M. von Bergen, T. Rattei, B. Bendinger, P. H. Nielsen and M. Wagner, Complete nitrification by Nitrospira bacteria, Nature, 2015, 528, 504–509 CrossRef CAS PubMed.
- A. Nocker, P. Sossa, M. Burr and A. K. Camper, Use of propidium monoazide for live-dead distinction in microbial ecology, Appl. Environ. Microbiol., 2007, 73, 5111–5117 CrossRef CAS PubMed.
- M. D. Burr, S. J. Clark, C. R. Spear and A. K. Camper, Denaturing gradient gel electrophoresis can readily display the bacterial diversity contained in 16S rDNA clone libraries, Appl. Environ. Microbiol., 2006, 51, 479–486 CAS.
- J. H. Rotthauwe, K. P. Witzel and W. Liesack, The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations, Appl. Environ. Microbiol., 1997, 63, 4704–4712 CAS.
- P. Junier, O. S. Kim, V. Molina, P. Limburg, T. Junier, J. K. Imhoff and K. P. Witzel, Comparative in silico analysis of PCR primers suited for diagnostics and cloning of ammonia monooxygenase genes from ammonia-oxidizing bacteria, FEMS Microbiol. Ecol., 2008, 64, 141–152 CrossRef CAS PubMed.
- S. Wertz, F. Poly, X. Le Roux and V. Degrange, Development and application of a PCR-denaturing gradient gel electrophoresis tool to study the diversity of Nitrobacter-like nxrA sequences in soil, FEMS Microbiol. Lett., 2008, 63, 261–271 CrossRef CAS PubMed.
- F. M. Maixner, The ecophysiology of nitrite-oxidizing bacteria in the genus Nitrospira: novel aspects and unique features, PhD Thesis, University of Vienna, 2009, http://othes.univie.ac.at/5088/1/2009-04-14_0409617.pdf, accessed January 2011 Search PubMed.
- C. A. Francis, K. J. Roberts, J. M. Beman, A. E. Santoro and B. B. Oakley, Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 14683–14688 CrossRef CAS PubMed.
- G. Muyzer, E. C. Dewall and A. G. Uitterlinden, Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S ribosomal RNA, Appl. Environ. Microbiol., 1993, 59(3), 695–700 CAS.
- M. J. Ferris, G. Muyzer and D. M. Ward, Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hotspring microbial mat community, Appl. Environ. Microbiol., 1996, 62, 340–346 CAS.
- E. O. Casamayor, H. Schäfer, L. Bañeras, C. Pedrós-Alió and G. Muyzer, Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis, Appl. Environ. Microbiol., 2000, 66, 499–508 CrossRef CAS PubMed.
- E. F. DeLong, Archaea in coastal marine environments, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 5685–5889 CrossRef CAS.
- C. Wuchter, B. Abbas, M. J. Coolen, L. Herfort, J. van Bleijswijk, M. Timmers, M. Strous, E. Teira, G. J. Herndl, J. J. Middelburg, S. Schouten and J. S. Sinninghe Damste, Archaeal nitrification in the ocean, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(3), 12317–12322 CrossRef CAS PubMed.
- V. J. McCracken, J. M. Simpson, R. I. Mackie and H. R. Gaskins, Molecular ecological analysis of dietary and antibiotic-induced alterations of the mouse intestinal microbiota, J. Nutr., 2001, 131, 1862–1870 CAS.
- A. Magurran, Diversity indices and species abundance models, Ecological diversity and its measurement, Princeton University Press, Princeton, NJ, 1988, pp. 8–45 Search PubMed.
- M. Maechler, P. Rousseeuw, A. Struyf and M. Hubert, Cluster analysis basics and extensions. Cited in R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, ISBN 3-900051-07-0, URL http://www.R-project.org/unpublished, 2005 Search PubMed.
- B. J. Haas, D. Gevers, A. M. Earl, M. Feldgarden, D. V. Ward, G. Giannoukos, D. Ciulla, D. Tabbaa, S. K. Highlander, E. Sodergren, B. Methé, T. Z. DeSantis, J. F. Petrosino, R. Knight and B. W. Birren, Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons, Genome Res., 2011, 21, 494–504 CrossRef CAS PubMed.
- T. Huber, G. Faulkner and P. Hugenholtz, Bellerophon: a program to detect chimeric sequences in multiple sequence alignment, Bioinformatics, 2004, 20, 2317–2319 CrossRef CAS PubMed.
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers and D. J. Lipman, Basic local alignment search tool, J. Mol. Biol., 1990, 215, 403–410 CrossRef CAS PubMed.
- J. Einen, I. H. Thorseth and I. Ovreås, Enumeration of Archaea and Bacteria in seafloor basalt using real-time quantitative PCR and fluorescence microscopy, FEMS Microbiol. Lett., 2008, 282, 182–187 CrossRef CAS PubMed.
