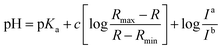Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A ratiometric two-photon probe for quantitative imaging of mitochondrial pH values†
Avik Ranjan
Sarkar‡
,
Cheol Ho
Heo‡
,
Lei
Xu
,
Hyo Won
Lee
,
Ho Young
Si
,
Ji Won
Byun
and
Hwan Myung
Kim
*
Department of Chemistry and Department of Energy Systems Research, Ajou University, Suwon 443-749, Korea. E-mail: kimhm@ajou.ac.kr
First published on 27th October 2015
Abstract
Mitochondrial pH (pHmito) is known to be alkaline (near 8.0) and has emerged as a potential factor for mitochondrial function and disorder. We have developed a ratiometric two-photon probe (CMP1) for quantitative analysis of pHmito in live cells and tissues. This probe is designed to function by controlling the intramolecular charge transfer from 2-naphthol, having an ideal pKa value (7.86 ± 0.05) in the cells to monitor pHmito. This transition results in a marked yellow to red emission color change in response to pH alterations from 6.0 to 9.0. CMP1 exhibits easy loading, selective and robust staining ability of mitochondria, low cytotoxicity, and bright two-photon excited fluorescence in situ, thereby allowing quantitative imaging of the pHmito in live cells and tissues. The ratiometric TPM imaging clearly reveals that subcellular distribution of the pHmito values is heterogeneous, with the pHmito values in the perinuclear region being higher than those at the periphery of the cells. The changes of pHmito values on carbonyl cyanide m-chlorophenyl hydrazone (CCCP) treatment and autophagic processes were also investigated along with their morphological alterations at specific subcellular positions. We also used CMP1 to visualize the pHmito values of Parkinson's disease model astrocytes as well as living hippocampal tissues. Our results demonstrate that CMP1 will be useful as a quantitative imaging probe to study pHmito in biomedical research.
Introduction
The mitochondrion is the primary organelle for oxygen utilization to generate bioenergy.1 While the cytosol and other organelles have neutral and acidic pH, the mitochondrial pH (pHmito) is alkaline (pH ∼ 8.0) in resting cells.2 Proton (H+) extrusion via the respiratory electron transport chain provides a proton motive force (ΨH+), which consists of a pH gradient across the mitochondrial membrane.3 Mitochondria have also been shown to dynamically change in their morphology, cellular location, and distribution. They constantly undergo remodeling through fission, fusion, biogenesis, and autophagy, as well as transport to specific sites with high energy demands.4 The disruption of mitochondrial dynamics could be directly linked to apoptosis, cancer, aging, and neurodegenerative disorders.5 As such, molecular imaging of mitochondrial dynamics could provide information in the study of human diseases and drug development.6 However, the variation of pHmito values along with their subcellular distribution, specific location, and morphological change has not been well established. Therefore, a method for quantitative imaging of pHmito in living cells and tissues holds promise to elucidate mitochondria-related physiology and diseases.To target the pHmito, pH-sensitive green fluorescent protein (GFP) mutants have been developed and successfully utilized to confirm the alkaline pH of the mitochondria.7 However, this approach suffers from variations in different cell types and technical difficulties when used with live tissues and animals.8 Recently, small-molecule probes for pHmito based on the fluorescence off-on response have been reported.9 However, these probes utilized a single detection window and/or double excitation source that limit quantitative measurements due to the presence of experimental artifacts such as probe distribution, incident laser power, detection sensitivity, and photobleaching. Moreover, the pKa values of these probes are in the acidic-neutral range (6.2–7.4), which is far from the ideal measurement of pHmito (pH ∼ 8.0). In addition, live cell imaging with these probes required short excitation wavelengths that can cause photodamage and the artificial production of reactive oxygen species (ROS).10
Two-photon microscopy (TPM) is a fast growing domain in biomedical research, because it utilizes two near IR photons and offers higher spatial resolution with minimum background emission, deeper tissue penetration depth, and longer observation time.11 In combination with TPM, an emission ratiometric probe with a single excitation wavelength would be an ideal method for quantitative imaging analysis of pHmito. The emission ratio simultaneously reflects the population of the protonated and deprotonated forms of the fluorescent emitter, which can cancel out the aforementioned experimental artefacts. Recently, a variety of small-molecule TP probes and their utilities in bioimaging applications have been reported.12–16 However, no ratiometric TP probe for pHmito have been exploited. Hence, there is a strong need to develop an emission ratiometric TP probe for pHmito with a pKa value near 8.0.
Recently, we have developed a benzimidazole-derived ratiometric TP probe (pKa = 5.82) for acidic pH detection, which has an electron donor–acceptor substituted dipolar character.17 The probe showed red-shifts in its spectrum upon protonation of the benzimidazole as a weak-base, owing to the increased electron withdrawing ability and the enhanced intramolecular charge transfer (ICT). However, this approach, utilizing protonation of a weak base, makes it difficult to modify the pKa to a value near 8.0. Thus, there is need to employ a weak acid for development of pHmito sensitive probe. In this work, we report a 2-naphthol-derived ratiometric TP probe (CMP1, Scheme 1) for pHmito that can quantitatively monitor subcellular pHmito values in living cells and tissues.
