Open Access Article
Open Access ArticleCarbohydrate microarrays for screening functional glycans†
Jaeyoung
Pai‡
a,
Ji Young
Hyun‡
a,
Jieun
Jeong
a,
Sohee
Loh
b,
Eun-Hee
Cho
b,
Young-Sun
Kang
b and
Injae
Shin
*a
aCenter for Biofunctional Molecules, Department of Chemistry, Yonsei University, Seoul 03722, Korea. E-mail: injae@yonsei.ac.kr
bDepartment of Biomedical Science and Technology, Department of Veterinary Pharmacology and Toxicology, College of Veterinary Medicine, Konkuk University, Seoul 05029, Korea
First published on 5th January 2016
Abstract
Carbohydrate microarrays have become robust and powerful tools for the rapid analysis of glycan-associated binding events. However, this microarray technology has rarely been applied in studies of glycan-mediated cellular responses. Herein we describe a carbohydrate microarray-based approach for the rapid screening of biologically active glycans that stimulate the production of reactive oxygen species (ROS) through binding to the cell-surface lectin. We employed a microarray assay and a fluorescent ROS probe to identify the functional glycans which enhance ROS production. Cells binding to glycans on the microarrays produced ROS, whose levels were decreased in the presence of a ROS scavenger or a NADPH oxidase inhibitor. The present study leads us to suggest that glycan microarrays are applicable to the simultaneous screening of various glycans whose binding to the cell-surface lectin elicits cellular response.
Introduction
Through binding to free glycans or glycoconjugates, animal cell surface lectins are involved in a wide range of biological processes.1 It is known that most animal lectins, including selectins, siglecs (sialic acid-binding immunoglobulin-type lectins), mannose receptors, mannose-6-phosphate receptors, asialoglycoprotein receptors and C-type lectins act as signal transducers after binding to glycans, although intracellular signaling events induced by lectins depend on the nature of glycans, cell type and/or species.2 In particular, cell-surface lectins in the immune system bind to glycans displayed on the exterior of pathogens and this recognition event stimulates an immune response.3 For example, mouse SIGN-R1 (SIGN-related 1), a homolog of human DC-SIGN (dendritic cell-specific ICAM-3-grabbing nonintegrin), is highly expressed on macrophages in the splenic marginal zone and the medullar lymph nodes.4 This lectin binds predominantly to mannose-rich or fucosylated glycans in a Ca2+-dependent manner.5 Once glycans on bacterial cells or viruses interact with SIGN-R1, glycan antigens elicit SIGN-R1 mediated cellular responses.One important area in glycoscience concerns studies aimed at the identification of functional glycans as ligands for lectins on the cell surface. For these efforts, analytical methods to study structure–function relationships of various glycans rapidly and on a large scale are in high demand. Since 2002, carbohydrate microarrays have proven their outstanding performance for the rapid analysis of glycan-mediated binding events.6–8 In addition, this microarray technology has been also demonstrated to be useful for the rapid assessment of acceptor specificities of glycosyltransferases and for measurements of binding affinities of glycans to proteins.9 In relation to cell interactions, glycan microarrays have been employed to evaluate the binding of bacteria, viruses and mammalian cells to glycans.9b,10
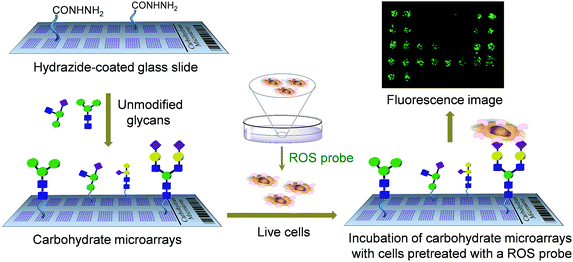
One potentially important application of carbohydrate microarrays is the rapid screening of glycans which stimulate cellular responses through interaction with cell-surface lectins. Although carbohydrate microarrays have been employed for studies of cell–glycan interactions,9b,10 microarray technology has seldom been used for the above described purpose.11 Recently, glycan arrays constructed by manually printing three heparin oligomers (0.5 μL) were applied for the identification of functional glycans which activated fibroblast growth factor signaling.11a Herein we describe a carbohydrate microarray-based approach for rapidly screening functional glycans which promote SIGN-R1-associated cellular responses. The efforts exploring this protocol were stimulated by the observation that the binding of glycan ligands to cell-surface SIGN-R1 triggers a cellular response that leads to the production of reactive oxygen species (ROS).12 In this investigation, we used a microarray assay and a ROS-sensitive fluorescent probe to identify glycans which induce ROS generation (Fig. 1). Cells which adhered to glycans on the microarrays generated ROS, whose levels were reduced in the presence of a ROS scavenger or a NADPH oxidase inhibitor. The findings suggest that carbohydrate microarrays can be employed to simultaneously screen glycans that elicit a cellular response by binding to the cell-surface lectins.
