Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microfluidic platforms for DNA methylation analysis
Ryoji
Kurita
* and
Osamu
Niwa†
Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology (AIST) and DAILAB, Tsukuba Central 6, 1-1-1 Higashi, Tsukuba, Ibaraki, 305-8566 Japan. E-mail: r.kurita@aist.go.jp; Fax: +029 861 6177; Tel: +029 861 6158
First published on 2nd August 2016
Abstract
In the field of genetics, epigenetics is the study of changes in gene expression without any change in DNA sequences. Chemical base modification in DNA by DNA methyltransferase, and specifically methylation, has been well studied as the main mechanism of epigenetics. Therefore, the determination of DNA methylation of, for example, 5′-methylcytosine in the CpG sequence in mammals has attracted attention because it should prove valuable in a wide range of research fields including diagnosis, drug discovery, and therapy. Methylated DNA bases and DNA methyltransferase activity are analyzed using conventional methods; however, these methods are time-consuming and require complex multiple operations. Therefore, new methods and devices for DNA methylation analysis are now being actively developed. Furthermore, microfluidic technology has also been applied to DNA methylation analysis because the microfluidic platform offers the promising advantage of making it possible to perform thousands of DNA methylation reactions in small reaction volumes, resulting in a high-throughput analysis with high sensitivity. This review discusses epigenetics and the microfluidic platforms developed for DNA methylation analysis.
1. Introduction
1.1 Epigenetics and DNA methylation
Epigenetics is the genetic study of cellular and physiological phenotypic trait variations and a mechanism for controlling gene expression that does not depend on DNA sequence changes.1,2 Namely, daughter cells inherit genetic characteristics through cell division, but epigenetics is an independent mechanism of change in DNA bases. Epigenetics is known to be a chemically stable modification in DNA; however, DNA is also known to be dynamically changed by environmental factors such as exposure to oxidative stress.3–5 DNA methylation and histone modification have received particular attention as the main mechanisms of epigenetics. In this review, we mainly focus on DNA methylation as indicated by our title.DNA methylation, especially the addition of a methyl group at the fifth position of the cytosine base (5′-methlcytosine) in mammalian cells, was first discovered6 in 1948 in thymus-derived bovine DNA. It became clear that the methylation of genomic DNA is related to various life phenomena7–9 that can be seen in a wide range of creatures, Escherichia coli, plants and vertebrates. In particular, cytosine methylation at CpG islands in mammals is becoming a crucially important study in ontogeny and cytodifferentiation. This is because the methylation of the CpG sequence has been revealed to relate to the genetic silence mechanism. The methylation of DNA is also known to be accompanied by a change in the chromatin structure. The methylation of DNA controls gene expression and functions as a storage system for tissue-specific expressed genes.
The determination of the DNA methylation information for various gene regions is important not only for basic biology areas such as cytogenesis and reproduction but also for nuclear cell transfer technology, tissue engineering and a range of diagnostic techniques. Furthermore, epigenetic drug discovery has been receiving a lot more attention lately. It is clear that epigenetics research is essential in the life sciences field. Epigenetics research is positioned between work on stable genome sequences and work on variable mRNA expression, and it represents a new life science paradigm.
1.2 Determination of 5′-methylcytosine
The methylation of the 5′ carbon of cytosine in DNA (5′-methylcytosine) is an epigenetic modification that regulates gene expression and plays crucial roles in embryonic development.10 5′-Methylcytosine at CpG islands has received particular attention as mentioned above because it is thought to be involved in controlling genetic expression, including that in cancer,9 genomic imprinting,11 cellular differentiation and Alzheimer's disease.12 5′-Methylcytosine is now recognized as the fifth DNA base containing heritable information. Therefore, highly sensitive, accurate and quantitative information concerning cytosine methylation in DNA would be valuable with respect to genetic disease diagnosis.Now, two major 5′-methylcytosine detection methods have been developed; bisulfite treatment followed by PCR and sequencing, and DNA restriction digests. A bisulfite-based determination method is widely used to distinguish between cytosine and 5′-methylcytosine.13–15 Treatment with bisulfite converts cytosine to uracil, while 5′-methylcytosine remains unaffected. Therefore, information about 5′-methylcytosine in DNA can be obtained at the single base level by determining the differences between the sequences of bisulfite-treated and untreated samples. For example, bisulfite-sequencing,16 combined bisulfite restriction analysis (COBRA),17 methylation-specific PCR15 and pyrosequencing18 provide the methylation status of a specific sequence with a single CpG level.
Methylation-sensitive restriction enzyme-based methods have also been used for the site-specific detection of DNA methylation.17,19 When nucleotides in the DNA recognition sequence are subjected to methylation, certain kinds of restriction enzymes are no longer able to cleave the DNA sequence. By combining the cleavage provided by a methylation-sensitive restriction enzyme and genetic engineering techniques such as real-time PCR, it is possible to perform a cytosine methylation analysis of the target DNA sequence.
1.3 Determination of DNA methyltransferase activity
DNA methylation is carried out by the catalysis of DNA methyltransferase. DNA methyltransferase can be divided into three different groups on the basis of the chemical reactions. These DNA methyltransferase groups generate N6-methyladenine, N4-methyladenine, and C5-methylcytosine (5′-methylcytosine). Riggs and Holliday first proposed the idea that heritable DNA methylation provides a mechanism for the developmental regulation of gene expression.20,21 They revealed the existence of a maintenance methyltransferase that does not add a methyl group to unmethylated bases; however, it promptly methylates hemi-methylated base pairs in a DNA duplex. Some research groups subsequently demonstrated the existence of such a maintenance methyltransferase activity by performing experiments showing the clonal inheritance of methylation patterns in mammalian cells.22,23 The first cloned mammalian DNA methyltransferase was DNA methyltranseferase-1 (Dnmt1).24,25 Purified Dnmt1 protein was confirmed to methylate a DNA duplex that contains hemi-methylated CpG sites more efficiently than unmethylated DNA in vitro.26 In the process of DNA methylation, a methyl group is transferred from a donor molecule to the target base in the unmethylated site. S-Adenosyl-L-methionine (SAM) is a well-known donor molecule that is used for various DNA methylation assays. Furthermore, new methyltransferases, namely the Dnmt2 and Dnmt3 families, were discovered and they exhibit de novo DNA methyltransferase activity. Specifically, the murine Dnmt3 family consists of two genes, Dnmt3a and Dnmt3b, which are highly expressed in undifferentiated ES cells but downregulated after differentiation and expressed at low levels in adult somatic tissues.25,27The traditional method for detecting DNA methyltransferase activity is radioactive labeling with [methyl-3H]-SAM or the separation of methylated fragments using high-performance liquid chromatography and gel electrophoresis.28 However, most of these methods have unavoidable disadvantages related to measurement time, complicated procedures and exclusive-use facilities for the radioactive materials. To avoid these disadvantages, alternative measurement techniques have recently been proposed, for example, fluorescence,29 colorimetry30 and electrochemical31 methods. Furthermore, microfluidic technology has been used to reduce the measurement time and sample volume as we will discuss later.