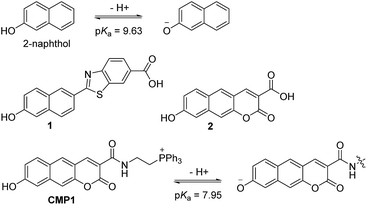 | ||
| Scheme 1 Structures of 2-naphthol, 1, 2, and CMP1, and proposed mechanism of the equilibrium on 2-naphthol and CMP1 and their deprotonated form. | ||
Results and discussion
We initiated this study using 2-naphthol, which has a pKa value of 9.63 and shows a red-shifted emission maximum with pH change from acidic to basic (357 to 409 nm).18 The spectral red-shift under basic conditions can be attribute to an enhanced ICT process due to the stronger electron donating ability of the deprotonated form (O−) than the OH (σ+ = −2.3 for O−vs. −0.92 for OH) (Scheme 1).19 We anticipated that the pKa value of 2-naphthol derivatives could be optimized to the mitochondrial pH by employing an electron withdrawing group and extended π-conjugated system. These translations would stabilize the conjugated base (O−), giving a lower pKa value than that of 2-naphthol, along with emission ratiometric character. Therefore, we firstly prepared compound 1 containing a benzothiazolyl electron withdrawing group (Scheme 1). Benzochromene-2-one derivative (2) was also synthesized as a linear π-conjugating system with the expectation of more effective ICT than 1.14b The synthetic route for these compounds is described in the ESI.†Photophysical properties of 1 and 2 were measured in buffer solutions of different pH. Under acidic conditions (4.0), 1 displayed an absorption maximum (λabs) at 344 nm and a fluorescence emission maximum (λfl) at 480 nm with a large Stokes shift of 136 nm (Table 1). When the pH was changed from acidic (4.0) to basic (10.0), the λabs and λfl of 1 were shifted to the longer wavelengths at 380 nm and 530 nm, respectively, and with an increased fluorescence quantum yield (Fig. 1a and Table 1). These red-shifts might reflect the presence of the phenolate form at basic pH, as a stronger electron donor than the phenol form, thereby boosting the ICT processes. The pKa value was calculated from the titration curve of the emission ratios (Ibasic/Iiso) at the isoemission point (Iiso) and λfl at pH 10.0 (Ibasic). The pKa value for 1 was 9.04 ± 0.07, which is a markedly reduced value as compared to 2-naphthol. At pH 4.0, 2 showed more red-shifted λabs and λfl than those for 1, presumably due to the enhanced ICT. As the pH of the solution was changed from acidic to basic, the intensity of fluorescence of 2 at 520 nm was gradually decreased with a simultaneous increase in intensity at 585 nm (Fig. 1b). The pKa value of 2 is found to be 8.41 ± 0.07, lower than the value for 1.
| Probe | pHa | λ (1)max (10−4ε) | λ flmax | Φ | pKae | λ (2)max | δ |
|---|---|---|---|---|---|---|---|
| a All the measurements were performed in buffer solution. 0.1 M KH phthalate buffer solution (pH 4.0) and 0.025 M sodium borate buffer solution (pH 10.0). b λ max of the one-photon absorption spectra in nm. The numbers in parentheses are molar extinction coefficients in M−1 cm−1. c λ max of the one-photon emission spectra in nm. d Fluorescence quantum yield. e pKa values measured by one-photon mode. f λ max of the two-photon excitation spectra in nm. g Two-photon absorption cross-section in 10−50 cm4 s per photon (GM). | |||||||
| 1 | 4.0 | 344 (1.14) | 479 | 0.29 | 9.04 ± 0.07 | 740 | 22 |
| 10.0 | 380 (1.61) | 530 | 0.40 | 750 | 69 | ||
| 2 | 4.0 | 366 (1.25) | 520 | 0.090 | 8.41 ± 0.07 | 750 | 24 |
| 10.0 | 412 (1.28) | 585 | 0.074 | 910 | 50 | ||
| CMP1 | 4.0 | 370 (1.35) | 542 | 0.12 | 7.95 ± 0.04 | 750 | 32 |
| 10.0 | 453 (1.64) | 604 | 0.084 | 910 | 62 | ||
Next, we utilized 2 to prepare the mitochondria targeting probe (CMP1) by attaching triphenylphosphonium salt (TPP), a well-known mitochondria staining anchor,20 through an amide linkage (Scheme 1). Upon changing the pH from acidic to basic, the spectral shifts for CMP1 were nearly identical to those of 2, except that both λabs and λfl of CMP1 and the phenolate form were shifted to the longer wavelength range (Fig. 1c, Table 1). These observations are likely due to the combined effects of the stronger electron withdrawing ability of the amide group than the carboxyl group (σ− = 0.31 for CO2−vs. 0.61 for CONH2)19 and the inductive effect of the phosphonium cation. Consistently, the pKa value of CMP1 is 7.95 ± 0.05, a lower value than for 2 and well-matched to mitochondrial pH (8.0). Further, the plot of the emission ratio with pH indicated that this probe would be suitable for detecting over the pH range of 6.0–9.0 (Fig. 1d). The ratio (I604/I540) of CMP1 was robustly cycled back and forth in the pH changes between pH 5.0 and 9.0 (Fig. S8, ESI†), indicating the reversible sensing ability. In addition, the ratio of CMP1 in buffer solutions was unperturbed in the presence of biologically relevant metal ions, thiol species, ROS, reactive nitrogen species, and hydrogen sulfide (Fig. S9, ESI†). Consequently, CMP1 showed a marked yellow-to-red emission color change in response to a change in pH from acidic to basic and an appropriate pKa value for assessing the change of pHmito without interference from other physiological species.
