Synthesis of a carboxymethylated guar gum grafted polyethyleneimine copolymer as an efficient gene delivery vehicle†
Piyali Janaa,
Kishor Sarkarc,
Tapas Mitraa,
Abhisek Chatterjeeb,
A. Gnanamanid,
Gopal Chakrabortib and
P. P. Kundu*a
aDepartment of Polymer Science & Technology, University of Calcutta, 92 A.P.C Road, Kolkata-700009, India. E-mail: ppk923@yahoo.com
bDepartment of Biotechnology & Dr. B.C. Guha Centre for Genetic Engineering, University of Calcutta, 35, Ballygung Circular Road, Kolkata 700019, India
cDepartment of Pharmaceutical Sciences, School of Pharmacy, University of Pittsburgh, PA-15261, USA
dMicrobiology Division, CSIR-Central Leather Research Institute, Adyar, Chennai 600020, Tamil Nadu, India
First published on 26th January 2016
Abstract
In the present study, a carboxymethylated guar gum-grafted-polyethyleneimine copolymer (CMGG-g-PEI) was synthesized and characterized by FT-IR, 1H NMR, XRD and zeta potential analyses. CMGG-g-PEI exhibited good binding ability with plasmid DNA (pDNA) and formed complexes with sizes ranging from 150 to 200 nm when the polymer/pDNA weight ratio was above 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. SEM analysis revealed a compact and spherical morphology of CMGG-g-PEI/pDNA complexes. The results from cytotoxicity and blood compatibility studies revealed the less toxic profile of CMGG-g-PEI. The in vitro gene transfection efficiency of the CMGG-g-PEI/pDNA complex was optimized in A549 cell and CMGG-g-PEI showed better transfection efficiency compared to the well-known standard polymer, polyethyleneimine (PEI). All the results suggested that the CMGG-g-PEI could find application as an alternative efficient gene delivery vehicle in the future.
1. SEM analysis revealed a compact and spherical morphology of CMGG-g-PEI/pDNA complexes. The results from cytotoxicity and blood compatibility studies revealed the less toxic profile of CMGG-g-PEI. The in vitro gene transfection efficiency of the CMGG-g-PEI/pDNA complex was optimized in A549 cell and CMGG-g-PEI showed better transfection efficiency compared to the well-known standard polymer, polyethyleneimine (PEI). All the results suggested that the CMGG-g-PEI could find application as an alternative efficient gene delivery vehicle in the future.
1. Introduction
Gene therapy is a modern clinical approach where a genetic material is transferred to specific cells to treat chronic diseases and genetic disorders. There are two types of carriers, which are used as a gene delivery vehicle i.e. viral and non-viral vectors. Now a days, uses of viral vectors are limited due to the fatal drawbacks such as immunogenicity, potential infectivity, complicated production and oncogenic effects.1–4Hence, non-viral gene delivery vehicles are becoming importance and are in the limelight. Among the non-viral vectors, cationic polymers are being used due to several advantages including low immunogenicity, high variability of structure and properties, ability to deliver large size genetic materials and large-scale production at low cost.5,6 Another decisive advantage of the cationic polymers is the ease of their protonation at physiological pH for endosomal buffering to prevent DNA from lysosomal degradation. As a macromolecular delivery agent, polycations can spontaneously condense poly-nucleotides such as plasmid DNA (pDNA), small interfering RNA (siRNA), and micro-RNA (miRNA) to deliver nucleic acids into different cell types.7–9 Amongst the non-viral vectors, polyethyleneimine (PEI) behaves as a “golden standard” in non-viral gene transfection application due to its high buffering capacity along with high transfection efficiency.10 This polymer is used to package plasmid DNA (pDNA) into nanoparticles for gene therapy due to its high charge density, membrane destabilization potential and ability to protect endocytosed pDNA from enzymatic degradation.11 The high transfection efficiency of PEI has been postulated and relate to its unique “proton sponge effect”.12–14 The high charge density of PEI and its strong interaction with cell membranes results in higher transfection efficiency.15,16 However, the cytocompatibility of PEI at higher doses in PEI/pDNA polyplex is not at satisfactory level.
Transfection efficiency and cytotoxicity of PEI are strongly related to the molecular weight of PEI. With low molecular weight branched-PEI (LMW b-PEI) molecular weight 2000 Da or less exhibits lower cytotoxicity and lower transfection efficiency, whereas PEI with high molecular weight (25 kDa) exhibits higher transfection efficiency and also possesses higher cytotoxicity.17,18
Currently, efforts are being undertaken to decrease cytotoxicity and improve the transfection efficiency of gene vectors by cross-linking low molecular weight units via linkages that are potentially degradable.5,19,20 However, toxicity is one of the major concerns for PEI to be used in gene delivery, and it increases with an increase in molecular weight17,18,21,22 as a non-biodegradable polymer, it will accumulate in the body especially for high molecular weight, leading to an unknown risk for long-term use.23 To overcome this drawback, several approaches have been explored to reduce the cytotoxicity of LMW b-PEI by grafting it with polyethylene glycol (PEG), other biocompatible and degradable polyesters24–26 or polysaccharides27,28 via disulfide bonds.29 Although, the cytotoxicity could be reduced significantly by the introduction of biodegradable bonds, the pDNA delivery efficiency is also reduced probably by the introduction of the biodegradability, causing a decrease in the amine density of the polymers.
By keeping all these in mind, we are in search for alternatives that should have similar pDNA-condensing capability, pH-buffering properties and potential transfection efficiency as comparable with native PEI. To prepare such kind of gene delivery vehicle with the above said properties, guar gum (GG) is selected for the present study to graft with LMW b-PEI to get efficient gene delivery system with reduced cytotoxicity. GG is a polygalactomannan derived from the seeds of a leguminacea plant, Cyamopsis tetragonolobus, and having a backbone of β-D-mannopyranoses linked 1 → 4 to which, on average, every alternate mannose and α-D-galactose is linked 1 → 6.26 Despite the wide spread application of GG as disintegrating agent, binding agent, film forming agent, matrix forming agent, release modifier, viscosity enhancing or gelling agent, emulsifier, suspending agent and bioadhesive agent,24 there is no report of GG as gene delivery system. Herein we synthesize GG grafted LMW b-PEI (800 Da) copolymer through conjugation of LMW b-PEI (800 Da) with carboxymethylated GG (CMGG) by carbodiimide chemistry. The synthesis of CMGG grafted PEI (CMGG-g-PEI) was characterized by Fourier transforms infrared (FTIR), X-ray diffractometer (XRD) and proton nuclear magnetic resonance (1H NMR). The complexation capability of CMGG-g-PEI with pDNA was confirmed by agarose gel electrophoresis assay followed by characterization of CMGG-g-PEI/pDNA complexes with dynamic light scattering (DLS), transmittance electron microscopy (TEM), scanning electron microscopy (SEM) and atomic force microscopy (AFM). Cytotoxicity and blood compatibility of CMGG-g-PEI/pDNA complexes were also evaluated. Finally, the gene transfection efficiency of CMGG-g-PEI copolymer was optimized in A549 lung carcinoma cell.