1.4 Microfluidics for epigenetics
Many researchers have reported integrated analysis systems called lab-on-a-chip or micro total analysis systems (micro-TAS) that are small, light, and capable of integrating all sample-handling steps in microfluidic channels on a chip. These techniques have made it possible to undertake various biochemical and clinical measurements simply and rapidly. These microfluidic devices for biochemical analysis have been reviewed by many researchers in relation to such applications as drug discovery,32,33 drug delivery,34–36 microbiology,37 immunosensors,38,39 PCR,40 single-cell analysis,41–43 point-of-care testing,44–47 cell separation48,49 proteomics,50 nucleic acids,51–53 and diagnostics.54,55 There is good chemistry between microfluidic technology and life science research because microfluidic technology enables us to deal with valuable small-volume samples for analysis with high sensitivity and a high throughput. Recently, microfluidic technology has also been applied to epigenome analysis. A microfluidic platform offers the advantage of making it possible to perform thousands of methylation reactions in nanoliter reaction volumes on a single device within isolated reaction units. A microfluidics-based methylation assay technique has been applied to the high-throughput screening of large-scale chemical/biological libraries for novel DNA methyltransferase activity or cellular proteins involved in DNA methylation regulation.56,572. Microfluidic DNA methylation analysis
2.1 Pretreatment devices (bisulfite conversion, DNA enrichment)
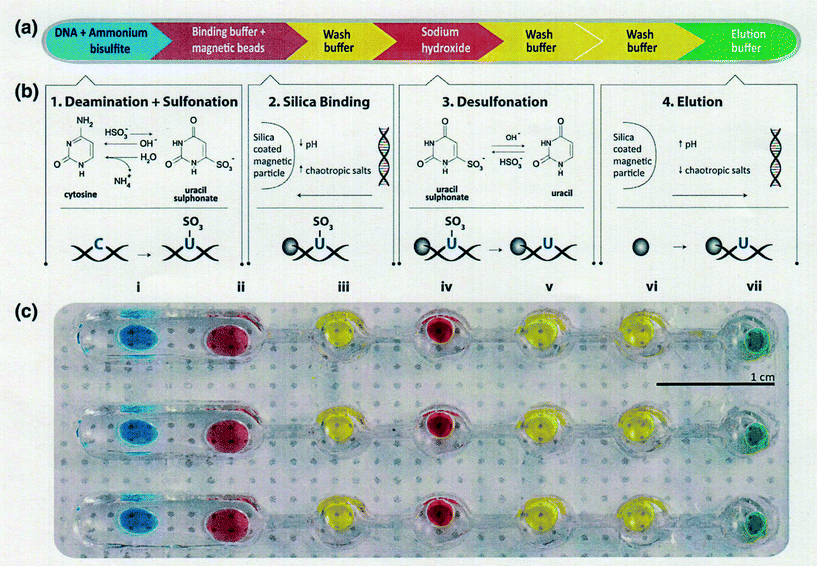
Bisulfite-based detection is the gold standard for DNA methylation analysis because it provides information about the methylation status across the entire PCR-amplified region with a single base level.15,58 However a significant limitation of all bisulfite-based approaches is the duration of the bisulfite treatment, which usually requires an overnight reaction and rigorous control for complete deamination.59,60 Microfluidic technology is known to be useful for reducing the total measurement time of various biomolecules because it allows manipulation with fast response times. Moreover, it can handle small fluid volumes, sense and control flows, and pattern substrates on small length scales.61 Furthermore, an arrayed microfluidic platform is a powerful tool for the high-throughput treatment and measurement of multiple samples. Stark and Wang et al.62 reported a parallelized microfluidic DNA bisulfite conversion module. Their module has three parallelized microchannels made of polydimethylsiloxane (PDMS), and each channel consists of one wide and five circular reservoir chambers, each containing aqueous reagent droplets methylated on beads that are isolated within topographic walls (Fig. 1). They used their module to successfully perform a simultaneous bisulfite conversion for all three channels with high reproducibility.A new pretreatment technique for biofluid samples such as blood or cell suspension is also critical in terms of reducing the total assay time of DNA methylation and not simply for bisulfite conversion. If it requires a lot of time to extract and purify DNA from a biological sample, the total epigenomic assay time will be extremely long. Phenol/chloroform extraction has generally been used for genomic DNA purification. Otherwise, anti-methylcytosine antibody and methyl-CpG binding domain (MBD) proteins are used to purify and preconcentrate genomic DNA in biofluids. There have been some reports of pretreatment with microfluidics before epigenomic measurement with a view to obtaining genomic DNA that can withstand the bisulfite reaction. Two research groups have reported unique solid-phase extraction techniques in a microchannel for epigenomic analysis. Shin and Park et al.63 reported a silicon microfluidic device that employs dimethyl adipimidate-based solid-phase extraction for the purification and extraction of nucleic acids from human body fluid samples for epigenetic analysis (Fig. 2). The silicon microfluidic chip has three components, including a pre-filtration part for cell separation, a micromixer consisting of a two-stage spiral mixer for cell lysis, and a meander-shaped microchannel for dimethyl adipimidate-based solid-phase extraction to maximize the SiO2 surface area. They confirmed that the device can be used to extract genomic DNA with higher purity from human blood and urine samples than other chaotropic methods. Furthermore, they showed that the device effectively captured and purified DNA, including methylated DNA, and improved the DNA amplification for the epigenetic analysis of disease-related DNA biomarkers.
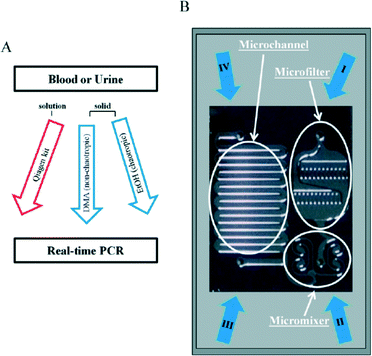 | ||
| Fig. 2 (A) Work-flow for DNA extraction with dimethyl adipimidate-based solid-phase method. (B) Photograph of a microfluidic chip. The microfluidic device consists of a microfilter, micromixer, and microchannel. (I–III) Inlets for addition of samples, lysis buffer, washing and elution buffer. (IV) Outlet for collection of extracted DNA. Reproduced from ref. 63 with permission from The Royal Society of Chemistry. | ||
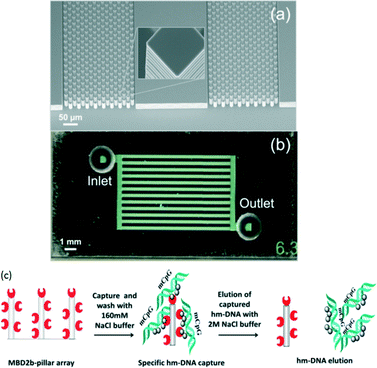
On the other hand, De and Carlen et al.64 reported a rapid microfluidic solid-phase extraction system for the capture and elution of low concentrations of hyper-methylated DNA, based on a methyl-binding domain protein modified surface, in small volumes using a passive microfluidic lab-on-a-chip platform (Fig. 3). They observed each assay step in Fig. 3 using a real-time surface plasmon resonance biosensor and undertook a quantitative characterization using fluorescence spectroscopy. The hyper-methylated DNA capture/elution process was completed in less than 5 min with efficiencies of 71% and 92% using elution volumes of 25 and 100 μL, respectively.
2.2 Bisulfite-based methylcytosine assay
A total system for measuring DNA methylation has been developed and not simply for the bisulfite reaction mentioned above. It is very difficult to integrate all required chemical and biological reactions into one chip, and at present a few pretreatment and detection chips are needed. For example, a two-module system for DNA methylation analysis was developed by Yoon and Shin et al.65 The system is based on bisulfite conversion, which couples a sample pretreatment module for on-chip bisulfite conversion and a label-free, real-time detection module for the rapid analysis of the DNA methylation status using an isothermal DNA amplification/detection technique. The system consists of two modules, one is a sample pretreatment module, and the other is a detection module (Fig. 4). The pretreatment module for on-chip DNA bisulfite conversion consists of a microchamber, a 3D micromixer, and a microchannel. A Peltier heater is also incorporated with a microfluidic device to maintain the temperature for the on-chip bisulfite reaction. The detection module employs an isothermal solid-phase amplification/detection technique after immobilization with either methyl- or non-methyl-specific primer to analyze the DNA methylation status. The methylation status of the RARβ gene in human genomic DNA extracted from MCF-7 cells was analyzed by the system within 80 min (excluding the 16 h needed for preparation). This is fast compared with a conventional methylation-specific PCR technique which takes 24 h. The authors also stated that the system is highly sensitive and can detect as little as 1% methylated DNA in a methylated/unmethylated cell mixture.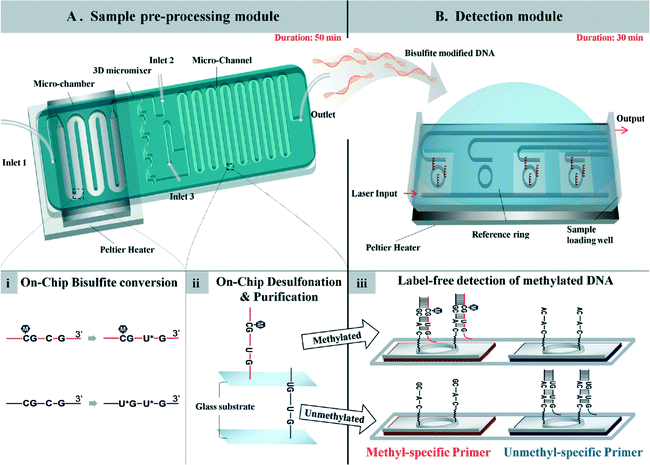 | ||
| Fig. 4 (A) Sample pre-processing module for on-chip DNA bisulfite conversion consisting of a microchamber, 3D micromixer, and microchannel. (i) Human gDNA with bisulfite solution was loaded into the module using inlet 1 and incubated at 70 °C for 20 min in a continuous flow passing through the microchamber region. (ii) Then, the bisulfite-converted DNA was mixed with chaotropic buffer through the 3D micromixer and bound to the surface of the microchannel for the desulfonation and purification steps. (B) Detection module employing the isothermal solid-phase amplification/detection technique after immobilization with either methyl- or non-methyl-specific primer for analysis of the DNA methylation status. The modified DNA was loaded onto the sensing window, and resonant wavelength shifts were observed during the reaction. (iii) Amplification of methylated DNA occurs on the sensor functionalized with the methyl-specific primer, while no amplification occurs on the chip with the non-methyl-specific primer. Reproduced from ref. 65 with permission from The Royal Society of Chemistry. | ||
Combined bisulfite restriction analysis (COBRA), which is a bisulfite-based technique, involves the PCR amplification of bisulfite converted DNA followed by the digestion of a restriction enzyme.17 COBRA is technically simple, and depending on the region being investigated, information on the DNA methylation status of several CpG sites can be explored in a single reaction. For these reasons, various research laboratories employ COBRA to screen large sample sets for DNA methylation.66 Microfluidic electrophoresis was employed by Brena and Plass et al. to confirm enzymatic digestion.66–68 They called their method Bio-COBRA, which is a modified COBRA protocol that incorporates an electrophoresis step in microfluidic chips. They used an Agilent 2100 Bioanalyzer, which provides quantitative results for DNA fragments by electrophoresis in microfluidic chips. A DNA methylation assay of 12 samples was completed within 1 h by using Bio-COBRA.