We then tested the ability of CMP1 as a TPM imaging probe. The maximum two-photon absorption cross section (δmax) values of 1, 2, and CMP1 at pH 4.0 and 10.0 were in the range of 20–70 GM, which are comparable to the values of existing small-molecule TP probes (Table 1 and Fig. S10, ESI†).12b Further, the TPM images of the CMP1-labeled HeLa cells and primary cultured astrocytes showed bright tubule-like networks, which might be the mitochondria. This probe displayed high photostability under the imaging conditions (Fig. S11, ESI†) and low cytotoxicity as determined by MTS and CCK-8 assays (Fig. 2).
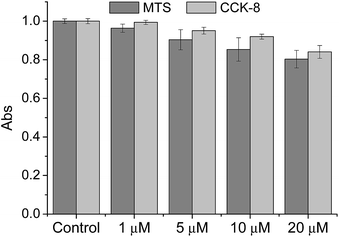 | ||
| Fig. 2 Viability of HeLa cells in the presence of CMP1 as measured by using MTS and CCK-8 assays. The cells were incubated with 0–20 μM CMP1 for 2 h. | ||
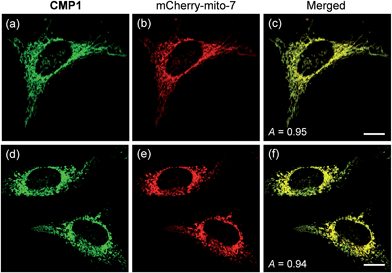
To demonstrate the mitochondrial staining ability of the probe, we conducted co-localization experiments with mCherry-mito-7, a well-known red-emissive protein for mitochondria.21 The TPM image of CMP1 merged well with the mCherry-mito-7 image (Fig. 3a–c). The Pearson's co-localization coefficient (A) was 0.95 ± 0.05, which indicated that CMP1 predominantly resided in the mitochondria. A similar result was also obtained when the co-localization experiment was performed with the molecular marker, MitoTracker Red (A = 0.88 ± 0.05) (Fig. S12, ESI†). Upon treatment of CCCP (carbonyl cyanide m-chlorophenyl hydrazone), a protonophore that promotes the release of intra-mitochondrial cations and causes rapid acidification by collapsing the mitochondrial membrane potential (ΔΨm),22 the A values between the images of CMP1 and mCherry-mito-7 were found to be 0.94 ± 0.05 (Fig. 3d–f). Because it has been well-established that the staining ability of mCherry-mito-7 does not depend on the ΔΨm,21 this result confirmed the robust staining ability of CMP1 for the mitochondria regardless of the alteration of ΔΨm (Fig. 3).
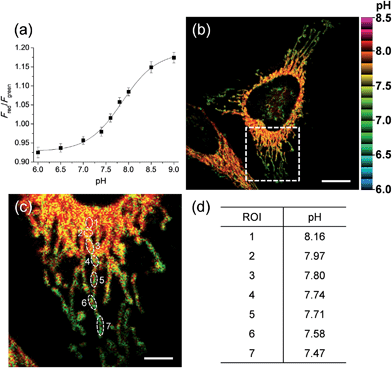
We next tested the probe to determine the pHmito value in live cells using TPM. Live cell imaging was carried out at 37 °C using an incubating chamber for maintaining humidity and pH. To find the optimal detection window for ratiometric imaging, the TP excited fluorescence (TPEF) spectra were collected from ionophore-treated HeLa cells labeled with CMP1 at pH 4.0 and 10.0. Upon excitation at 820 nm, the TPEF spectra at pH 4.0 and pH 10.0 were similar to those measured in buffer solution, in which the λfl maxima (505 and 569 nm at pH 4.0 and 10.0) are slightly blue-shifted from those measured in buffer solution (Fig. S13, ESI†). These results indicated that the polarity of the probe environment within the cells was rather homogeneous and slightly more hydrophobic than in the buffer solution.17 This consequence allowed ratiometric imaging (Fred/Fgreen) for pHmito by using 450–500 nm (Fgreen) and 550–600 nm (Fred) as most sensitive windows. The pH calibration curve was obtained by using Fred/Fgreen of ionophore-treated and CMP1-labeled HeLa cells (Fig. 4a). The pKa of CMP1 in cells was 7.86 ± 0.05, a nearly identical value as that measured in buffer solution. In addition, the plots of Fred/Fgreenvs. pH value indicated that CMP1 is suitable for measuring the pHmito in the range of pH 6.0–9.0.