2. Experimental section
2.1. Materials
Guar gum and chloroacetic acid were obtained from Merck (India) and picrylsulfonic acid [2,4,6-trinitrobenzene sulfonic acid (TNBS)] was obtained from Sigma-Aldrich (USA). 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), 1-ethyl-3(3 dimethyl aminopropyl)carbodiimide (EDC) and N-hydroxy succinamide (NHS), 2-(N-morpholino)ethane sulfonic acid (MES buffer), Dulbecco's modified Eagle's medium (DMEM), penicillin–streptomycin, trypsin and fetal bovine serum (FBS) were purchased from HiMedia (India). DNase I was purchased from Thermo Scientific, India. Agarose was purchased from Sisco Research Laboratories Pvt. Ltd., India. pEGFP-N1 control vector (4.7 kb containing SV-40 promoter) was kindly donated by Dr Gopal Chakroborty, Department of Biotechnology, University of Calcutta. The plasmids were propagated in Escherichia coli (E. coli) and the plasmid DNA (pDNA) from E. coli was isolated with QIAGEN Midi prep pDNA isolation Kit (USA) according to the manufacturer's instructions. All other reagents were analytical grade and were used directly without further modification.2.2. GG purification procedure
The commercial GG was purified according to the procedure reported earlier25 without any modifications.2.3. Synthesis of CMGG
CMGG was synthesized by following the previously reported method.27 In brief, 6 g of purified GG was dispersed in 160 ml of isopropanol and water mixture (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
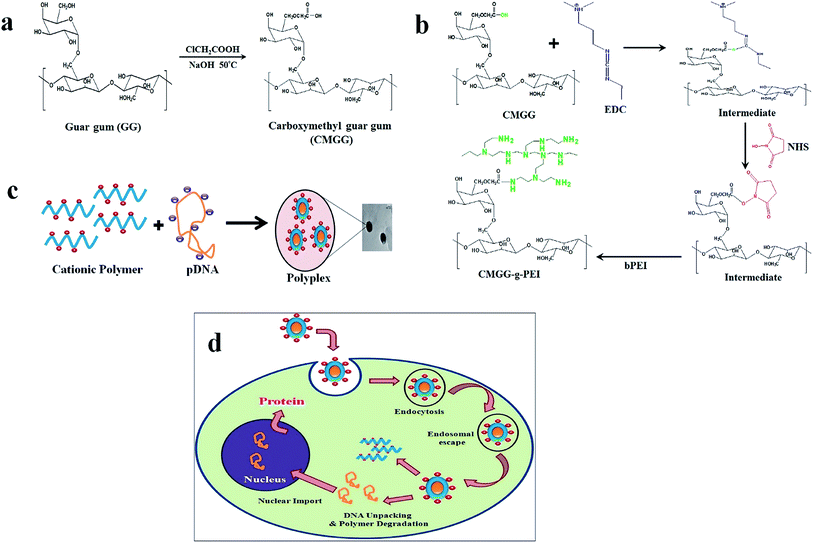
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 v/v ratio), in a 250 ml round bottom flask connected to an oil bath, and equipped with magnetic stirrer. To that 18 g of NaOH was added and stirred for an hour at 50 °C. 15 g chloroacetic acid dissolved in 10 ml isopropanol was then added drop wise to the reaction mixture over a period of 30 min. The reaction mixture was heated at 50 °C with continuous stirring for 4 h. The reaction was stopped by addition of excess cold ethanol and the reaction product was repeatedly extracted with ethanol and separated by centrifugation. After the third extraction, the pH was adjusted to 7 with several drops of glacial acetic acid and then washed with water and 80% ethanol and finally vacuum-dried. The schematic diagram for the synthesis of CMGG is shown in Fig. 1a.
2 v/v ratio), in a 250 ml round bottom flask connected to an oil bath, and equipped with magnetic stirrer. To that 18 g of NaOH was added and stirred for an hour at 50 °C. 15 g chloroacetic acid dissolved in 10 ml isopropanol was then added drop wise to the reaction mixture over a period of 30 min. The reaction mixture was heated at 50 °C with continuous stirring for 4 h. The reaction was stopped by addition of excess cold ethanol and the reaction product was repeatedly extracted with ethanol and separated by centrifugation. After the third extraction, the pH was adjusted to 7 with several drops of glacial acetic acid and then washed with water and 80% ethanol and finally vacuum-dried. The schematic diagram for the synthesis of CMGG is shown in Fig. 1a.
2.4. Determination of degree of substitution of CMGG
The degree of substitution (DS) of CMGG was estimated using titration method as reported elsewhere.28 CMGG (1.5 g) was dispersed in 50 ml of 2 M HCl (using 70% methanol as solvent) and the suspension was stirred continuously for 2 h. During this process, the sodium form of CMGG (Na-CMGG) was converted to its hydrogen form (H-CMGG) and then washed with 95% (v/v) ethanol and filtered. The filtrate was then dried in a vacuum oven at 60 °C for 2 h. The dried 0.5 g of H-CMGG was dissolved in 50 ml of 0.1 M NaOH solution and stirred for 2 h and the excess of NaOH was back titrated with 0.1 M HCl solution using phenolphthalein as an indicator. The DS was calculated using the following equation:| WA = CNaOHVNaOH − CHCLVHCL/m |
| DS = 162WA/5900 − 58WA |
2.5. Synthesis of CMGG-g-PEI
CMGG-g-PEI was synthesized through conjugation of LMW b-PEI (800 Da) with GG by carbodiimide chemistry using 1-ethyl-3(3 dimethyl aminopropyl)carbodiimide (EDC) and N-hydroxy succinamide (NHS) as coupling agent. At first, 3 g of CMGG was dissolved in 50 ml 0.1 M MES buffer to maintain pH at 6.5. Then, EDC and NHS were added to the solution at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (CMGG
4 (CMGG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EDC
EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NHS) and the reaction mixture was stirred at room temperature for 24 h to activate the carboxyl group of CMGG. Secondly, 15 g of LMW b-PEI (800 Da) dissolved in water was added to the activated CMGG solution. Then, the pH of the solution was maintained at 6.5 and the reaction continued for another 48 h at room temperature. The resultant solution was purified by exhaustive dialysis (MWCO-10 kDa) against double distilled water for 48 h followed by lyophilization to get CMGG-g-PEI. The reaction scheme of CMGG-g-PEI is shown in Fig. 1b.
NHS) and the reaction mixture was stirred at room temperature for 24 h to activate the carboxyl group of CMGG. Secondly, 15 g of LMW b-PEI (800 Da) dissolved in water was added to the activated CMGG solution. Then, the pH of the solution was maintained at 6.5 and the reaction continued for another 48 h at room temperature. The resultant solution was purified by exhaustive dialysis (MWCO-10 kDa) against double distilled water for 48 h followed by lyophilization to get CMGG-g-PEI. The reaction scheme of CMGG-g-PEI is shown in Fig. 1b.