A unique COBRA-based method utilizing an electrophoretic feature has also been reported for analyzing electrophoretic separation differences in an ssDNA conformation with a self-complementary strand. Chen and Chang et al.69 reported the combination of COBRA and electrophoresis with laser-induced fluorescence for determining the heterogeneity of DNA methylation. Chang's group further reported70 a screening method for DNA methylation based on single-strand conformation polymorphisms and electrophoresis with laser-induced fluorescence. PCR products that were amplified from bisulfite-treated genomic DNA were denatured, followed by immediate chilling in ice water to form ssDNA. The ssDNA was separated with poly(ethylene oxide) in the presence of an electroosmotic flow according to the different conformations represented by their methylation states. The method does not require a restriction endonuclease or specific saturating dye; thus it would be suitable for the large-scale screening of DNA methylation.
The analysis of bisulfite-based methylation by comparing differences between the target sequences of bisulfite-converted and unconverted samples is one of the most frequently used methods in conventional epigenomic research. An important point is to find a simple way of distinguishing the sequence differences in microfluidics on a chip because it is very hard to integrate all the functions of a DNA sequencer onto a single chip with the current technology. COBRA-based methods are considered to be suitable for microfluidics. This is because assay results obtained by digestion with a restriction enzyme can be quickly observed by employing electrophoretic separation or a hybridization assay with microfluidic technology. The conventional COBRA technique is unsuitable for making multiple simultaneous measurements of DNA methylation. Microfluidic technology will provide a high-throughput assay by employing parallel processing for multiple measurements. In fact, a DNA methylation assay of 12 samples has been completed within 1 h on a microchip,66–68 which is much faster than the conventional polyacrylamide gel electrophoresis (PAGE) technique.
2.3 Bisulfite-free methylcytosine assay
The main drawbacks of the bisulfite-based determination methods are the degradation of the sample DNA and the treatment time. More than 99% of the original DNA is reportedly destroyed after a 16 h standard bisulfite treatment.71 This is mainly caused by depurination under the required acidic and thermal conditions. Therefore, to avoid misleading results, the quality of the bisulfite-treated DNA must be assessed before a detection assay is undertaken. Moreover, the bisulfite treatment makes the analyte DNA thymine-rich since unmethylated cytosine is converted to thymine, and this complicates the design of specific probes for PCR amplification.72 Therefore, bisulfite-free techniques have been proposed such as chemical or protein modification. These techniques are simple because assay results can be obtained by measuring the interaction between the target DNA and the methylcytosine recognition molecules. This allows us to design the microfluidics very simply. New recently reported methods and devices for DNA methylation analysis are listed in Table 1.| Discrimination molecule | Target | Response range | Detection limit | Biological application | Detection principle | Ref. |
|---|---|---|---|---|---|---|
| 5-mC, 5′-methylcytosine; 5-hmC, 5′-hydroxymethylcytosine; 5-fC, 5′-formylcytosine; 5-caC, 5-carboxylcytosine; SRP, surface plasmon resonance; MBD, methyl-CpG binding domain; m6A, N6-methyladenosine. | ||||||
| Antibody | 5-mC | 0.5–10 nM | 0.5 nM | — | Electrochemical | 73 |
| 5-mC | 1 × 10−14–5 × 10−9 M | 2 × 10−15 M | Spiked test in serum | Electrochemical | 74 | |
| m6A | 0.01–10 nM | 2.57 pM | Rice seeding | Electrochemical | 75 | |
| 5-mC | 50–3200 fM | 50 fM | Spiked test in serum | SPR | 76 | |
| 5-mC | 0.1–10 pM | 6 amol | Human cancer cell | SPR | 77 | |
| 5-mC | 0.5–3 nM | 0.5 nM | λDNA | Absorbance | 78 | |
| 5-mC, 5-hmC | 3 × 10−13–5 × 10−11 M | 4.2 × 10−13 M | — | Optical microcavity | 79 | |
| Chromatographic or electrophoretic separation | 5-mC, m6A | — | Single cell | Circulating tumor cells | MS | 80 |
| m6A | 0.00005–0.002% in DNA | 0.42 fmol | Human cell lines and plants | MS | 81 | |
| 5-mC, 5-hmC, 5-fC, and 5-caC | — | 0.10, 0.06, 0.11, and 0.23 fmol, respectively | Human cancer cell | MS | 82 | |
| Various RNA modifications | 0.21–4.0 fmol | 63 amol–1.2 fmol | Human ES cells | MS | 83 | |
| 5-mC, 5-hmC | 2–64 nM | 50 pM, 100 pM | Mouse stem cell | MS | 84 | |
| 5-mC | 1–50 μM | 0.02 pmol | Blood | Fluorescence | 85 | |
| β-Glucosyltransferase | 5-hmC | 0–0.1087% in DNA | 0.0012% (0.489 pg) | Mouse tissues and cancer cell lines | Electrochemical | 86 |
| 5-hmC | 0.01–50 nM | 1.43 pM | — | Electrochemical | 87 | |
| MBD | 5-mC | — | 200 pg (input DNA) | Cancer cell lines | SERS | 88 |
| Oxidation potential | 5-mC | 0.6–400 μM | 0.23 μM | — | Electrochemical | 89 |
| Silver nanocluster | 5-mC | 2.0 × 10−9–6.3 × 10−7 M | 9.4 × 10−10 M | Spiked test in serum | Fluorescence | 90 |
Two proteins have mainly been used to label the methylated regions of a genome; one is an anti-methylcytosine antibody and the other is a methyl-CpG binding domain (MBD) protein.59,91 The anti-methylcytosine antibody recognizes single-stranded molecules containing one or more methylated CpG sites. In contrast, the MBD protein recognizes double-stranded methylated CpG sites in DNA fragments. MBD has a unique characteristic whereby different methylation densities can be analyzed depending on the salt fractionation employed;92 lower salt fractions contain hypomethylated DNA fragments, while higher salt fractions contain hypermethylated DNA fragments.93
Small artificial molecules such as osmium complexes71,94 and vanadium complexes95 were also designed for labeling methylcytosine. Unfortunately, thymine in DNA as well as methylcytosine is labeled via OsO4. Therefore, it is difficult to distinguish methylcytosine from thymine in a methylcytosine determination of natural DNA sequences. Mixtures of V2O5 or NaIO4 and LiBr were used in an anaerobic condition to differentiate methylcytosine from both cytosine and thymine followed by a hot piperidine treatment and electrophoretic analysis.95 A sequence-selective cleavage assay technique with a metal complex at a DNA bulge has been reported;96 however, this approach requires a hot piperidine treatment and electrophoresis. The main drawbacks of the chemical modification methods as regards methylcytosine assay are the relatively long detection time and the need for a high concentration (around μM) DNA sample, and in many cases this is insufficient to detect DNA methylation in genomic DNA without PCR amplification.
Recently, several bisulfite-free methylcytosine assays were further integrated with microfluidics. Heimer and Sikes et al.97 reported a microfluidic device for detecting methylated DNA fragments from the MGMT gene promoter. Target oligonucleotides from the test sample hybridize directly to capture probes printed in 300 μm diameter spots on a chip without the bisulfite conversion. They detected methylated DNA duplexes using an MBD protein by interaction between the MBD protein and the methylated DNA using either fluorescence or photo-polymerization-based signal amplification (Fig. 5). They also fabricated a reusable PDMS-based microfluidic device so that they could use a recirculating mixing method to improve DNA hybridization efficiency and provide an assay format suitable for automation. They stated that signals in the microfluidic device were enhanced by about one-third compared with those obtained with static DNA hybridization.