More importantly, the ratiometric TPM image (Fred/Fgreen) of CMP1-labeled HeLa cells clearly displayed various pHmito values ranging from 7.4 to 8.2, indicating the heterogeneity of pHmito (Fig. 4b and c). In addition, there was a denser population of mitochondria around the nucleus than in the periphery. Interestingly, Fig. 4b–d shows that the mitochondria in the perinuclear position had higher pH values in comparison to mitochondria at the periphery of cells. This difference might reflect a mitochondrial region-specific function.23 It has been well established that perinuclear mitochondria are mainly involved in mitochondria-metabolized ATP generation by regulating nuclear function by the assimilated phosphotransfer network between mitochondria and the nucleus.23b The higher pH values in the perinuclear region may be linked to the local metabolic state, thus indicating to the need for future studies to elucidate its physiological function. Similar results were observed with primary cultured astrocytes (Fig. 7).
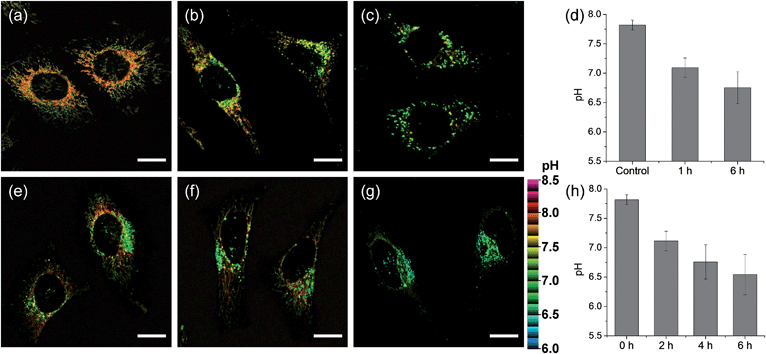
We next monitored the change of pHmito with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) treatments and under starvation conditions. The average value of pHmito of CMP1-labeled HeLa cells was 7.82 ± 0.08. Upon treatment with CCCP for 1 h and 6 h, the average values of pHmito decreased to 7.09 ± 0.16 and 6.75 ± 0.27, respectively (Fig. 5a–d). In addition, the ratio TPM imaging clearly displayed that the perinuclear mitochondria preferentially adopted a globular morphology with acidic pH, compared to the peripheral mitochondria (Fig. 5b and c) and the process extended to the periphery. Moreover, higher magnification images clearly showed the pH values of globular structures ranged from 7.40 to 6.36 (Fig. 5c), which were significantly decreased values by 0.7–1.8 pH units as compared to those for long and highly interconnected structures. Similar results were detected in the cell starvation process. It is well-established that the damaged mitochondria can be eliminated by cell degradation processes, i.e., autophagy, a process in which mitochondria that have attained an acidic pH are entrapped in acidic lysosomes followed by hydrolytic digestion.24 Upon nutrient deprivation for 2–6 h (Fig. 5e–h), a globular morphology was noted in mitochondria near the nucleus appeared and this population of altered mitochondria increased with time; during this time, the pH values ranged from 7.1 to 6.2 (Fig. 5e–g).
To confirm these processes, HeLa cells were co-labeled with CMP1 and BLT-blue,25 a TP LysoTracker that emits separate TPEF from CMP1 in the cells (Fig. S14, ESI†). The dual-color TPM images (Fig. 6) clearly exhibited the autophagy processes. The Pearson's co-localization coefficient (A) was found to be 0.14 ± 0.5 before stimulating the cells (Fig. 6a, e and i). When the cells were starved in nutrient-deprived buffer solution, the A value increased to 0.24 to 0.38 to 0.55 after starvation for 2 h, 4 h and 6 h, respectively (Fig. 6). Hence, these outcomes clearly demonstrated that CMP1 is capable of monitoring the change of pHmito values in live cells while defining the dynamic shapes of mitochondria at specific positions.
We further utilized CMP1 to measure the pHmito values in Parkinson's disease (PD) model astrocytes such as DJ-1, Parkin, and Pink gene knockout (KO) astrocytes. These astrocytes contain a PD-related gene that has multiple functions. The ratiometric TPM imaging with CMP1 exhibited the average pHmito value of 7.78 ± 0.09 in wild-type (WT) astrocytes (Fig. 7a). Moreover, the distribution of pHmito were similar between PD models and WT astrocytes, with the exception that the average pHmito values of PD models are slightly lower (ca. 0.1–0.2 pH unit) than those for WT astrocytes (Fig. 7b–d). This feature suggests the potential use of CMP1 in clinical applications.
Finally, we investigated the utility of CMP1 in live tissue imaging. The ratiometric TPM images were obtained from a fresh slice of rat hippocampus, a region of the brain that is important for learning and memory. The accumulated 360 TPM images collected at 90–210 μm depths provided the overall pHmito distribution through the regions of CA1, CA3, and DG (dentate gyrus) (Fig. 8). They showed the average pHmito values of these regions in 7.86–7.88 (Fig. 8), in which higher pH values of approximately 8.2 (red spots) are mainly appeared in the DG (Fig. 8a and d). Moreover, the higher magnification images at these regions revealed the heterogeneous pHmito distribution at the tissue level.