Results and discussion
To date a number of immobilization methods of glycans on the solid surface have been developed to prepare carbohydrate microarrays.7b Most immobilization techniques include the attachment of modified glycans on a properly derivatized surface. However, the synthesis of modified glycans containing functional group appendages, which are required for immobilization, is time-consuming and sometimes difficult. To avoid this task, we previously developed a facile immobilization protocol which relied on the attachment of unmodified glycans to a hydrazide-coated glass slide.13 In this method, mono-, di-, oligo- and polysaccharides can be site-specifically and covalently attached to the solid surface. Thus, we used this method to prepare the glycan microarrays employed in the present study.Carbohydrate microarrays were constructed by printing thirty unmodified glycans (2.6 nL, mono (1–7), di (8–11), oligo (12–15, 18) and polysaccharides (16 and 17, 19–30)) and a high-mannose glycan containing invertase (31) (Table 1 and Fig. S1†) on a hydrazide-coated glass slide by using an automatic pin-type microarrayer (spot size: ca. 200 μm in diameter).13 To initially test the successful attachment of glycans on the surface, glycan microarrays were probed with several Cy3-labeled lectins, including A. aurantia (AA), wheat germ agglutinin (WGA), R. communis agglutinin I (RCA120) and Concanavalin A (ConA). The results of the microarray data analysis show that the glycan binding properties of lectins match those observed in previous studies (Fig. S2†).13 This finding indicates that carbohydrate probes become attached to the hydrazide-modified glass slide.
| No | Glycan | No | Glycan | No | Glycan |
|---|---|---|---|---|---|
| 1 | Man | 2 | ManNAc | 3 | 6-P-Man |
| 4 | GlcNAc | 5 | Fuc | 6 | Gal |
| 7 | GalNAc | 8 | Manα1,2Man | 9 | Manα1,4Man |
| 10 | GlcNAcβ1,4GlcNAc | 11 | Galβ1,4Glc | 12 | Lea |
| 13 | Leb | 14 | Lex | 15 | Ley |
| 16 | Mannan (S. cerevisiae) | 17 | Mannan (C. albicans) | 18 | Mannan oligosaccharide (ivory nuts) |
| 19 | Mannan polysaccharide (ivory nuts) | 20 | Glucomannan (konjac) | 21 | Galactomannan (guar) |
| 22 | 500 kDa dextran | 23 | 2000 kDa dextran | 24 | Heparin |
| 25 | Fucoidan | 26 | Hyaluronic acid | 27 | Colominic acid |
| 28 | Chondroitin sulfate A | 29 | Dermatan sulfate | 30 | Chondroitin sulfate C |
| 31 | Invertase (S. cerevisiae) | 32 | Buffer |
Studies were then conducted to determine if the glycans on the microarrays are recognized by cells expressing the lectin. To this end, DCEK cells expressing SIGN-R1 on the surface (DCEK-SIGN-R1 cells, Fig. S3†) were grown under adherent culture conditions16 and then detached from the culture dish by treatment with a mixture of EDTA and trypsin. After centrifugation and washing to remove the remaining EDTA and trypsin, cells in culture media were applied to glycan microarrays washed with 70% ethanol for sterilization. Microscopy image analysis of microarrays incubated for 1 h at 37 °C reveals that the cells do not adhere to glycans immobilized on the surface.