- J. D. Thompson, D. G. Higgins and T. J. Gibson, CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice, Nucleic Acids Res., 1994, 22, 4673–4680 CrossRef CAS PubMed.
- N. Saitou and M. Nei, The neighbor joining method—a new method for reconstructing phylogenetic trees, Mol. Biol. Evol., 1987, 4, 406–425 CAS.
- T. H. Jukes and C. R. Cantor, Evolution of protein molecules, in Mammalian Protein Metabolism, ed. H. N. Munro, Academic Press, New York, 1969, pp. 21–132 Search PubMed.
- M. S. Rahman, Nitrification in premise plumbing systems, Ph.D. thesis, Montana State University, Bozeman, MT., 2008 Search PubMed.
- M. J. McGuire, J. N. Lieu and M. S. Pearthree, Using chlorite ion to control nitrification, J. - Am. Water Works Assoc., 1999, 91(10), 52–62 CAS.
- M. J. McGuire, M. S. Pearthree, N. K. Blute, K. F. Arnold and T. Hoogerwerf, Nitrification control by chlorite ion at pilot scale, J. - Am. Water Works Assoc., 2006, 98(1), 95–105 CAS.
- K. Bal Krishna, A. Sathasivan and M. Ginige, Microbial community changes with decaying chloramine residuals in a lab-scale system, Water Res., 2013, 47(13), 4666–4679 CrossRef CAS PubMed.
- W. Lin, Z. Yu, X. Chen, R. Liu and H. Zhang, Molecular characterization of natural biofilms from household taps with different materials: PVC, stainless steel, and cast iron in drinking water distribution system, Appl. Microbiol. Biotechnol., 2013, 97(18), 8393–8401 CrossRef CAS PubMed.
- D. Li, Z. Li, J. Yu, N. Cao, R. Liu and M. Yang, Characterization of bacterial community structure in a drinking water distribution system during an occurrence of red water, Appl. Environ. Microbiol., 2010, 76(21), 7171–7180 CrossRef CAS PubMed.
- M. Williams, J. Domingo, M. Meckes, C. Kelty and H. Rochon, Phylogenetic diversity of drinking water bacteria in a distribution system simulator, J. Appl. Microbiol., 2004, 96(5), 954–964 CrossRef CAS PubMed.
- H. Jang, Y. J. Choi and J. Ka, Effects of diverse water pipe materials on bacterial communities and water quality in the annular reactor, J. Microbiol. Biotechnol., 2011, 21, 115–123 CrossRef CAS PubMed.
- E. V. Lebedeva, R. Hatzenpichler, E. Pelletier, N. Schuster, S. Hauzmayer, A. Bulaev, N. V. Grigor'eva, A. Galushko, M. Schmid, M. Palatinszky, D. Le Paslier, H. Daims and M. Wagner, Enrichment and genome sequence of the Group 1.1a ammonia-oxidizing archaeon “Ca. Nitrosotenius uzonensis” representing a clade globally distributed in thermal habitats, PLoS One, 2013, 8(11), e80835, DOI:10.1371/journal.pone.0080835.
- B.-J. Park, S.-J. Park, D.-Y. Yoon, S. Schouten, J. S. Sinninghe Damaste and S.-K. Rhee, Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria, Appl. Environ. Microbiol., 2010, 76, 7575–7587 CrossRef CAS PubMed.
- J. A. Kapplenbach, P. R. Saxman, J. R. Cole and T. M. Schmidt, rrndb: ribosomal RNA operon copy number database, Nucleic Acids Res., 2001, 29, 181–184 CrossRef.
- M. Mußmann, I. Brito, A. Pitcher, J. S. S. Damste, R. Hatzenpichler, A. Richter, J. L. Nielsen, P. H. Nielsen, A. Muller, H. Daimes, M. Wagner and I. M. Head, Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16771–16776 CrossRef PubMed.
- T. J. Mincer, M. J. Church, L. T. Taylor, C. Preston, D. M. Karl and E. F. DeLong, Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific subtropical gyre, Environ. Microbiol., 2007, 9, 1162–1175 CrossRef CAS PubMed.
- W. Martens-Habbena, P. M. Berube, H. Urakawa, J. R. de la Torre and D. A. Stahl, Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria, Nature, 2009, 461(7266), 976–979 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supporting information includes the original DGGE image, primer sequences and thermal cycler conditions for PCR, probe sequences for FISH, and the sequences deposited in the GenBank NCBI database as well as identities of DGGE bands from copper and PVC coupon biofilms. See DOI: 10.1039/c5ew00273g |
| ‡ Current address: Department of Plant Pathology, North Carolina State University, Raleigh, NC 27695, USA. |
| This journal is © The Royal Society of Chemistry 2016 |