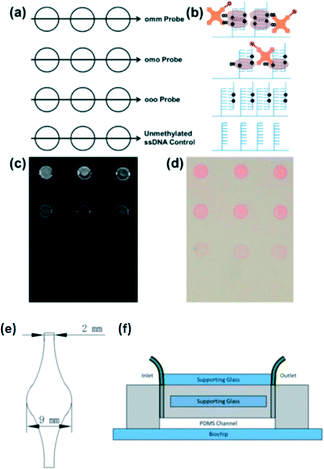 | ||
| Fig. 5 (a) Biochips were spotted with capture probe ssDNA having two, one or no methylated CpG in order to epigenotype the target DNA. Unmethylated ssDNA served as the negative control. (b) Schematic representation of the area within each group of spots following hybridization with 100 nM doubly methylated target ssDNA and detection. Fluorescence (c) and colorimetric (d) readout of MBD binding to methylated DNA. (e) Diagram of the microfluidic channel etched in PDMS. (f) Supporting glass was added to the unetched side of the microfluidic device, and it was clamped to the biochip. Inlet and outlet tubing was connected to each end of the channel, fed into a microcentrifuge tube reservoir, and passed through a peristaltic pump for recirculation. Reproduced from ref. 97 with permission from The Royal Society of Chemistry. | ||
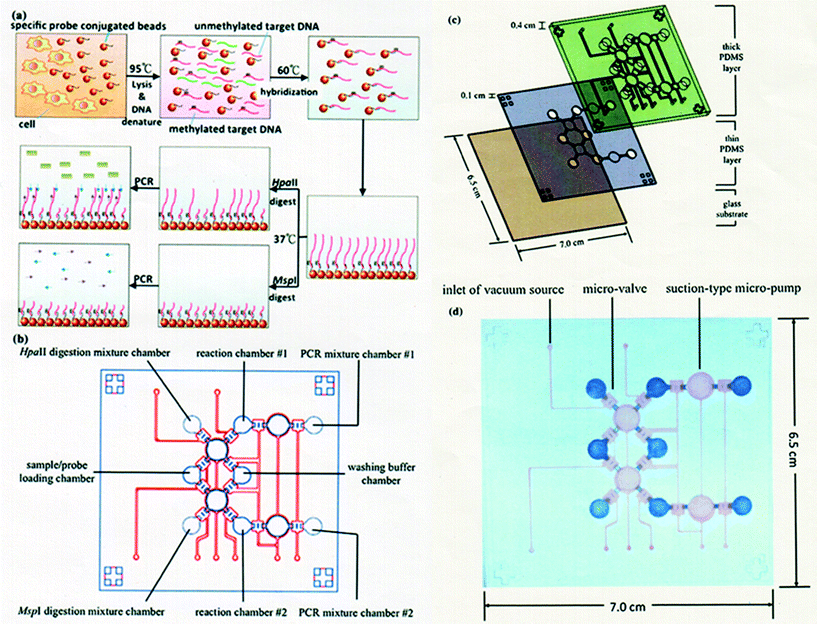
A microfluidic system integrating the entire experimental process for a DNA methylation assay was reported by Wang and Lee et al.98 The system includes target DNA isolation, HpaII/MspI endonuclease digestion, and nucleic acid amplification (Fig. 6). Instead of employing a bisulfite reaction, they attempt to shorten the entire process using endonuclease digestion. In their system, all the genomic DNA from the cultured cell lines was directly extracted and purified with a specific nucleotide probe conjugated on the surface of magnetic beads. Methylated DNAs of tumor suppressor genes, HAAO, HOXA9 and SFRP5, were chosen as candidates for the detection of ovarian cancer cells. The detection limit of their microfluidic system was found to be 102 cells per reaction. Three hours were required to complete the entire process from sample loading to analysis, which is much faster than the conventional protocols. They concluded that different sources of biosamples, for example, other cell lines, ascites and serum, would be applicable to the detection of DNA methylation, indicating that the developed microfluidic system will be useful for clinical use.
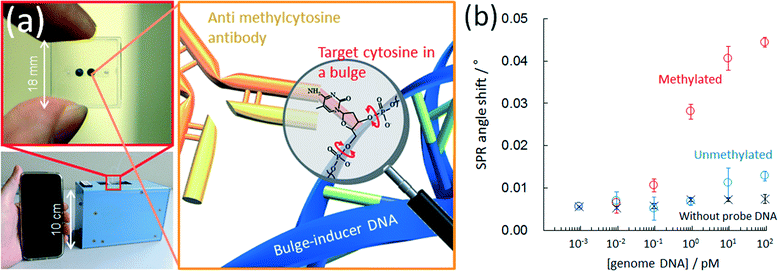
An anti-methylcytosine antibody has been used for epigenetics analysis; however, its use has been limited to the immunoprecipitation or pre-concentration of methyl-CpG regions in a DNA sample.99–101 Recently, immunochemical methods for detecting methylcytosine with an anti-methylcytosine antibody have been reported for analyzing the methylation level. The reported methods employ a microtiter plate,102 capillary electrophoresis,103 magnetic particles,104 microspheres,105 a nitrocellulose membrane106 and a DNA microarray.107 However, previously reported immunochemical methods for detecting methylcytosine have no sequence specificity and so only the total amount of methylcytosine in the analyte DNA was quantified. One of the authors found that an anti-methylcytosine antibody can recognize mismatched methylcytosine especially in a bulge region but cannot recognize methylcytosine in a pair.78,108 This is because methylcytosine at a single-base bulge is predominantly in a looped-out state due to the π–π stacking formation between the flanking bases of bulged methylcytosine. In contrast, methylcytosine paired with guanine is in a stacked state in a duplex. This makes it possible to perform a site-specific methylation analysis of genome DNA on a conventional microtiter plate with a biotinylated probe DNA that has a sequence to form a single base bulge at the target cytosine. We further reported77 a sequence-specific microfluidic chip for DNA methylation assessment by surface plasmon resonance detection (Fig. 7a). This was achieved by utilizing an affinity measurement involving the target, (methyl-) cytosine, in a single-base bulge region and an anti-methylcytosine antibody in a microchannel, following hybridization with a biotinylated bulge-inducing DNA probe. The probe alters the target cytosine in a looped-out state because of the π–π stacking between flanking bases of the target. The probe design is simple and consists of the elimination of guanine paired with the target cytosine from a fragmented full-match sequence. The single methylation status in 6 attomoles (48 femtograms) of DNA was obtained within 45 min, which is the fastest DNA methylation assessment yet reported (Fig. 7b). The discrimination of the methylation status of single cytosine in genomic HCT116 human colon cancer cells was also carried out with the microfluidic device.
Some quick bisulfite-conversion kits are commercially available; however, the degradation of the input DNA remains an unavoidable problem. Ultimately, bisulfite-free assay is considered to be ideal; therefore the development of a new bisulfite-free methylation assay technique is being actively studied as mentioned in this section. Affinity measurements with an antibody to methylated bases or MDB proteins are a valid approach to a bisulfite-free assay; however, these affinity measurements cannot be combined with PCR amplification, i.e. in many cases the sensitivity is insufficient to detect DNA methylation in genomic DNA. Microfluidic technology is known to be useful for improving sensitivity with small samples and should prove to be a powerful tool for DNA methylation analysis. In fact, we showed that the methylation status of less than a pM of genomic DNA could be evaluated within 1 h by utilizing microfluidic technology.