2.6. Estimation of primary amine groups in CMGG-g-PEI (TNBS assay)
The percentage of primary amine groups presented in CMGG-g-PEI was quantified using TNBS assay according to the procedure summarized by Mitra et al.302.7. Characterization of the polymers
FTIR analysis was carried out with FTIR spectroscopy (model-Alpha, Bruker, Germany).XRD of the polymers was performed by a wide angle X-ray diffractometer (Panalytical X-Ray Diffractometer, model-X'pert Powder) with CuKα radiation (λ = 1.544) in the range of 5–50° (2θ) at 40 kV and 30 mA.
1H NMR spectra were determined on a Bruker NMR system (Germany) at 400 MHz using D2O as solvent. Chemical shifts were reported in ppm using tetramethyl silane (TMS) as an internal reference.
The average molecular weight of pure GG, CMGG, and CMGG-g-PEI was determined by gel-permeation chromatographic technique. Standard dextran T-250, T-200, T-70 and T-40 were passed through a Sepharose 6B column, and the elution volumes were plotted against the logarithms of their respective molecular weights.31 The elution volume of GG was then plotted in the same graph, and molecular weight of GG was determined. In the same way CMGG and CMGG-g-PEI molecular weight were determined.
Zeta potential value of GG, CMGG, and CMGG-g-PEI were measured by Zetasizer Nano ZS (Malvern Instrument, UK). For investigating the influence of pH, the zeta potential value for native GG and modified GG were measured at different pH values of 4, 5, 6 and 7.
ESI file ES1† detailed the thermal stability studies on the CMGG-g-PEI copolymer in comparison with the individual reactants.
2.8. Preparation of polymer/pDNA complexes
GG and CMGG-g-PEI were dissolved in acetic acid/sodium acetate buffer at pH 5.5 with a concentration of 1 mg ml−1 and 0.2 mg ml−1 respectively and the solutions were filtered by a millipore (0.45 μm) filter paper. pDNA was dissolved separately (100 μg ml−1) in 25 mM of sodium sulphate solution. Prior to the polymer/pDNA complex (polyplex) formation, the polymer and pDNA solutions were preheated separately at 55 °C for 10 min. Then, polyplexes at different weight ratios (polymer/pDNA weight ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 10
1, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 15
1, 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 20
1, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 25
1, 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 30
1 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) containing 0.5 μg pDNA in each weight ratio were prepared by immediate mixing of equal volume of polymer and pDNA solution as given details in Table 2 and vortexed for 15–30 s with cyclomixer (REMI, India). The resulting mixtures were then incubated at room temperature for 30 min to complete the polyplex formation. A schematic representation on the preparation of polymer/DNA complex shown in Fig. 1c.
1) containing 0.5 μg pDNA in each weight ratio were prepared by immediate mixing of equal volume of polymer and pDNA solution as given details in Table 2 and vortexed for 15–30 s with cyclomixer (REMI, India). The resulting mixtures were then incubated at room temperature for 30 min to complete the polyplex formation. A schematic representation on the preparation of polymer/DNA complex shown in Fig. 1c.
2.9. Agarose gel retardation assay
The binding efficiency of GG and CMGG-g-PEI with pDNA was determined by agarose gel (0.8%, w/w) electrophoresis (100 V for 45 min) and the gel image was subsequently captured by BIOTOP gel doc system (Shanghai, China). Agarose gel (0.8%, w/w) was prepared in TAE buffer (40 mmol l−1 Tris acetate, 1 mmol l−1 EDTA) and ethidium bromide (10 μg ml−1) was added to the gel as a pDNA visualizer.2.10. DNase I digestion assay
The digestion of naked pDNA and polyplexes with DNase I (1 U per μg of pDNA) was assayed in 10 mM phosphate buffered saline (PBS) containing 5 mM MgCl2 at 37 °C. At first, freshly prepared polyplexes and naked pDNA were incubated with 1 μl of DNase I (1 U μl−1, Fermentas, USA) for 10 min at 37 °C and then DNase I was inactivated by the addition of 0.5 M of EDTA. The degradation of pDNA was monitored by 0.8% agarose gel electrophoresis and subsequently photographed by using Bio-Rad Chemi Doc XRS+ molecular imager.2.11. Characterization of polyplexes
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was observed using TEM (JEM 1010, JEOL, Japan).
1 was observed using TEM (JEM 1010, JEOL, Japan).| % Cell viability = [100 − (At/As)] × 100 | (1) |
At and As indicated the absorbance of the test substances and solvent control, respectively.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 30
1 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 according to the conditions described above (containing 1 μg pDNA in each weight ratio) prior to the transfection. Two hours before transfection, the complete growth medium was replaced with fresh media without FBS and antibiotics. To maintain the pH of the transfection media at ∼pH 6.5, 5 mM MES buffer (pH 6.5) was added to DMEM. The polymer/pDNA complexes containing 1 μg pDNA at different w/w ratios were added into each well and were incubated at 37 °C in a CO2 incubator under humidified condition for another 4 h. After 4 h, the transfection media was replaced with complete growth media and incubated for 24 h. All transfections were carried out in triplicate. Naked pDNA and b-PEI (25 kDa)/pDNA at N/P ratio (nitrogen to phosphate ratio) of 10 were used as negative and positive control, respectively. Following 24 h of incubation, the cells were analyzed for green fluorescence protein (pEGFP-N1) expression with a fluorescence microscope (OLYMPUS IX70, Japan).
1 according to the conditions described above (containing 1 μg pDNA in each weight ratio) prior to the transfection. Two hours before transfection, the complete growth medium was replaced with fresh media without FBS and antibiotics. To maintain the pH of the transfection media at ∼pH 6.5, 5 mM MES buffer (pH 6.5) was added to DMEM. The polymer/pDNA complexes containing 1 μg pDNA at different w/w ratios were added into each well and were incubated at 37 °C in a CO2 incubator under humidified condition for another 4 h. After 4 h, the transfection media was replaced with complete growth media and incubated for 24 h. All transfections were carried out in triplicate. Naked pDNA and b-PEI (25 kDa)/pDNA at N/P ratio (nitrogen to phosphate ratio) of 10 were used as negative and positive control, respectively. Following 24 h of incubation, the cells were analyzed for green fluorescence protein (pEGFP-N1) expression with a fluorescence microscope (OLYMPUS IX70, Japan).For flow cytometric analysis, pEGFP-N1 transfected A549 cells were harvested by trypsinization, washed twice with PBS, pH 7.4 and the cell suspensions were then transferred to 5 ml flow cytometry tubes and EGFP expression in the transfected cells were quantified using a Becton-Dickinson FACS Calibur flow cytometer. The obtained data were analyzed using Cell Quest program from Becton-Dickinson. For each sample, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were counted and the green fluorescence of EGFP was detected by FL1 channel of the flow cytometer.