Conclusions
We have developed a ratiometric two-photon probe (CMP1) for quantitative measurement of pHmito in live cells and tissues. This probe, designed with the aim of controlling ICT in 2-naphthol, has a pKa value of 7.86 ± 0.05 in the cells, and shows a marked yellow to red emission color change in response to pH alterations from 6.0 to 9.0. CMP1 exhibits easy cell loading, robust staining ability of mitochondria, low cytotoxicity, and bright TPEF in situ, thereby allowing quantitative detection of the pHmito in live cells and tissues. The ratiometric TPM imaging studies by using CMP1 clearly reveal the heterogeneity of pHmito values with respect to the specific location of mitochondria within the cells, along with their different shapes. Further, we found that the pHmito values of mitochondria in the perinuclear region are higher than those at the periphery of cells. The average pHmito values of PD model astrocytes are slightly more acidic than those for WT astrocytes. These findings demonstrate that CMP1 will be useful as a quantitative imaging probe to study pHmito in biomedical research.Experimental sections
Spectroscopic measurements
Absorption spectra were recorded on S-3100 UV-Vis spectrophotometer and fluorescence spectra were obtained with FluoroMate FS-2 fluorescence spectrophotometer with a 1 cm standard quartz cell. The fluorescence quantum yield was determined by using Coumarin 307 (Φ = 0.95 in MeOH) and Rhodamine 6G (Φ = 0.95 in MeOH) as the reference by the literature method.26pKa value
Fluorescence pH titrations were performed in buffer solution [pH 3.0, pH 4.0, pH 5.0 (0.1 M KH phthalate buffer solution), pH 6.0, pH 6.5, pH 7.0, pH 7.2, pH 7.5, pH 7.8, pH 8.0, pH 8.2, pH 8.5 and pH 8.8 (0.1 M KH2PO4 buffer solution), pH 9.0, pH 9.2, pH 9.5, pH 9.8 and pH 10.0 (0.025 M sodium borate buffer solution), pH 11.0 (0.05 M NaHCO3 buffer solution)]. For 1, 2 and CMP1, a 3.0 μL of the stock solution of probe in DMSO (1 × 10−3 M) was added to cuvette containing 3.0 mL of buffer solution to prepared 1.0 μM of probe solution and the spectral change in the fluorescence were measured as a function of pH (3.0–11.0), pKa values of 1, 2 and CMP1 were calculated by linear regression analysis fluorescence data to fit the following equation.27where R is the observed ratios (I529/Iiso for 1, I587/Iiso for 2 and I604/Iiso for CMP1, respectively) at given pH. Rmax and Rmin are the maximum and minimum limiting values of R and c is the slope. Ia/Ib is the ratio of the fluorescence intensity in acid (pH 3.0) to the intensity in the base (pH 11.0) at the wavelength chosen for the denominator of R. In these cases, this correction vanishes by using the sharp isoemissive point (472 nm for 1, 554 nm for 2 and 572 nm for CMP1).
Measurement of two-photon cross section
The two-photon cross section (δ) was determined by using femto second (fs) fluorescence measurement technique as described.281, 2 and CMP1 (1.0 × 10−6 M) were dissolved in buffer solution (pH 4.0 and 10.0) and the two-photon induced fluorescence intensity was measured at 720–950 nm by using Rhodamine 6G as the reference, whose two-photon property has been well characterized in the literature.29 The intensities of the two-photon induced fluorescence spectra of the reference and sample emitted at the same excitation wavelength were determined. The TPA cross section was calculated by using following equationwhere the subscripts s and r stand for the sample and reference molecules. The intensity of the signal collected by a CCD detector was denoted as S. Φ is the fluorescence quantum yield. φ is the overall fluorescence collection efficiency of the experimental apparatus. The number density of the molecules in solution was denoted as c. δr is the TPA cross section of the reference molecule. Fig. S9, ESI† represents the two-photon spectra of 1, 2 and CMP1 in different buffer solution conditions.
Cell culture
All the cells were passed and plated on glass-bottomed dishes (NEST) before imaging for two days. They were maintained in a humidified atmosphere of 5/95 (v/v) of CO2/air at 37 °C. The cells were treated and incubated with 2 μM CMP1 at 37 °C under 5% CO2 for 30 min, washed three times with phosphate buffered saline solution (PBS; Gibco), and then imaged after further incubation in colorless serum-free media for 30 min. The culture mediums for each cells are as below.HeLa human cervical carcinoma cells (ATCC, Manassas, VA, USA): MEM (WelGene Inc, Seoul, Korea) supplemented with 10% FBS (WelGene), penicillin (100 units per mL), and streptomycin (100 μg mL−1). Primary astrocytes were cultured from the cortex of DJ-1-KO, Parkin-KO or WT mice brains. In brief, cortexes were removed and triturated in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% FBS (HyClone, Logan, UT, USA), plated in 75 cm2 T-flasks (0.5 hemisphere per flask), and incubated for 2–3 weeks. Microglia were detached from flasks by mild shaking, filtered through a nylon mesh to remove cell clumps, and cultured in DMEM containing 10% FBS.30 Astrocytes remaining in the flask were harvested with 0.1% trypsin and cultured in DMEM containing 10% FBS.