2.4 DNA methyltransferase activity
The determination of DNA methyltransferase activity has been attracting attention. One reason for this is that DNA methyltransferase and its inhibitors are reported to be a novel family of pharmacological targets for the treatment of tumors.141 Therefore, there is a strong need for sensitive, selective and high-throughput methods for performing DNA methyltransferase activity assays.142 However, traditional methods for DNA methyltransferase activity assay rely on radioisotope materials.109 Therefore, radioisotope-free approaches have been developed, including electrochemical,109–119 electrochemiluminescence,120–122 photo-electrochemical,123 fluorometric,124–135 surface-enhanced Raman scattering (SERS),136–138 and circular dichroism (CD) spectroscopy,139 and surface plasmon resonance140 based DNA methyltransferase measurement techniques have recently been proposed as summarized in Table 2 to realize simple, quick and easy monitoring.| Detection principle | Target | Response range (U mL−1) | Detection limit (U mL−1) | Biological application | Signal generator | Ref. |
|---|---|---|---|---|---|---|
| Dam, deoxyadenosine methylase; M.SssI, CpG methyltransferase. | ||||||
| Electrochemical | Dam | 0.1–20 | 0.04 | — | 1-Naphthyl phosphate | 109 |
| Dam | 0.075–30 | 0.02 | Human serum | Methylene blue | 110 | |
| Dam | 0.27–60 | 0.27 | Human serum | Methylene blue | 111 | |
| Dam | 0.05–40 | 0.031 | — | Ferricyanide | 112 | |
| Dam | 1–40 | 0.96 | — | Hydroquinone | 113 | |
| M.SssI | 0.5–0.6 | 0.12 | Human serum | Aniline | 114 | |
| Dam | 1–60 | 0.31 | — | Hydroquinone | 115 | |
| M.SssI | 0.28–50 | 0.28 | — | Methylene blue | 116 | |
| Dam | 0.04–4 | 0.004 | — | Methylene blue | 117 | |
| M.SssI | 0.05–200 | 0.025 | — | Ascorbic acid | 118 | |
| Dam | 0.12–20 | 0.04 | — | Methylene blue | 119 | |
| Electrochemiluminescence | M.SssI | 1–120 | 0.05 | Cancer human serum | CdS quantum dot | 120 |
| Dam | 0.1–100 | 0.03 | — | Tris (2,2′-bipyridine) ruthenium | 121 | |
| Dam | 0.1–50 | 0.03 | — | Luminol | 122 | |
| Photo-electrochemical | M.SssI | 0.01–150 | 0.0042 | — | CdSe quantum dot | 123 |
| Fluorometric | Dam | 0.1–8 | 0.1 | — | Thioflavin | 124 |
| M.SssI | 0.02–40 | 0.0082 | Human serum | Molecular beacon/FAM | 125 | |
| M.SssI | 0.01–50 | 0.0024 | HeLa cells | Sybr green I | 126 | |
| Dam | 1.2–10 | 0.57 | LB medium | Molecular beacon/FAM | 127 | |
| Dam | 0.0005–50 | 2 × 10−4 | E. coli cells | Molecular beacon | 128 | |
| Dam | 0–50 | 0.0025 | E. coli cells | FAM | 129 | |
| Dam | 0.05–10 | 0.015 | LB medium | FAM | 130 | |
| Dam | 0.0005–0.01 | 1.5 × 10−4 | E. coli cells | Molecular beacon/FAM | 131 | |
| Dam | 0–15 | 0.1 | — | FAM | 132 | |
| Dam | 1–100 | 1 | — | Fluorescent silver nanocluster | 133 | |
| Dam | 0.2–20 | 0.14 | — | FAM | 134 | |
| Dam | 0.0002–20 | 8.6 × 10−5 | Human serum | Zinc protoporphyrin | 135 | |
| SERS | Dam | 0.001–10 | 2.57 × 10−4 | Human serum | Au nanoparticles | 136 |
| M.SssI | 0.1–10 | 0.067 | — | Silver nanoparticles | 137 | |
| Dam | 0.1–10 | 0.02 | — | Mesoporous silica nanoparticels | 138 | |
| CD spectroscopy | M.SssI | 0.5–150 | 0.27 | Human serum | Gold nanoparticles dimer | 139 |
| Surface plasmon resonance | Dam | 0.5–120 | 0.2 | HeLa cells | Gold nanorod | 140 |
By combining these DNA methyltransferase biosensors with microfluidic technology, they have the potential to reduce both the assay time and the required sample volume. Microfluidic DNA methyltransferase activity measurement is a challenge for the future. However, Ronen and Gerber et al.56 have published a preliminary report on a microfluidic-based fluorometric assay technique for studying DNA methylation in vitro. The microfluidic device consists of a 64 × 16 reaction unit array in a flow channel, which is accessed through several input holes and drained into a single output (Fig. 8). The microfluidic device was compartmentalized with micromechanical valves into 16 separate reaction conditions on a single device within isolated columns. For the methylation reaction, a mixture solution containing DNA methyltransferase and SAM was injected into the microfluidic device, and the immobilized DNA substrate was incubated in the mixture solution. After washing, the endonuclease containing reaction buffer was injected and incubated. Finally, the cleaved DNA fragments were washed away and the fluorescence intensity of Cy5-modified DNA was measured. The same platform was then used to demonstrate a two-step approach for the high-throughput in vitro identification and characterization of small-molecule inhibitors of DNA methylation. The microfluidic device enabled the authors to perform thousands of simultaneous DNA methylation reactions on a one-chip device.
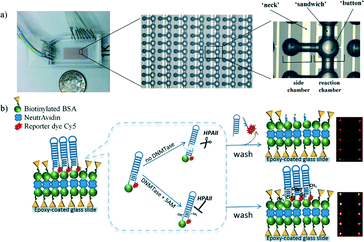 | ||
| Fig. 8 a) Photograph and schematic representation of a microfluidic device consisting of flow and control layers. b) Schematic representation of microfluidic-based methylation assay using biotinylated hairpin-shaped DNA probe substrate. The DNA probe is immobilized to the surface through biotin–avidin interactions. The recognition site becomes resistant to HpaII activity only when there is a methylation event. Otherwise, the unprotected DNA probe is digested and the unbound Cy5-containing DNA piece is washed out leading to an overall reduction in fluorescence signal. Reproduced from ref. 56 with permission from The Royal Society of Chemistry. | ||
Various enzymatic activities have been measured with microfluidic technology to obtain quick and highly sensitive results by utilizing a large surface-to-volume ratio. Microfluidic technology is also considered a promising approach for measuring DNA methyltransferase activity. Moreover, in epigenetic research, huge numbers of samples must be evaluated during, for example, epigenetic drug screening. This is a challenge; however, it will be realized in the near future because the simultaneous monitoring of 64 × 16 reactions has already been achieved on a chip.56
2.5 Single cell epigenetics
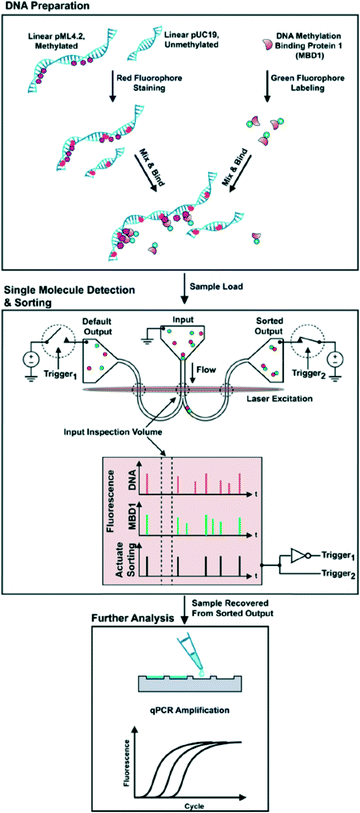
The goal of single cell epigenetics is to analyze information from a single cell to obtain a holistic understanding of the cell population. This reductionist approach allows researchers to unravel the way in which molecular events within a single cell link to the behaviour of tissues, organs, and eventually entire organisms.143 The small dimensions of microfluidic systems offer a great advantage as regards single cell epigenomics because minimal dilution is required, resulting in a highly sensitive epigenomic analysis. Furthermore, microfluidic systems offer several potential advantages for the study of single cells including facile automation, parallelization and reagent reduction.144As mentioned above, the damage to DNA that occurs during bisulfite conversion is serious. Therefore, direct DNA methylation analysis in a single cell with a bisulfite-based assay requires great skill. ChIP-on-chip, which is a technology that combines chromatin immunoprecipitation with a DNA microarray, is currently used for single cell epigenetics. The results of epigenetic modifications of chromatin by traditional methods usually include blended responses from many cells in a tissue; however, such bulk measures miss the spatial and temporal differences that occur from cell to cell and cannot uncover novel or rare populations of cells.145 As regards combining micro- and nanofluidic technology, Cipriany and Craighead et al.146 reported a method using nanofluidics and multicolor fluorescence microscopy to detect DNA and histones in individual chromatin fragments at about 10 Mbp min−1. They demonstrated its utility for epigenetic analysis by identifying DNA methylation on individual molecules. They further reported147 a nanofluidic device that combines real-time detection and the automated sorting of individual molecules based on their epigenetic state. Fluorescently labeled antibodies or proteins were bound to epigenetic modifications located on histone proteins or DNA, and then the mixture solutions were injected into the nanofluidic device (Fig. 9) to identify these molecules and their corresponding modifications by their fluorescence color signature. Each molecule with a color signature that matches the criteria for collection is then sorted during a brief actuation, or pulse, that redirects the flow to the sorted output. Sorted molecules are collected downstream, and then the collected samples are analyzed by quantitative PCR. They stated that up to 98% accuracy was achieved in sorting molecules from femtogram quantities of genetic material.