000 events were counted and the green fluorescence of EGFP was detected by FL1 channel of the flow cytometer.
3. Result and discussion
In the present study, the purified commercial product GG was used for carboxymethylation. By Williamson ether synthesis procedure, carboxymethylation of GG was synthesized by consecutive two-step reaction. A strong base, such as sodium hydroxide was added to the GG solution to deprotonate the free hydroxyl groups (particularly, the hydroxyl group of –CH2OH groups in GG) to form alkoxides, thereby increasing their nucleophilicity. Carboxymethyl groups are then formed in a reaction between guar alkoxides and chloroacetic acid as shown schematically (Fig. 1a).The synthesis of CMGG and CMGG-g-PEI was confirmed by FTIR. The spectrum of pure GG shows absorption bands around 3386, 2928 and 1150 cm−1 due to the presence of O–H, C–H and C–O stretching vibrations. The absorption band around 1025 cm−1 was due to the glycosidic linkage of pyranose ring of GG.33 Whereas, in CMGG spectrum, two new bands are appeared at 1604, and 1324 cm−1, corresponding to the COO− asymmetric and symmetric stretching vibration due to carboxyl group of the carboxymethyl moiety of CMGG. PEI was characterized by the broad band of primary, secondary and tertiary amine groups at around 3200–3500 cm−1 and absorptions at 1640, 1546 and 1464 cm−1 were assigned to the primary amine end groups (N–H and C–N). FTIR spectrum of CMGG-g-PEI displayed significant changes compared to the parent GG due to grafting of PEI with CMGG. The peak at 1604 cm−1 in CMGG disappeared in CMGG-g-PEI and the intensity of the peak at 1454 cm−1 was reduced as compared to CMGG. The peak observed at 1640 cm−1 in PEI was shifted to 1576 cm−1 in CMGG-g-PEI resulting from the –CONH due to the reaction between the carboxyl group of the CMGG and amine group of PEI. The FTIR spectra Fig. 2a confirm that the monochloroacetic acid was successfully immobilized on the GG backbone as well as PEI (800 Da) was successfully grafted on CMGG back bone.
 | ||
| Fig. 2 (a) FTIR spectrum of GG, CMGG, PEI (800 Da) and CMGG-g-PEI. (b) X-ray diffraction patterns of GG, CMGG and CMGG-g-PEI. (C) 1H NMR spectrum of (a) CMGG and (b) CMGG-g-PEI in D2O. | ||
The wide angle XRD of native GG, CMGG and CMGG-g-PEI were represented in Fig. 2b. It is observed that the native GG exhibits low crystallinity similar to the observations made by Pal et al.34 After carboxymethylation, a typical reduction in crystallinity was observed in CMGG. This loss in crystallinity may be due to destruction of hydrogen bonding through the carboxymethylation of the hydroxyl groups of native GG. The intermolecular hydrogen bonding in GG is responsible for the higher crystallinity in GG; when the interaction is disrupted, it leads to the reduction in the crystallinity of CMGG. However, in the case of CMGG-g-PEI, the crystallinity increased after grafting of PEI to CMGG. This could be due to the formation of hydrogen bond between amine group of PEI and carboxyl group of CMGG of CMGG-g-PEI.
Fig. 2c illustrates the 1H NMR spectra details of GG, CMGG and CMGG-g-PEI. The characteristic peaks of pure GG in D2O solvent appeared at δ 4.65 (s) was due to anomeric protons and at δ 3.5–3.9 (m) and 2.05–2.06 (d) were due to sugar protons. While in CMGG spectrum, peaks at δ = 3.8, 3.9 and 4.08 ppm, were attributed to the methylene protons of carboxymethoxy substituents of α-D-galactose unit and β-D-mannose unit of C-6 and C-3 positions, respectively. The peak at δ = 4.6 and 4.95 was due to anomeric proton of CMGG overlaid by the solvent peak (D2O-d), and the other peaks located around δ = 3–4.1 were due to sugar protons of CMGG.35 CMGG-g-PEI shows the presence of either peaks resonating at 3.4–2.6 ppm belonging to PEI36 or those of CMGG at 4.0–3.6 ppm and the smaller signal of anomeric protons around 5.0 ppm was clearly decreased by increasing the amount of PEI grafted onto the CMGG. The grafting of PEI on CMGG backbone has also been confirmed by the following results.
The change in peak position and molecular weight of GG before modification and after modification also suggested the synthesis of CMGG-g-PEI copolymer as shown in Fig. 3. The average molecular weight of unmodified GG was 218.07 kDa. After reaction of GG with chloroacetic acid, the molecular weight slightly increased from 218.07 kDa to 218.78 kDa due to incorporation of small molecule of carboxymethyl group in CMGG. But, after conjugation of PEI (800 Da) with CMGG the molecular weight significantly increased to 239.83 kDa indicating the synthesis of CMGG-g-PEI copolymer.
The zeta (ζ) potential is an important parameter to examine the possibility of the existence of a positive or negative surface charge, depending on the pH of the solution. At low pH, most particles exhibit a positive charge; as the pH is raised, a negatively charged surface is formed. Table 1 depicts the zeta potential values for GG, CMGG and CMGG-g-PEI. It is observed that GG show negative zeta potential at pH range from −4.0 to 7.0. But the negative zeta potential increased significantly after carboxymethylation of GG attributed to the incorporation of anionic carboxylate groups in CMGG. The zeta potential became positive in CMGG-g-PEI due to conjugation of cationic PEI (800 Da) with CMGG. CMGG-g-PEI showed higher positive zeta potential in the acidic pH region due to the protonation of the amino groups of PEI, leading to higher positive charges and hence higher positive zeta potential value. On the other hand, at the higher pH range, CMGG-g-PEI shows less positive zeta potential value due to the presence of less number of positively charged ions. The results suggest the conjugation of PEI with CMGG through amide linkage. The amine groups on the surface of CMGG-g-PEI could then be beneficial for the formation of bio conjugates.
| pH | GG | CMGG | CMGG-g-PEI |
|---|---|---|---|
| 7 | −10.8 | −29.8 | +0.345 |
| 6 | −10.2 | −19.8 | +2.09 |
| 5 | −4.49 | −18.5 | +7.59 |
| 4 | −3.57 | −15.9 | +15.9 |
| Stock polymer solution concentration (mg ml−1) | Polymer/DNA weight ratio (w/w) | DNA stock solution (μl) (stock concentration 0.25 μg μl−1) | Sodium sulphate solution (μl) | Polymer solution (μl) | Acetate buffer solution (μl) | Total volume of solution (μl) |
|---|---|---|---|---|---|---|
| 0.2 | 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2 | 18 | 0 | 0 | 20 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2 | 8 | 1 | 9 | 20 | |
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2 | 8 | 5 | 5 | 20 | |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2 | 8 | 10 | 0 | 20 | |
| 1 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2 | 8 | 3 | 7 | 20 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2 | 8 | 4 | 6 | 20 | |
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2 | 8 | 5 | 5 | 20 | |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2 | 8 | 6 | 4 | 20 |
Though, all the spectral studies have proved the successful carboxymethylation on native GG and the grafting through covalent bonding between the free –NH2 group of PEI and the –COOH group of CMGG, the percentage of degree of substitution by TNBS assay corroborates well with the results obtained and the percentage degree of substitution was calculated as 43.22%, calculated by the following equation.