Two-photon fluorescence microscopy
Two-photon fluorescence microscopy images of CMP1 labeled cells and tissues were obtained with spectral confocal and multiphoton microscopes (Leica TCS SP8 MP) with ×10 dry, ×40 oil and ×100 oil objectives, numerical aperture (NA) = 0.30, 1.30, and 1.30. The two-photon fluorescence microscopy images were obtained with a DMI6000B Microscope (Leica) by exciting the probes with a mode-locked titanium-sapphire laser source (Mai Tai HP; Spectra Physics, 80 MHz, 100 fs) set at wavelength 820 nm and output power 2901 mW, which corresponded to approximately 6.26 × 108 mW cm−2 average power in the focal plane. Live cell imaging was performed using the live cell incubator systems (Chamlide IC; Live Cell Instrument) for stable cell environment by maintaining proper temperature, humidity and pH over the long term. To obtain images at 450–500 nm (Fgreen) and 550–600 nm (Fred) range (Fig. S13a, ESI†) internal PMTs were used to collect the signals in an 8 bit unsigned 512 × 512 and 1024 × 1024 pixels at 400 and 200 Hz scan speed, respectively. Ratiometric image processing and analysis was carried out using MetaMorph software.Cell viability
To evaluate the cytotoxic effect of CMP1 in HeLa cells, MTS (cell Titer 96H; Promega, Madison, WI, USA) and CCK-8 kit (Cell Counting Kit-8; Dojindo, Japan) assay were performed according to the manufacture's protocol. The results are shown in Fig. 2, which revealed that the CMP1 has low cytotoxicity at its different concentration in our incubation condition.Co-localization experiments
Co-localization experiments were conducted by co-staining the HeLa cells and astrocytes with appropriate combinations of CMP1 (2 μM), MTR (1 μM), mCherry-mito-7 and BLT-blue (0.5 μM) for 30 min. TPM and OPM images were obtained by collecting the emissions at 400–450 (BLT-blue), 450–500 and 550–600 (CMP1) and 650–700 (MTR, mCherry-mito-7), respectively. The two different detection windows for CMP1 were adopted upon requirement for colocalization experiments. The background images were corrected, and the distribution of pixels in the OPM and TPM images acquired in the green and red channels, respectively, was compared by using scatter gram. The Pearson's co-localization coefficients (A) were calculated by using AutoQuant X2 program.Cell calibration
A pH calibration curve was generated by Fred/Fgreen of ionophores-treated and CMP1-labeled HeLa cells. The cells were treated and incubated with 2 μL of CMP1 in DMSO sock solution (1.0 mM) at 37 °C under 5% CO2 for 30 min, and then the extracellular media was replaced with 1 mL of calibration buffer (125 mM KCl, 20 mM NaCl, 0.5 mM CaCl2, 0.5 mM MgCl2, 5 μM nigericine, 5 μM monensin, and 25 mM buffer; MES for pH 6.0 and 6.5, HEPES for pH 6.5–8.0 and Tris for pH 8.5–9.0).7a The cells were treated with the calibration buffer for 15–20 min at room temperature. The TPEF intensity at 450–500 nm (Fgreen) and 550–600 nm (Fred) of CMP1 (Fig. S13a, ESI†) well changed with pH, and we obtained pH calibration curve the plots of Fred/Fgreenversus pH values.Preparation and staining of fresh rat hippocampal slices
Rat Hippocampal slice were prepared from the hippocampi of 2 weeks-old rat (SD) according to an approved institutional review board protocol. Coronal slices were cut into 400 μM-thicks using a vibrating-blade microtome in artificial cerebrospinal fluid (ACSF; 138.6 mM NaCl, 3.5 mM KCl, 21 mM NaHCO3, 0.6 mM NaH2PO4, 9.9 mM D-glucose, 1 mM CaCl2, and 3 mM MgCl2). Slices were incubated with 20 μM CMP1 in ACSF bubbled with 95% O2 and 5% CO2 for 1 h at 37 °C. Slices were then washed three times with ACSF and transferred to glass-bottomed dishes (NEST) and observed in a spectral confocal multiphoton microscope. The TPM images obtained at about 90–210 μm depth.Acknowledgements
We thank Prof. Sang Myun Park (Department of Pharmacology, Ajou University School of Medicine) for the supply of PD model astrocytes. This work was supported by National Research Foundation (NRF) grants funded by the Korean Government (2012R1A2A1A03670456).Notes and references
- V. G. Antico Arciuch, M. E. Elguero, J. J. Poderoso and M. C. Carreras, Antioxid. Redox Signaling, 2012, 16, 1150 CrossRef CAS PubMed.
- J. R. Casey, S. Grinstein and J. Orlowski, Nat. Rev. Mol. Cell Biol., 2010, 11, 50 CrossRef CAS PubMed.
- (a) J. M. Herrmann and W. Neupert, Curr. Opin. Microbiol., 2000, 3, 210 CrossRef CAS PubMed; (b) N. Kim, M. O. Ripple and R. Springett, Biophys. J., 2012, 102, 1194 CrossRef CAS PubMed; (c) J. Santo-Domingo and N. Demaurex, J. Gen. Physiol., 2012, 139, 415 CrossRef CAS PubMed.