3. Conclusions and future prospects
In this review article, we have presented and discussed epigenomic research undertaken with microfluidic devices. Epigenetics is a biological application that can take advantage of the features included in microfluidic technology, and it continues to be a fascinating research area. Many examples of application to bio-sensing devices have been reported for measuring DNA methyltransferase activity. This is because DNA methyltransferase activity can be detected by using relatively simple chemical and biological reactions with a methylation-sensitive restriction enzyme. The cleavage with the methylation sensitive restriction enzyme was monitored with electrochemical and optical measurements and involved a relatively minor change in existing biosensors. By employing nanomaterials such as carbon nanotubes, graphene, quantum dots and metal nanoparticles, sensitivity was improved and these results have been reported. Quick and highly sensitive measurements with small sample volumes can be expected by integrating the methods and materials with microfluidics. In the future, microfluidic devices will be reported that employ the above materials, and their device performance is promising. Because most of them are being developed to obtain high sensitivity with a similar approach to conventional affinity biosensors, there have been some problems as regards non-specific adsorption and selectivity when measuring real samples. Unless these problems are overcome, industrial application will be difficult; therefore it might also be useful to integrate pretreatment systems utilizing microfluidic technology.When measuring base methylation in DNA, the detection principle is somewhat more complicated than that used for methyltransferase activity; therefore coming up with a bio-sensing device for DNA methylation analysis remains a challenging proposition. There have been several reports on PCR-free measurement of DNA methylation. However, the sensitivity and selectivity are insufficient for genomic DNA measurement; therefore results with genomic DNA are limited. It is difficult to integrate all the complicated chemical and biological reactions into a single chip. Therefore, each elemental technology, for example, genomic extraction, pretreatment, and detection, is currently being developed. Epigenomic analysis in a single cell, which is difficult the conventional analytical technique, may be realized by integrating these devices. This work does not relate solely to biomolecular researchers; it is certain that the need to detect DNA methylation is high in the fields of diagnosis and drug development. The microfluidic approach will provide a promising way to realize a DNA methylation sensor which can obtain epigenomic information quickly and with a high throughput.
Acknowledgements
Our study was financially supported by JSPS KAKENHI, Grant No. 26410168. We thank Mr. D. Meacock for revising the language of the manuscript.References
- R. Jaenisch and A. Bird, Nat. Genet., 2003, 33, 245–254 CrossRef CAS PubMed.
- P. A. Jones and S. B. Baylin, Nat. Rev. Genet., 2002, 3, 415–428 CrossRef CAS PubMed.
- R. R. Kanherkar, N. Bhatia-Dey and A. B. Csoka, Front. Cell Dev. Biol., 2014, 2, 49 Search PubMed.
- V. K. Cortessis, D. C. Thomas, A. J. Levine, C. V. Breton, T. M. Mack, K. D. Siegmund, R. W. Haile and P. W. Laird, Hum. Genet., 2012, 131, 1565–1589 CrossRef CAS PubMed.
- L. Hou, X. Zhang, D. Wang and A. Baccarelli, Int. J. Epidemiol., 2012, 41, 79–105 CrossRef PubMed.
- R. D. Hotchkiss, J. Biol. Chem., 1948, 175, 315–332 CAS.
- M. Esteller, Annu. Rev. Pharmacol. Toxicol., 2005, 45, 629–656 CrossRef CAS PubMed.
- K. D. Robertson and A. P. Wolffe, Nat. Rev. Genet., 2000, 1, 11–19 CrossRef CAS PubMed.
- P. A. Jones, Cancer Res., 1996, 56, 2463–2467 CAS.
- J. A. Yoder, C. P. Walsh and T. H. Bestor, Trends Genet., 1997, 13, 335–340 CrossRef CAS PubMed.
- M. Monk, Dev. Genet., 1995, 17, 188–197 CrossRef CAS PubMed.
- S. Ledoux, J. Nalbantoglu and N. R. Cashman, Mol. Brain Res., 1994, 24, 140–144 CrossRef CAS PubMed.
- S. J. Clark, J. Harrison, C. L. Paul and M. Frommer, Nucleic Acids Res., 1994, 22, 2990–2997 CrossRef CAS PubMed.
- S. J. Docherty, O. S. P. Davis, C. M. A. Haworth, R. Plomin and J. Mill, Methods, 2010, 52, 255–258 CrossRef CAS PubMed.
- J. G. Herman, J. R. Graff, S. Myohanen, B. D. Nelkin and S. B. Baylin, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 9821–9826 CrossRef CAS.
- C. Bock, S. Reither, T. Mikeska, M. Paulsen, J. Walter and T. Lengauer, Bioinformatics, 2005, 21, 4067–4068 CrossRef CAS PubMed.
- Z. G. Xiong and P. W. Laird, Nucleic Acids Res., 1997, 25, 2532–2534 CrossRef CAS PubMed.
- A. S. Yang, M. R. H. Estecio, K. Doshi, Y. Kondo, E. H. Tajara and J. P. J. Issa, Nucleic Acids Res., 2004, 32, e38 CrossRef PubMed.
- L. J. Rush and C. Plass, Anal. Biochem., 2002, 307, 191–201 CrossRef CAS PubMed.
- A. D. Riggs, Cytogenet. Genome Res., 2002, 99, 17–24 CrossRef CAS PubMed.
- R. Holliday and J. E. Pugh, Science, 1975, 187, 226–232 CAS.
- M. Wigler, D. Levy and M. Perucho, Cell, 1981, 24, 33–40 CrossRef CAS PubMed.
- R. Stein, Y. Gruenbaum, Y. Pollack, A. Razin and H. Cedar, Proc. Natl. Acad. Sci. U. S. A., 1982, 79, 61–65 CrossRef CAS.
- T. Bestor, A. Laudano, R. Mattaliano and V. Ingram, J. Mol. Biol., 1988, 203, 971–983 CrossRef CAS PubMed.
- M. Okano, D. W. Bell, D. A. Haber and E. Li, Cell, 1999, 99, 247–257 CrossRef CAS PubMed.
- T. H. Bestor, EMBO J., 1992, 11, 2611–2617 CAS.
- M. Okano, S. P. Xie and E. Li, Nat. Genet., 1998, 19, 219–220 CrossRef CAS PubMed.
- Q. Q. Lai, M. D. Liu, C. C. Gu, H. G. Nie, X. J. Xu, Z. H. Li, Z. Yang and S. M. Huang, Analyst, 2016, 141, 1383–1389 RSC.
- Y. Zhao, F. Chen, Y. Wu, Y. Dong and C. Fan, Biosens. Bioelectron., 2013, 42, 56–61 CrossRef CAS PubMed.
- T. Liu, J. Zhao, D. Zhang and G. Li, Anal. Chem., 2010, 82, 229–233 CrossRef CAS PubMed.
- N. B. Muren and J. K. Barton, J. Am. Chem. Soc., 2013, 135, 16632–16640 CrossRef CAS PubMed.
- P. S. Dittrich and A. Manz, Nat. Rev. Drug Discovery, 2006, 5, 210–218 CrossRef CAS PubMed.
- B. H. Weigl, R. L. Bardell and C. R. Cabrera, Adv. Drug Delivery Rev., 2003, 55, 349–377 CrossRef CAS PubMed.
- J. K. Oh, R. Drumright, D. J. Siegwart and K. Matyjaszewski, Prog. Polym. Sci., 2008, 33, 448–477 CrossRef CAS.
- W. M. Saltzman and W. L. Olbricht, Nat. Rev. Drug Discovery, 2002, 1, 177–186 CrossRef CAS PubMed.
- A. Nisar, N. AftuIpurkar, B. Mahaisavariya and A. Tuantranont, Sens. Actuators, B, 2008, 130, 917–942 CrossRef CAS.
- D. B. Weibel, W. R. DiLuzio and G. M. Whitesides, Nat. Rev. Microbiol., 2007, 5, 209–218 CrossRef CAS PubMed.
- A. Bange, H. B. Halsall and W. R. Heineman, Biosens. Bioelectron., 2005, 20, 2488–2503 CrossRef CAS PubMed.
- M.-I. Mohammed and M. P. Y. Desmulliez, Lab Chip, 2011, 11, 569–595 RSC.
- C. S. Zhang, J. L. Xu, W. L. Ma and W. L. Zheng, Biotechnol. Adv., 2006, 24, 243–284 CrossRef CAS PubMed.
- C. E. Sims and N. L. Allbritton, Lab Chip, 2007, 7, 423–440 RSC.
- D. Wang and S. Bodovitz, Trends Biotechnol., 2010, 28, 281–290 CrossRef CAS PubMed.
- S. Lindstrom and H. Andersson-Svahn, Lab Chip, 2010, 10, 3363–3372 RSC.
- A. J. Tudos, G. A. J. Besselink and R. B. M. Schasfoort, Lab Chip, 2001, 1, 83–95 RSC.
- F. B. Myers and L. P. Lee, Lab Chip, 2008, 8, 2015–2031 RSC.
- A. M. Foudeh, T. F. Didar, T. Veres and M. Tabrizian, Lab Chip, 2012, 12, 3249–3266 RSC.
- T. Tian, J. Li, Y. Song, L. Zhou, Z. Zhu and C. J. Yang, Lab Chip, 2016, 16, 1139–1151 RSC.
- D. R. Gossett, W. M. Weaver, A. J. Mach, S. C. Hur, H. T. K. Tse, W. Lee, H. Amini and D. Di Carlo, Anal. Bioanal. Chem., 2010, 397, 3249–3267 CrossRef CAS PubMed.