3.1. Complexation of the copolymer with pDNA
Electrostatic interactions between the negatively charged nucleic acids and the cationic CMGG-g-PEI to form CMGG-g-PEI/pDNA complexes were investigated by agarose gel electrophoresis assay. The gene condensing capabilities of CMGG-g-PEI were evaluated by conducting a gel retardation assay to determine whether the gene was shielded by polycations or exposed on the surface of the polyplex. The ability of CMGG-g-PEI to condense pDNA was evaluated by agarose gel electrophoresis with CMGG-g-PEI/DNA at different weight ratios ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 30
1 to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as shown in Fig. 4a and b. Fig. 4 shows that free pDNA displays two distinct fluorescent bands, corresponding to the super coiled and circular forms of the plasmid. Due to negative zeta potential of GG (Table 1), GG did not show any pDNA complexation capability at any weight ratios from 1
1 as shown in Fig. 4a and b. Fig. 4 shows that free pDNA displays two distinct fluorescent bands, corresponding to the super coiled and circular forms of the plasmid. Due to negative zeta potential of GG (Table 1), GG did not show any pDNA complexation capability at any weight ratios from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 30
1 to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as shown in Fig. 4a. But, the complexation capability of GG drastically improved after conjugation of PEI (800 Da) and GG-g-PEI started to complex all pDNA at very low weight ratio of 10
1 as shown in Fig. 4a. But, the complexation capability of GG drastically improved after conjugation of PEI (800 Da) and GG-g-PEI started to complex all pDNA at very low weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 where no migration of pDNA was observed (Fig. 4b). After conjugation of PEI (800 Da) with GG, CMGG-g-PEI showed positive zeta potential and as a result CMGG-g-PEI complexed with negatively charged pDNA through electrostatic interaction compared to unmodified GG. The results suggest, positive zeta potential would be good for DNA binding within our experimental conditions.
1 where no migration of pDNA was observed (Fig. 4b). After conjugation of PEI (800 Da) with GG, CMGG-g-PEI showed positive zeta potential and as a result CMGG-g-PEI complexed with negatively charged pDNA through electrostatic interaction compared to unmodified GG. The results suggest, positive zeta potential would be good for DNA binding within our experimental conditions.
3.2. DNase I digestion assay
For successful gene delivery, it is a prerequisite for the carrier in the carrier/pDNA complexes to protect the pDNA against degradation by cellular nucleases.2 Since mammalian cells contain several nucleases which are capable to degrade the DNA in their extracellular space and also in their cytoplasm, so it is necessary for the carrier to protect the DNA from these nucleases for successful transfection. Here, we performed DNase I protection assay to evaluate whether the synthesized CMGG-g-PEI can protect the pDNA from nucleases during transfection or not.Fig. 4c demonstrates the capability of GG and CMGG-g-PEI copolymers to protect pDNA against DNase I digestion. As shown in Fig. 4c, naked pDNA was completely digested by DNase within 15 min of incubation, confirming the activity of nuclease. As observed from the Fig. 4c GG did not show any protection of pDNA against DNase I irrespective of all weight ratios, but CMGG-g-PEI copolymers efficiently protected the pDNA from DNase I digestion at or above weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. From agarose gel electrophoresis assay, it was found that GG was not able to bind pDNA and resulted no protection of pDNA against DNase I whereas CMGG-g-PEI protected pDNA efficiently from weight ratio of 10
1. From agarose gel electrophoresis assay, it was found that GG was not able to bind pDNA and resulted no protection of pDNA against DNase I whereas CMGG-g-PEI protected pDNA efficiently from weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 because CMGG-g-PEI started complex efficiently with pDNA from this weight ratio. These results showed that CMGG-g-PEI could protect DNA efficiently against digestion by DNase I, which is one of the prerequisite for efficient gene delivery inside the mammalian cells.
1 because CMGG-g-PEI started complex efficiently with pDNA from this weight ratio. These results showed that CMGG-g-PEI could protect DNA efficiently against digestion by DNase I, which is one of the prerequisite for efficient gene delivery inside the mammalian cells.
3.3. Particle size and zeta potential
The particle size and zeta potential of carrier/DNA complex for efficient transfection are another two important parameters.37,38 Positive charge of carrier/DNA complex is thought to be helpful for its absorption to negatively charged cellular membrane, also leading to efficient intracellular trafficking.5 It is reported that the cells typically uptake particles ranging from about 50 to several hundred nano meters.39 Fig. 5a and b illustrate the average diameters measured by dynamic light scattering (DLS) and overall surface charge of CMGG-g-PEI/pDNA complexes at different weight ratios in PBS buffer (pH 7.4), respectively. As GG did not show any complexation capability with pDNA, we have not shown here the particle size and zeta potential of GG/pDNA complexes. It is found that the particle size of the complexes tends to decrease with the increase in weight ratio of CMGG-g-PEI/pDNA from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 30
1 to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. From the figure, it is observed that the particle sizes of CMGG-g-PEI/pDNA complex at weight ratios of 1
1. From the figure, it is observed that the particle sizes of CMGG-g-PEI/pDNA complex at weight ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 5
1 and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were around 170–190 nm but the size increased at weight ratio of 10
1 were around 170–190 nm but the size increased at weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The particle size at low weight ratios may be responsible either uncomplexed CMGG-g-PEI or pDNA. Agarose gel electrophoresis assay (Fig. 4b) showed CMGG-g-PEI started to complex with pDNA at weight ratio of 10
1. The particle size at low weight ratios may be responsible either uncomplexed CMGG-g-PEI or pDNA. Agarose gel electrophoresis assay (Fig. 4b) showed CMGG-g-PEI started to complex with pDNA at weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and resulted larger particle size due to loosely complexation. The particle size was further decreased with increase in CMGG-g-PEI/pDNA weight ratio beyond 10
1 and resulted larger particle size due to loosely complexation. The particle size was further decreased with increase in CMGG-g-PEI/pDNA weight ratio beyond 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 because of strong complexation. CMGG-g-PEI formed smallest complex with pDNA at weight ratio of 30
1 because of strong complexation. CMGG-g-PEI formed smallest complex with pDNA at weight ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the particle size of the complex at this weight ratio was around 185 nm. CMGG-g-PEI/pDNA complexes also showed positive zeta potential having the value around 10–20 mV beyond the weight ratio of 10
1 and the particle size of the complex at this weight ratio was around 185 nm. CMGG-g-PEI/pDNA complexes also showed positive zeta potential having the value around 10–20 mV beyond the weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The positive zeta potential of CMGG-g-PEI/pDNA complex above the weight ratio of 10
1. The positive zeta potential of CMGG-g-PEI/pDNA complex above the weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 may be beneficial for better cellular uptake as well as enhanced transfection efficiency. Similar results were observed in previous studies.32
1 may be beneficial for better cellular uptake as well as enhanced transfection efficiency. Similar results were observed in previous studies.32
Further analysis using transmission electronic microscope (TEM) revealed that the particle size of CMGG-g-PEI/pDNA complex at weight ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 varied from 136.8–142.2 nm Fig. 5c. It was also observed from the figure that CMGG-g-PEI/pDNA complex was spherical in shape.