- (a) D. A. Rube and A. M. van der Bliek, Mol. Cell. Biochem., 2004, 256–257, 331 CrossRef PubMed; (b) D. T. Chang, A. S. Honick and I. J. Reynolds, J. Neurosci., 2006, 26, 7035 CrossRef CAS PubMed; (c) C. M. Peterson, D. L. Johannsenand and E. Ravussin, Journal of Aging Research, 2012, 2012, 194821 CrossRef PubMed.
- (a) D. F. Suen, K. L. Norris and R. J. Youle, Genes Dev., 2008, 22, 1577 CrossRef CAS PubMed; (b) D. C. Wallace, Nat. Rev. Cancer, 2012, 12, 685 CrossRef CAS PubMed; (c) A. Y. Seo, A. M. Joseph, D. Dutta, J. C. Hwang, J. P. Aris and C. Leeuwenburgh, J. Cell Sci., 2010, 123, 2533 CrossRef CAS PubMed; (d) M. T. Lin and M. F. Beal, Nature, 2006, 443, 787 CrossRef CAS PubMed; (e) P. H. Reddy and M. F. Beal, Trends Mol. Med., 2008, 14, 45 CrossRef CAS PubMed; (f) B. C. Dickinson and C. J. Chang, Nat. Chem. Biol., 2011, 7, 504 CrossRef CAS PubMed.
- (a) J. S. Armstrong, Br. J. Pharmacol., 2007, 151, 1154 CrossRef CAS PubMed; (b) A. M. Walters, G. A. Porter and P. S. Brookes Jr, Circ. Res., 2012, 111, 1222 CrossRef CAS PubMed.
- (a) J. Llopis, J. M. McCaffery, A. Miyawaki, M. G. Farquhar and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6803 CrossRef CAS PubMed; (b) S. Matsuyama, J. Llopis, Q. L. Deveraux, R. Y. Tsien and J. C. Reed, Nat. Cell Biol., 2000, 2, 318 CrossRef CAS PubMed; (c) M. F. Abad, G. Di Benedetto, P. J. Magalhaes, L. Filippin and T. Pozzan, J. Biol. Chem., 2004, 279, 11521 CrossRef CAS PubMed; (d) R. de Michele, F. Carimi and W. B. Frommer, Int. J. Biochem. Cell Biol., 2014, 48, 39 CrossRef CAS PubMed.
- M. Zimmer, Chem. Rev., 2002, 102, 759 CrossRef CAS PubMed.
- (a) M. H. Lee, N. Park, C. Yi, J. H. Han, J. H. Hong, K. P. Kim, D. H. Kang, J. L. Sessler, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2014, 136, 14136 CrossRef CAS PubMed; (b) P. Li, H. Xiao, Y. Cheng, W. Zhang, F. Huang, W. Zhang, H. Wang and B. Tang, Chem. Commun., 2014, 50, 7184 RSC; (c) Y. Chen, C. Zhu, J. Cen, Y. Bai, W. He and Z. Guo, Chem. Sci., 2015, 6, 3187 RSC.
- M. J. Jou, S. B. Jou, M. J. Guo, H. Y. Wu and T. I. Peng, Ann. N. Y. Acad. Sci., 2004, 1011, 45 CrossRef CAS PubMed.
- (a) W. R. Zipfel, R. M. Williams and W. W. Webb, Nat. Biotechnol., 2003, 21, 1369 CrossRef CAS PubMed; (b) F. Helmchen and W. Denk, Nat. Methods, 2005, 2, 932 CrossRef CAS PubMed.
- (a) H. M. Kim and B. R. Cho, Chem. Rev., 2015, 115, 5014 CrossRef CAS PubMed; (b) H. M. Kim and B. R. Cho, Acc. Chem. Res., 2009, 42, 863 CrossRef CAS PubMed.
- (a) Z. Mao, L. Hu, X. Dong, C. Zhong, B. F. Liu and Z. Liu, Anal. Chem., 2014, 86, 6548 CrossRef CAS PubMed; (b) L. Zhou, X. Zhang, Q. Wang, Y. Lv, G. Mao, A. Luo, Y. Wu, Y. Wu, J. Zhang and W. Tan, J. Am. Chem. Soc., 2014, 136, 9838 CrossRef CAS PubMed.
- (a) X. Wang, D. M. Nguyen, C. O. Yanez, L. Rodriguez, H. Y. Ahn, M. V. Bondar and K. D. Belfield, J. Am. Chem. Soc., 2010, 132, 12237 CrossRef CAS PubMed; (b) A. R. Sarkar, C. H. Heo, H. W. Lee, K. H. Park, Y. H. Suh and H. M. Kim, Anal. Chem., 2014, 86, 5638 CrossRef CAS PubMed.
- (a) L. Li, C. W. Zhang, G. Y. Chen, B. Zhu, C. Chai, Q. H. Xu, E. K. Tan, Q. Zhu, K. L. Lim and S. Q. Yao, Nat. Commun., 2014, 5, 3276 Search PubMed; (b) H. Zhang, J. Fan, J. Wang, S. Zhang, B. Dou and X. Peng, J. Am. Chem. Soc., 2013, 135, 11663 CrossRef CAS PubMed; (c) H. W. Lee, C. H. Heo, D. Sen, H. O. Byun, I. H. Kwak, G. Yoon and H. M. Kim, Anal. Chem., 2014, 86, 10001 CrossRef CAS PubMed.