- C. W. Shields, C. D. Reyes and G. P. Lopez, Lab Chip, 2015, 15, 1230–1249 RSC.
- N. Lion, T. C. Rohner, L. Dayon, I. L. Arnaud, E. Damoc, N. Youhnovski, Z. Y. Wu, C. Roussel, J. Josserand, H. Jensen, J. S. Rossier, M. Przybylski and H. H. Girault, Electrophoresis, 2003, 24, 3533–3562 CrossRef CAS PubMed.
- P. A. Auroux, Y. Koc, A. deMello, A. Manz and P. J. R. Day, Lab Chip, 2004, 4, 534–546 RSC.
- L. Chen, A. Manz and P. J. R. Day, Lab Chip, 2007, 7, 1413–1423 RSC.
- C. W. Price, D. C. Leslie and J. P. Landers, Lab Chip, 2009, 9, 2484–2494 RSC.
- B. Weigl, G. Domingo, P. LaBarre and J. Gerlach, Lab Chip, 2008, 8, 1999–2014 RSC.
- P. Y. Liu, L. K. Chin, W. Ser, H. F. Chen, C. M. Hsieh, C. H. Lee, K. B. Sung, T. C. Ayi, P. H. Yap, B. Liedberg, K. Wang, T. Bourouina and Y. Leprince-Wang, Lab Chip, 2016, 16, 634–644 RSC.
- M. Ronen, D. Avrahami and D. Gerber, Lab Chip, 2014, 14, 2354–2362 RSC.
- D. Gerber, S. J. Maerkl and S. R. Quake, Nat. Methods, 2009, 6, 71–74 CrossRef CAS PubMed.
- X. Lu, C.-X. Song, K. Szulwach, Z. Wang, P. Weidenbacher, P. Jin and C. He, J. Am. Chem. Soc., 2013, 135, 9315–9317 CrossRef CAS PubMed.
- Y. Yu, S. Blair, D. Gillespie, R. Jensen, D. Myszka, A. H. Badran, I. Ghosh and A. Chagovetz, Anal. Chem., 2010, 82, 5012–5019 CrossRef CAS PubMed.
- L. S. Kristensen and L. L. Hansen, Clin. Chem., 2009, 55, 1471–1483 CAS.
- H. A. Stone and S. Kim, AIChE J., 2001, 47, 1250–1254 CrossRef CAS.
- A. Stark, D. J. Shin, T. Pisanic, II, K. Hsieh and T.-H. Wang, Biomed. Microdevices, 2016, 18, 5 CrossRef PubMed.
- Y. Shin, A. P. Perera, C. C. Wong and M. K. Park, Lab Chip, 2014, 14, 359–368 RSC.
- A. De, W. Sparreboom, A. van den Berg and E. T. Carlen, Biomicrofluidics, 2014, 8, 054119 CrossRef PubMed.
- J. Yoon, M. K. Park, T. Y. Lee, Y.-J. Yoon and Y. Shin, Lab Chip, 2015, 15, 3530–3539 RSC.
- R. M. Brena, H. Auer, K. Kornacker, B. Hackanson, A. Raval, J. C. Byrd and C. Plass, Nucleic Acids Res., 2006, 34, e17 CrossRef PubMed.
- R. M. Brena, H. Auer, K. Kornacker and C. Plass, Nat. Protoc., 2006, 1, 52–58 CrossRef PubMed.
- R. M. Brena and C. Plass, Methods Mol. Biol., 2009, 507, 257–269 CAS.
- H.-C. Chen, Y.-S. Chang, S.-J. Chen and P.-L. Chang, J. Chromatogr., 2012, 1230, 123–129 CrossRef CAS PubMed.
- M.-H. Yu, Y.-C. Huang and P.-L. Chang, Electrophoresis, 2014, 35, 2378–2385 CrossRef CAS PubMed.
- K. Tanaka, K. Tainaka, T. Umemoto, A. Nomura and A. Okamoto, J. Am. Chem. Soc., 2007, 129, 14511–14517 CrossRef CAS PubMed.
- DNA Methylation - From Genomics to Technology, ed. T. Tatarinova and O. Kerton, Rijeka, 2012 Search PubMed.
- H. Yanagisawa, R. Kurita, T. Yoshida, T. Kamata and O. Niwa, Sens. Actuators, B, 2015, 221, 816–822 CrossRef CAS.
- M. Daneshpour, L. S. Moradi, P. Izadi and K. Omidfar, Biosens. Bioelectron., 2016, 77, 1095–1103 CrossRef CAS PubMed.
- H. Yin, Y. Zhou, Z. Yang, Y. Guo, X. Wang, S. Ai and X. Zhang, Sens. Actuators, B, 2015, 221, 1–6 CrossRef CAS.
- A. H. Nguyen and S. J. Sim, Biosens. Bioelectron., 2015, 67, 443–449 CrossRef CAS PubMed.
- R. Kurita, H. Yanagisawa, K. Yoshioka and O. Niwa, Anal. Chem., 2015, 87, 11581–11586 CrossRef CAS PubMed.
- R. Kurita, H. Yanagisawa, K. Yoshioka and O. Niwa, Biosens. Bioelectron., 2015, 70, 366–371 CrossRef CAS PubMed.
- R. M. Hawk and A. M. Armani, Biosens. Bioelectron., 2015, 65, 198–203 CrossRef CAS PubMed.
- W. Huang, C.-B. Qi, S.-W. Lv, M. Xie, Y.-Q. Feng, W.-H. Huang and B.-F. Yuan, Anal. Chem., 2016, 88, 1378–1384 CrossRef CAS PubMed.
- W. Huang, J. Xiong, Y. Yang, S.-M. Liu, B.-F. Yuan and Y.-Q. Feng, RSC Adv., 2015, 5, 64046–64054 RSC.
- Y. Tang, S.-J. Zheng, C.-B. Qi, Y.-Q. Feng and B.-F. Yuan, Anal. Chem., 2015, 87, 3445–3452 CrossRef CAS PubMed.
- M. Basanta-Sanchez, S. Temple, S. A. Ansari, A. D'Amico and P. F. Agris, Nucleic Acids Res., 2016, 44, e26 CrossRef PubMed.
- F. Yuan, X.-H. Zhang, J. Nie, H.-X. Chen, Y.-L. Zhou and X.-X. Zhang, Chem. Commun., 2016, 52, 2698–2700 RSC.
- M. Giel-Pietraszuk, M. Insinska-Rak, A. Golczak, M. Sikorski, M. Barciszewska and J. Barciszewski, Acta Biochim. Pol., 2015, 62, 281–286 CrossRef CAS PubMed.
- S. Chen, Y. Dou, Z. Zhao, F. Li, J. Su, C. Fan and S. Song, Anal. Chem., 2016, 88, 3476–3480 CrossRef CAS PubMed.
- Y. Zhou, Z. Yang, X. Li, Y. Wang, H. Yin and S. Ai, Electrochim. Acta, 2015, 174, 647–652 CrossRef CAS.
- Y. Wang, E. J. H. Wee and M. Trau, Chem. Commun., 2016, 52, 3560–3563 RSC.
- P. Wang, P. Han, L. Dong and X. Miao, Electrochem. Commun., 2015, 61, 36–39 CrossRef CAS.
- M. Dadmehr, M. Hosseini, S. Hosseinkhani, M. R. Ganjali and R. Sheikhnejad, Biosens. Bioelectron., 2015, 73, 108–113 CrossRef CAS PubMed.
- L. G. Acevedo, A. Sanz and M. A. Jelinek, Epigenomics, 2011, 3, 93–101 CrossRef CAS PubMed.
- D. Serre, B. H. Lee and A. H. Ting, Nucleic Acids Res., 2010, 38, 391–399 CrossRef CAS PubMed.
- S. S. Nair, M. W. Coolen, C. Stirzaker, J. Z. Song, A. L. Statham, D. Strbenac, M. D. Robinson and S. J. Clark, Epigenetics, 2011, 6, 34–44 CrossRef CAS PubMed.
- K. Tanaka, K. Tainaka, T. Kamei and A. Okamoto, J. Am. Chem. Soc., 2007, 129, 5612–5620 CrossRef CAS PubMed.
- S. Bareyt and T. Carell, Angew. Chem., Int. Ed., 2008, 47, 181–184 CrossRef CAS PubMed.
- A. Okamoto, K. Tainaka and T. Kamei, Org. Biomol. Chem., 2006, 4, 1638–1640 CAS.
- B. W. Heimer, T. A. Shatova, J. K. Lee, K. Kaastrup and H. D. Sikes, Analyst, 2014, 139, 3695–3701 RSC.
- C.-H. Wang, H.-C. Lai, T.-M. Liou, K.-F. Hsu, C.-Y. Chou and G.-B. Lee, Microfluid. Nanofluid., 2013, 15, 575–585 CrossRef CAS.