1 varied from 136.8–142.2 nm Fig. 5c. It was also observed from the figure that CMGG-g-PEI/pDNA complex was spherical in shape.
The size and morphology of CMGG-g-PEI/pDNA complexes were further observed by SEM as well as AFM as shown in Fig. S1 and S2,† respectively. SEM and AFM images also showed that the morphology of CMGG-g-PEI/pDNA complex was round and spherical, and there was no obvious aggregated polyplexes in the field of vision as shown in Fig. S1;† the polyplexes were found to have spherical shape and compact structure. Moreover, the AFM images demonstrated that the size of the nanoparticles was roughly within the range of 130–145 nm at weight ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. These results are well collaborated with DLS data. The particle size in Fig. 5a was hydrodynamic size of CMGG-g-PEI/pDNA complex at weight ratio of 10
1. These results are well collaborated with DLS data. The particle size in Fig. 5a was hydrodynamic size of CMGG-g-PEI/pDNA complex at weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the size was around 300 nm which was larger than the size obtained from AFM. The TEM image was taken at dried condition. Definitely the size at dried condition will be smaller than hydrodynamic diameter.40,41
1 and the size was around 300 nm which was larger than the size obtained from AFM. The TEM image was taken at dried condition. Definitely the size at dried condition will be smaller than hydrodynamic diameter.40,41
3.4. Enzymatic degradation
The wet weight of the film was slightly increased initially due to swelling; and then decreased due to the disintegration of the film in presence of the enzyme. The time required for complete disappearance of the film was used to assess their degradability. Results revealed that the complete disappearance of CMGG-g-PEI was observed after eight hours of incubation with enzyme solution whereas GG film was completely disappeared within two hours of time.3.5. Buffering capacity
According to the proton sponge hypothesis, an appropriate buffering capacity is important to cationic polymeric vectors because the buffering effect causes an increase in osmotic pressure in the endosome, leading to the disruption of the endosomal membrane to facilitate the carrier/DNA complex transport into the cytoplasm.42 One of the primary causes of low transfection efficiency of any non-viral vector is inefficient release carrier/DNA complex from endosomes to cytoplasm. The buffering capacities of CMGG-g-PEI, LMW b-PEI (800 Da) and GG were examined by acid–base titration method as shown in Fig. 5d. The polymer with a high buffering ability would undergo a small change in pH when the same amount of HCl was added into the polymer solution during titration.43 PEI (800 Da) had a strong buffering capacity. GG showed very poor buffering capacity due to absence of proton accepting functional groups. After grafting of PEI (800 Da) with GG, the buffering capacity of CMGG-g-PEI significantly improved as compared with GG but still lower than that of PEI. The lower buffer capacity of CMGG-g-PEI could be attributed to lower number of amine group in copolymer compared to PEI only. According to Gabrielson & Pack, (2006) (ref. 44) an increase in the acetylation degree of PEI resulted in a reduction of the polymer buffering capacity. However, the titration curves of these copolymers showed a decrease in pH at a faster rate than that of PEI (800 Da) due to the following two reasons; (i) in comparison with PEI (800 Da), the number of primary, secondary, and tertiary amines groups which may be protonated in the copolymers is less, resulting in lower buffer capacity; (ii) the unreacted carboxyl groups of the copolymer would inhibit the protonation of the amine groups of the oligoamine. This neighbouring effect of carboxyl groups of copolymers might also result in the lower buffer capacity.3.6. Blood compatibility
In vitro erythrocyte-induced hemolysis is considered to be a simple and reliable measure for estimating blood compatibility of materials. The haemolytic assay is a significant index of the material for the application in the biomedical field because the material is usually exposed to blood environment and damaged the erythrocytes in a certain degree. In the present study, the assay was carried out to evaluate the blood compatibility of CMGG-g-PEI. The results as shown in Fig. 6a suggest there was no damage on the erythrocytes due to the addition of CMGG-g-PEI and confirm the blood compatibility property of the polymer.3.7. Cytotoxicity of the polymers
For eventual application in vivo, it is important that our synthesized copolymer should have minimal cytotoxicity. So, to investigate the cytotoxicity of the CMGG-g-PEI copolymers, cell viability was measured in A549 cells using MTT colorimetric assay. Fig. 6b illustrates that CMGG-g-PEI showed significantly lower cytotoxicity than that of, LMW b-PEI (800 Da) and, b-PEI (25 kDa) in A549 cells. Thus, Fig. 6b showed that unmodified GG exhibited very little cytotoxicity towards A549 cells and almost 80% of the cells were viable in the presence of GG even at very high concentration (500 μg ml−1). However, Fig. 6b also demonstrates that the cell viability of A549 cells in presence of CMGG-g-PEI copolymers over the concentration range studied (0–500 μg ml−1) were significantly higher than that of both, LMW b-PEI (800 Da) and b-PEI (25 kDa) suggesting significantly lower cytotoxicity. The cytotoxicity data also demonstrated that though CMGG-g-PEI showed a little higher cytotoxicity than normal GG but it has significantly lower cytotoxicity that of the corresponding PEI dose.3.8. In vitro transfection
To estimate the transfection efficiency of synthesized polymer, CMGG-g-PEI/pDNA complexes were prepared at different weight ratios from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 30
1 to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with plasmid pEGFP-N1 according to the conditions described previously. The transfection studies were carried out in A549 cells and compared with GG and LMW b-PEI (800 Da) alone. PEI (25 kDa)/pDNA complex weight ratio of 30
1 with plasmid pEGFP-N1 according to the conditions described previously. The transfection studies were carried out in A549 cells and compared with GG and LMW b-PEI (800 Da) alone. PEI (25 kDa)/pDNA complex weight ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was used as a positive control. Typical fluorescence images of the transfected A549 cells were shown in Fig. 7a. It is observed that GG did not show any transfection efficiency at any weight ratios as expected although some GFP positive cells were found which may be resulted by only pDNA as obtained in only pDNA transfection. But the transfection efficiency significantly increased after conjugation of LMW b-PEI (800 Da) with GG. However, the transfection efficiency of CMGG-g-PEI/pDNA complex at weight ratio of 30