- (a) L. Yuan, W. Lin, H. Chen, S. Zhu and L. He, Angew. Chem., Int. Ed., 2013, 52, 10018 CrossRef CAS PubMed; (b) C. S. Lim, G. Masanta, H. J. Kim, J. H. Han, H. M. Kim and B. R. Cho, J. Am. Chem. Soc., 2011, 133, 11132 CrossRef CAS PubMed; (c) H. Y. Ahn, K. E. Fairfull-Smith, B. J. Morrow, V. Lussini, B. Kim, M. V. Bondar, S. E. Bottle and K. D. Belfield, J. Am. Chem. Soc., 2012, 134, 4721 CrossRef CAS PubMed; (d) P. Li, W. Zhang, K. Li, X. Liu, H. Xiao, W. Zhang and B. Tang, Anal. Chem., 2013, 85, 9877 CrossRef CAS PubMed; (e) Y. Hu, C. H. Heo, G. Kim, E. J. Jun, J. Yin, H. M. Kim and J. Yoon, Anal. Chem., 2015, 87, 3308 CrossRef CAS PubMed; (f) Q. Xu, C. H. Heo, G. Kim, H. W. Lee, H. M. Kim and J. Yoon, Angew. Chem., Int. Ed., 2015, 54, 4890 CrossRef CAS PubMed.
- H. J. Kim, C. H. Heo and H. M. Kim, J. Am. Chem. Soc., 2013, 135, 17969 CrossRef CAS PubMed.
- (a) D. R. Lide, The Handbook of Chemistry and Physics, CRC Press, Boca Raton, FL, 89th edn, 2008 Search PubMed; (b) J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, New York, 3rd edn, 2006 Search PubMed.
- (a) C. Hansch, A. Leo and R. W. Taft, Chem. Rev., 1991, 91, 165 CrossRef CAS; (b) J. Hines, Structural Effects on Equilibria in Organic Chemistry, John & Wiley, New York, 1975 Search PubMed.
- (a) M. P. Murphy and R. A. Smith, Annu. Rev. Pharmacol. Toxicol., 2007, 47, 629 CrossRef CAS PubMed; (b) G. F. Kelso, C. M. Porteous, C. V. Coulter, G. Hughes, W. K. Porteous, E. C. Ledgerwood, R. A. Smith and M. P. Murphy, J. Biol. Chem., 2001, 276, 4588 CrossRef CAS PubMed; (c) L. F. Yousif, K. M. Stewart and S. O. Kelley, ChemBioChem, 2009, 10, 1939 CrossRef CAS PubMed.
- (a) N. C. Shaner, R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer and R. Y. Tsien, Nat. Biotechnol., 2004, 22, 1567 CrossRef CAS PubMed; (b) T. D. MacVicar and J. D. Lane, J. Cell Sci., 2014, 127, 2313 CrossRef CAS PubMed.
- T. Caporale, D. Ciavardelli, C. Di Ilio, P. Lanuti, D. Drago and S. L. Sensi, Exp. Neurol., 2009, 218, 228 CrossRef CAS PubMed.
- (a) A. V. Kuznetsov and R. Margreiter, Int. J. Mol. Sci., 2009, 10, 1911 CrossRef CAS PubMed; (b) P. P. Dzeja, R. Bortolon, C. Perez-Terzic, E. L. Holmuhaedov and A. Terzic, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 10156 CrossRef CAS PubMed.
- R. J. Youle and D. P. Narendra, Nat. Rev. Mol. Cell Biol., 2009, 12, 9 CrossRef PubMed.
- J. H. Han, S. K. Park, C. S. Lim, M. K. Park, H. J. Kim, H. M. Kim and B. R. Cho, Chem.–Eur. J., 2012, 18, 15246 CrossRef CAS PubMed.
- (a) J. N. Demas and G. A. Crosby, J. Phys. Chem., 1971, 75, 991 CrossRef; (b) A. M. Brouwer, Pure Appl. Chem., 2011, 83, 2213 CrossRef CAS.
- (a) T. Myochin, K. Kiyose, K. Hanaoka, H. Kojima, T. Terai and T. Nagano, J. Am. Chem. Soc., 2011, 133, 3401 CrossRef CAS PubMed; (b) J. E. Whitaker, R. P. Haugland and F. G. Prendergast, Anal. Biochem., 1991, 194, 330 CrossRef CAS PubMed.
- S. K. Lee, W. J. Yang, J. J. Choi, C. H. Kim, S. J. Jeon and B. R. Cho, Org. Lett., 2005, 7, 323–326 CrossRef CAS PubMed.
- N. S. Makarov, M. Drobizhev and A. Rebane, Opt. Express, 2008, 16, 4029 CrossRef CAS PubMed.
- H. Pyo, I. Jou, S. Jung, S. Hong and E. H. Joe, NeuroReport, 1998, 9, 871 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis, additional methods, and figures (Fig. S1–S14). See DOI: 10.1039/c5sc03708e |
| ‡ These authors equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2016 |