- C. Bock, E. M. Tomazou, A. B. Brinkman, F. Mueller, F. Simmer, H. Gu, N. Jaeger, A. Gnirke, H. G. Stunnenberg and A. Meissner, Nat. Biotechnol., 2010, 28, 1106–U1196 CrossRef CAS PubMed.
- J. Borgel, S. Guibert, Y. Li, H. Chiba, D. Schuebeler, H. Sasaki, T. Forne and M. Weber, Nat. Genet., 2010, 42, 1093–1100 CrossRef CAS PubMed.
- L. Zhang, K. E. Szulwach, G. C. Hon, C.-X. Song, B. Park, M. Yu, X. Lu, Q. Dai, X. Wang, C. R. Street, H. Tan, J.-H. Min, B. Ren, P. Jin and C. He, Nat. Commun., 2013, 4, 1517 CrossRef PubMed.
- B. Chowdhury, I.-H. Cho, N. Hahn and J. Irudayaraj, Anal. Chim. Acta, 2014, 852, 212–217 CrossRef CAS PubMed.
- X. L. Wang, Y. L. Song, M. Y. Song, Z. X. Wang, T. Li and H. L. Wang, Anal. Chem., 2009, 81, 7885–7891 CrossRef CAS PubMed.
- Z. Wang, X. Wang, S. Liu, J. Yin and H. Wang, Anal. Chem., 2010, 82, 9901–9908 CrossRef CAS PubMed.
- C. Ge, Z. Fang, J. Chen, J. Liu, X. Lu and L. Zeng, Analyst, 2012, 137, 2032–2035 RSC.
- D. D. Deobagkar, C. Panikar, S. N. Rajpathak, N. S. Shaiwale and S. Mukherjee, Methods, 2012, 56, 260–267 CrossRef CAS PubMed.
- A. Kelkar and D. Deobagkar, Epigenetics, 2009, 4, 311–316 CrossRef.
- R. Kurita and O. Niwa, Anal. Chem., 2012, 84, 7533–7538 CrossRef CAS PubMed.
- H. Wu, S. Liu, J. Jiang, G. Shen and R. Yu, Chem. Commun., 2012, 48, 6280–6282 RSC.
- L. Hong, J. Wan, X. Zhang and G. Wang, Talanta, 2016, 152, 228–235 CrossRef CAS PubMed.
- P. Liu, D. Wang, Y. Zhou, H. Wang, H. Yin and S. Ai, Biosens. Bioelectron., 2016, 80, 74–78 CrossRef CAS PubMed.
- X. Li, Z. Xie, W. Wang, Y. Zhou, H. Yin, Z. Yang and S. Ai, Anal. Methods, 2016, 8, 2771–2777 RSC.
- P. Liu, M. Liu, H. Yin, Y. Zhou and S. Ai, Sens. Actuator, B, 2015, 220, 101–106 CrossRef CAS.
- L. Zhang, M. Wei, C. Gao, W. Wei, Y. Zhang and S. Liu, Biosens. Bioelectron., 2015, 73, 188–194 CrossRef CAS PubMed.
- Z. Yang, L. Xie, H. Yin, Y. Zhou and S. Ai, Microchim. Acta, 2015, 182, 2607–2613 CrossRef CAS.
- P. Liu, J. Pang, H. Yin and S. Ai, Anal. Chim. Acta, 2015, 879, 34–40 CrossRef CAS PubMed.
- W. Li, X. Liu, T. Hou, H. Li and F. Li, Biosens. Bioelectron., 2015, 70, 304–309 CrossRef CAS PubMed.
- J. Zhou, X. Zhang, E. Xiong, P. Yu and J. Chen, Chem. Commun., 2015, 51, 5081–5084 RSC.
- X. Wang, X. Liu, T. Hou, W. Li and F. Li, Sens. Actuators, B, 2015, 208, 575–580 CrossRef CAS.
- H. Zhou, T. Han, Q. Wei and S. Zhang, Anal. Chem., 2016, 88, 2976–2983 CrossRef CAS PubMed.
- X. Luo, Y. Li, J. Zheng, H. Qi, Z. Liang and X. Ning, Chem. Commun., 2015, 51, 9487–9490 RSC.
- H.-F. Zhao, R.-P. Liang, J.-W. Wang and J.-D. Qiu, Biosens. Bioelectron., 2015, 63, 458–464 CrossRef CAS PubMed.
- Q. Shen, L. Han, G. Fan, E. S. Abdel-Halim, L. Jiang and J.-J. Zhu, Biosens. Bioelectron., 2015, 64, 449–455 CrossRef CAS PubMed.
- C. Ma, H. Liu, W. Li, H. Chen, S. Jin, J. Wang and J. Wang, Mol. Cell. Probes, 2016, 30, 118–121 CrossRef CAS PubMed.
- W. Cui, L. Wang and W. Jiang, Biosens. Bioelectron., 2016, 77, 650–655 CrossRef CAS PubMed.
- H. Zhao, L. Wang and W. Jiang, Chem. Commun., 2016, 52, 2517–2520 RSC.
- W. Zhang, X. Zu, Y. Song, Z. Zhu and C. J. Yang, Analyst, 2016, 141, 579–584 RSC.
- Q. Xue, Y. Zhang, S. Xu, H. Li, L. Wang, R. Li, Y. Zhang, Q. Yue, X. Gu, S. Zhang, J. Liu and H. Wang, Analyst, 2015, 140, 7637–7644 RSC.
- F. Tang, X.-W. Xing, J.-M. Chu, Q. Yuan, X. Zhou, Y.-Q. Feng and B.-F. Yuan, Analyst, 2015, 140, 4636–4641 RSC.
- Y. Zhang, W.-j. Xu, Y.-p. Zeng and C.-y. Zhang, Chem. Commun., 2015, 51, 13968–13971 RSC.
- Q. Xue, L. Wang and W. Jiang, Chem. Commun., 2015, 51, 13538–13541 RSC.
- Y. Ma, L. Chen, L. Zhang, S. Liao and J. Zhao, Analyst, 2015, 140, 4076–4082 RSC.
- W. Liu, H. Lai, R. Huang, C. Zhao, Y. Wang, X. Weng and X. Zhou, Biosens. Bioelectron., 2015, 68, 736–740 CrossRef CAS PubMed.
- H. Deng, X. Yang and Z. Gao, Analyst, 2015, 140, 3210–3215 RSC.
- Q. Xue, Y. Lv, S. Xu, Y. Zhang, L. Wang, R. Li, Q. Yue, H. Li, X. Gu, S. Zhang and J. Liu, Biosens. Bioelectron., 2015, 66, 547–553 CrossRef CAS PubMed.
- Y. Li, C. Yu, H. Han, C. Zhao and X. Zhang, Biosens. Bioelectron., 2016, 81, 111–116 CrossRef CAS PubMed.
- P. P. Hu, H. Liu, S. J. Zhen, C. M. Li and C. Z. Huang, Biosens. Bioelectron., 2015, 73, 228–233 CrossRef CAS PubMed.
- X. Wang, M. Cui, H. Zhou and S. Zhang, Chem. Commun., 2015, 51, 13983–13985 RSC.
- Y. Liu, M. Wei, L. Zhang, W. Wei, Y. Zhang and S. Liu, Chem. Commun., 2015, 51, 14350–14353 RSC.
- X. Li, T. Song and X. Guo, Analyst, 2015, 140, 6230–6233 RSC.
- M. Esteller, Oncogene, 2002, 21, 5427–5440 CrossRef CAS PubMed.
- W. Li, P. Wu, H. Zhang and C. Cai, Anal. Chem., 2012, 84, 7583–7590 CrossRef CAS PubMed.
- S. J. Hosic, S. K. Murthy and A. N. Koppes, Anal. Chem., 2016, 88, 354–380 CrossRef PubMed.
- A. K. White, K. A. Heyries, C. Doolin, M. VanInsberghe and C. L. Hansen, Anal. Chem., 2013, 85, 7182–7190 CrossRef CAS PubMed.
- M. Dhar, R. Khojah, A. Tay and D. Di Carlo, Lab Chip, 2015, 15, 4109–4113 RSC.
- B. R. Cipriany, R. Zhao, P. J. Murphy, S. L. Levy, C. P. Tan, H. G. Craighead and P. D. Soloway, Anal. Chem., 2010, 82, 2480–2487 CrossRef CAS PubMed.
- B. R. Cipriany, P. J. Murphy, J. A. Hagarman, A. Cerf, D. Latulippe, S. L. Levy, J. J. Benitez, C. P. Tan, J. Topolancik, P. D. Soloway and H. G. Craighead, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 8477–8482 CrossRef CAS PubMed.
Footnote |
| † Present address: Advanced Science Research Laboratory, Saitama Institute of Technology, Fukaya, Saitama 369-0293, Japan. |
| This journal is © The Royal Society of Chemistry 2016 |