1 was used as a positive control. Typical fluorescence images of the transfected A549 cells were shown in Fig. 7a. It is observed that GG did not show any transfection efficiency at any weight ratios as expected although some GFP positive cells were found which may be resulted by only pDNA as obtained in only pDNA transfection. But the transfection efficiency significantly increased after conjugation of LMW b-PEI (800 Da) with GG. However, the transfection efficiency of CMGG-g-PEI/pDNA complex at weight ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was much higher than the weight ratio of 10
1 was much higher than the weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The increased transfection efficiency of CMGG-g-PEI/pDNA complex at weight ratio of 30
1. The increased transfection efficiency of CMGG-g-PEI/pDNA complex at weight ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 might be related to the smaller particle size compared to that of 10
1 might be related to the smaller particle size compared to that of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio. The transfection efficiency of the CMGG-g-PEI/pDNA complexes gradually increased and optimal transfection efficiency was obtained at a w/w ratio of 1
1 weight ratio. The transfection efficiency of the CMGG-g-PEI/pDNA complexes gradually increased and optimal transfection efficiency was obtained at a w/w ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30. Further increase in the weight ratio resulted in gradual decrease in transfection efficiency with an increase in cell death (data not shown). The transfection efficiency of CMGG-g-PEI/pDNA complex at weight ratio of 30
30. Further increase in the weight ratio resulted in gradual decrease in transfection efficiency with an increase in cell death (data not shown). The transfection efficiency of CMGG-g-PEI/pDNA complex at weight ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was higher than that of LMW b-PEI (800 Da)/pDNA complex but lower than PEI (25 kDA)/pDNA complex at the same weight ratio. Although b-PEI (25 kDa) was much more toxic compared to that of CMGG-g-PEI at the equivalent concentration to the weight ratio of 30
1 was higher than that of LMW b-PEI (800 Da)/pDNA complex but lower than PEI (25 kDA)/pDNA complex at the same weight ratio. Although b-PEI (25 kDa) was much more toxic compared to that of CMGG-g-PEI at the equivalent concentration to the weight ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The increased transfection efficiency of the copolymer may be ascribed to the higher amine content from LMW b-PEI (800 Da). After the grafting of PEI to CMGG, the PEI moieties acted as a sponge and enhanced the release of CMGG-g-PEI/pDNA complexes from endosome and leading to improve transfection efficiency. Though CMGG-g-PEI copolymers have little less transfection ability than that of b-PEI (25 kDa), it was found to be of much lower cytotoxicity than that of b-PEI (25 kDa); this less cytotoxicity is a prerequisite for successful gene transfection.
1. The increased transfection efficiency of the copolymer may be ascribed to the higher amine content from LMW b-PEI (800 Da). After the grafting of PEI to CMGG, the PEI moieties acted as a sponge and enhanced the release of CMGG-g-PEI/pDNA complexes from endosome and leading to improve transfection efficiency. Though CMGG-g-PEI copolymers have little less transfection ability than that of b-PEI (25 kDa), it was found to be of much lower cytotoxicity than that of b-PEI (25 kDa); this less cytotoxicity is a prerequisite for successful gene transfection.
The transfection efficiency of CMGG-g-PEI/DNA complexes was further quantitatively determined through flow cytometric analysis of the transfected A549 cells. Fig. 7b demonstrated the respective dot plots of transfection efficiency with GG and the copolymer at weight ratios of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 30
1 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and LMW b-PEI (800 Da and 25 kDa)/pDNA complexes at weight ratio of 30
1 and LMW b-PEI (800 Da and 25 kDa)/pDNA complexes at weight ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. B-PEI (25 kDa)/pDNA complex at weight ratio of 30
1. B-PEI (25 kDa)/pDNA complex at weight ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 served as a positive control and only pDNA as negative control. Fig. 7b shows that grafting of LMW b-PEI (800 Da) with GG back bone significantly increased its pDNA binding ability and consequently the transfection efficiency. While unmodified GG showed comparable transfection ability with pDNA only. CMGG-g-PEI copolymer showed almost 40% gene transfection efficiency, which was higher than that of LMW b-PEI (800 Da) Fig. 7c. This is probably due to increase in surface charge density of CMGG by the grafting of LMW b-PEI (800 Da) molecule. The higher surface charge of the copolymer resulted in the more efficient pDNA binding and enhanced complexation ability of the copolymer with pDNA and thereby showed increased transfection efficiency. The overall transfection process of the prepared CMGG-g-PEI/pDNA complex has been described in the Fig. 1d.
1 served as a positive control and only pDNA as negative control. Fig. 7b shows that grafting of LMW b-PEI (800 Da) with GG back bone significantly increased its pDNA binding ability and consequently the transfection efficiency. While unmodified GG showed comparable transfection ability with pDNA only. CMGG-g-PEI copolymer showed almost 40% gene transfection efficiency, which was higher than that of LMW b-PEI (800 Da) Fig. 7c. This is probably due to increase in surface charge density of CMGG by the grafting of LMW b-PEI (800 Da) molecule. The higher surface charge of the copolymer resulted in the more efficient pDNA binding and enhanced complexation ability of the copolymer with pDNA and thereby showed increased transfection efficiency. The overall transfection process of the prepared CMGG-g-PEI/pDNA complex has been described in the Fig. 1d.
4. Conclusion
We successfully conjugated PEI to the carboxyl groups of CMGG which was prepared from GG to synthesize CMGG-g-PEI copolymer via Williamson's ether synthesis and carbodiimide chemistry. The buffering capacity of CMGG-g-PEI was significantly enhanced compared to that of GG. The synthesized copolymer displayed strong pDNA condensation capability by forming positively charged polyplexes and protected pDNA degradation against DNase I digestion as well as displayed its capacity to release pDNA into the cell due to its high proton sponge effect. In vitro experiments indicated that the transfection efficiency of CMGG-g-PEI/pDNA complexes was significantly higher than that of LMW b-PEI (800 Da)/pDNA but lower than that of b-PEI (25 kDa)/pDNA complexes in A549 cell. On the other hand, the graft copolymer showed much lower cytotoxicity compared to both PEI. The results suggested that CMGG-g-PEI could be a safe and efficient non-viral vector for gene therapy application.Acknowledgements
One of the authors Dr Tapas Mitra acknowledges UGC, New Delhi for financial assistance provided in the form of Dr D. S. Kothari Post Doctoral Fellowship.References
- M. J. Tiera, Q. Shi, F. M. Winnik and J. C. Fernandes, Curr. Gene Ther., 2011, 11, 288–306 CrossRef CAS PubMed.
- H.-L. Jiang, T.-H. Kim, Y.-K. Kim, I.-Y. Park, M.-H. Ch and C.-S. Cho, Biomed. Mater., 2008, 3, 025013 CrossRef PubMed.
- M. R. Rekha and C. P. Sharma, Recent Pat. DNA Gene Sequences, 2012, 6, 98–107 CrossRef CAS.
- R. H. Simon, J. F. Engelhardt, Y. Yang, M. Zepeda, S. Weber-Pendleton, M. Grossman and J. M. Wilson, Hum. Gene Ther., 1993, 4, 771–780 CrossRef CAS PubMed.
- D. W. Pack, A. S. Hoffman, S. Pun and P. S. Stayton, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS PubMed.
- T. G. Park, J. H. Jeong and S. W. Kim, Adv. Drug Delivery Rev., 2006, 58, 467–486 CrossRef CAS PubMed.
- Y. Fujita, K. Kuwano and T. Ochiya, Int. J. Mol. Sci., 2015, 16, 5254–5270 CrossRef CAS PubMed.
- D. Schaffert, C. Troiber and E. Wagner, Bioconjugate Chem., 2012, 23, 1157–1165 CrossRef CAS PubMed.
- H. Wang, Y. Jiang, H. Peng, Y. Chen, P. Zhu and Y. Huang, Adv. Drug Delivery Rev., 2015, 81, 142–160 CrossRef CAS PubMed.
- H. Lu, Y. Dai, L. Lv and H. Zhao, PLoS One, 2014, 9, 1–12 CrossRef CAS.
- M. Bezanilla, B. Drake, E. Nudler, M. Kashlev, P. K. Hansma and H. G. Hansma, Biophys. J., 1994, 67, 2454–2459 CrossRef CAS PubMed.
- O. Boussif, F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix and J.-P. Behr, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 7297–7301 CrossRef CAS.
- Y. Xu and F. C. Szoka, Biochemistry, 1996, 35, 5616–5623 CrossRef CAS PubMed.
- K. Zhu, C. Guo, H. Lai, W. Yang, Y. Xia, D. Zhao and C. Wang, J. Mater. Sci.: Mater. Med., 2011, 22, 2477–2485 CrossRef CAS PubMed.
- A. Akinc, M. Thomas, A. M. Klibanov and R. Langer, J. Gene Med., 2005, 7, 657–663 CrossRef CAS PubMed.
- A. Zintchenko, A. Philipp, A. Dehshahri and E. Wagner, Bioconjugate Chem., 2008, 19, 1448–1455 CrossRef CAS PubMed.
- D. Fischer, T. Bieber, Y. Li, H.-P. Elsasser and T. Kissel, Pharm. Res., 1999, 16, 1273–1279 CrossRef CAS.
- W. T. Godbey, K. K. Wu and A. G. Mikos, J. Biomed. Mater. Res., 1999, 45, 268–275 CrossRef CAS PubMed.
- H. Tian, Z. Tang, X. Zhuang, X. Chen and X. Jing, Prog. Polym. Sci., 2012, 37, 237–280 CrossRef CAS.
- E. Wagner and J. Kloeckner, in Polymer Therapeutics I, Springer, 2006, vol. 192, pp. 135–173 Search PubMed.
- C.-S. Cho, ISRN Mater. Sci., 2012, 2012, 1–24 CrossRef.
- G. Grandinetti, N. P. Ingle and T. M. Reineke, Mol. Pharm., 2011, 8, 1709–1719 CrossRef CAS PubMed.
- I. Teasdale and O. Bruggemann, Polymers, 2013, 5, 161–187 CrossRef PubMed.
- M. Ahuja, A. Kumar and K. Singh, Int. J. Biol. Macromol., 2012, 51, 1086–1090 CrossRef CAS PubMed.
- P. J. Manna, T. Mitra, N. Pramanik, V. Kavitha, A. Gnanamani and P. P. Kundu, Int. J. Biol. Macromol., 2015, 75, 437–446 CrossRef CAS PubMed.
- G. Panariello, R. Favaloro, M. Forbicioni, E. Caputo and R. Barbucci, Macromol. Symp., 2008, 266, 68–73 CrossRef CAS.
- X.-G. Chen and H.-J. Park, Carbohydr. Polym., 2003, 53, 355–359 CrossRef CAS.
- Z. Stojanovic, K. Jeremic, S. Jovanovic and M. D. Lechner, Starch-Starke, 2005, 57, 79–83 CrossRef CAS.
- L. Zhang, Z. Chen and Y. Li, Int. J. Nanomed., 2013, 8, 3689 CrossRef PubMed.
- T. Mitra, G. Sailakshmi, A. Gnanamani and A. B. Mandal, J. Appl. Polym. Sci., 2012, 125, E490–E500 CrossRef CAS.
- C. Hara, T. Kiho, Y. Tanaka and S. Ukai, Carbohydr. Res., 1982, 110, 77–87 CrossRef CAS PubMed.
- K. Sarkar and P. P. Kundu, Carbohydr. Polym., 2013, 98, 495–504 CrossRef CAS PubMed.
- H. Gong, M. Liu, J. Chen, F. Han, C. Gao and B. Zhang, Carbohydr. Polym., 2012, 88, 1015–1022 CrossRef CAS.
- S. Pal, D. Mal and R. P. Singh, J. Appl. Polym. Sci., 2007, 105, 3240–3245 CrossRef CAS.
- G. Dodi, D. Hritcu and M. I. Popa, Cellul. Chem. Technol., 2011, 45, 171–176 CAS.
- S.-C. Park, J.-P. Nam, Y.-M. Kim, J.-H. Kim, J.-W. Nah and M.-K. Jang, Int. J. Nanomed., 2013, 8, 3663–3677 Search PubMed.
- H. C. Kang, H.-J. Kang and Y. H. Bae, Biomaterials, 2011, 32, 1193–1203 CrossRef CAS PubMed.
- X. Zhao, Z. Li, H. Pan, W. Liu, M. Lv, F. Leung and W. W. Lu, Acta Biomater., 2013, 9, 6694–6703 CrossRef CAS PubMed.
- Y. Liu and T. M. Reineke, J. Am. Chem. Soc., 2005, 127, 3004–3015 CrossRef CAS PubMed.
- F.-J. Xu, H. Li, J. Li, Z. Zhang, E.-T. Kang and K.-G. Neoh, Biomaterials, 2008, 29, 3023–3033 CrossRef CAS PubMed.
- C. Yang, H. Li, S. H. Goh and J. Li, Biomaterials, 2007, 28, 3245–3254 CrossRef CAS PubMed.
- S. K. Tripathi, R. Goyal, P. Kumar and K. C. Gupta, Nanomedicine, 2012, 8, 337–345 CrossRef CAS PubMed.
- J.-P. Behr, Int. J. Chem., 1997, 51, 34–36 CAS.
- N. P. Gabrielson and D. W. Pack, Biomacromolecules, 2006, 7, 2427–2435 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra23447f |
| This journal is © The Royal Society of Chemistry 2016 |