We assumed that this phenomenon might be a consequence of cell-surface lectin damage caused by trypsin. Thus, in order to avoid the use of trypsin during the cell harvesting process, DCEK-SIGN-R1 cells were cultured in an uncoated plastic dish under non-adherent conditions. The cell aggregates in the suspension culture were then disassembled by cell shearing with pipetting. After centrifugation, various densities of cells (105 to 5 × 107 cells per mL) were applied to carbohydrate microarrays washed with 70% ethanol, which were then incubated for 1 h at 37 °C. Notably, cells cultured under the non-adherent conditions recognize glycans on the microarrays in a cell density-dependent manner (Fig. S4†). A cell density of 107 cells per mL was found to be appropriate for generating reproducible and reliable microarray data. Specifically, DCEK-SIGN-R1 cells reproducibly adhere to α-mannose containing glycans including 1, mannobiose (8, 9), a high-mannose conjugated invertase (31), S. cerevisiae mannan (16) and C. albicans mannan (17).14 Also, these cells bind to terminal fucose (5, 12–15)5 and GlcNAc bearing glycans (4, 10).15 However, DCEK-SIGN-R1 cells do not recognize β-mannose bearing glycans (18–21) and N-acetylated (2) and 6-phosphorylated mannose (3), a phenomenon which is consistent with previous results.14 The invertase containing a high-mannose glycan and C. albicans mannan among glycans tested were found to reproducibly bind to DCEK-SIGN-R1 cells with the strongest affinity.
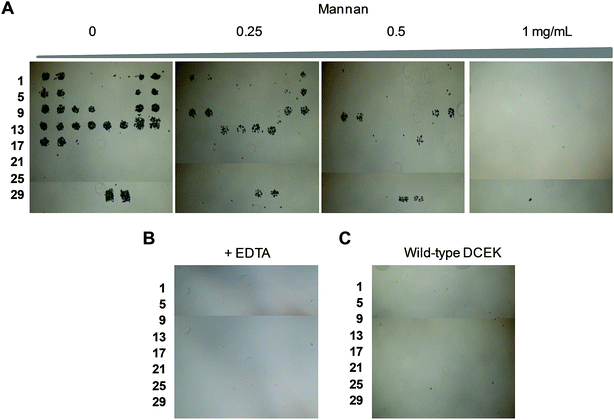
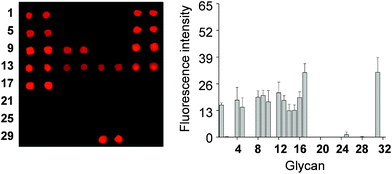
To determine if the nonselective adhesion of DCEK-SIGN-R1 cells to glycans occurs, competition experiments using S. cerevisiae mannan, a tight binding ligand of SIGN-R1,16 were performed. We observed that the binding ability of DCEK-SIGN-R1 cells, which were pre-incubated with mannan, to glycans on carbohydrate microarrays was markedly attenuated in a mannan concentration dependent manner (Fig. 2A). SIGN-R1 is known to recognize glycans in a Ca2+-dependent fashion.5 To examine the effect of calcium ions on the binding of SIGN-R1 to glycans, DCEK-SIGN-R1 cells were pretreated with EDTA, a calcium ion chelator, and then applied to carbohydrate microarrays. The results show that the cells do not adhere to glycans on the microarrays (Fig. 2B). Moreover, glycans on microarrays are not recognized by DCEK cells that lack SIGN-R1 (Fig. S3† and 2C). To further explore the binding properties of SIGN-R1 displayed on the cell surface, the carbohydrate microarrays were probed with a Cy3-labeled SIGN-R1 lectin. Microarray data analysis shows that the free lectin binds to glycans that are recognized by DCEK-SIGN-R1 cells (Fig. 3).
In the next step, we tested the practicability of glycan microarrays for screening functional glycans which stimulate the SIGN-R1 associated cellular response. It is known that glycan binding to SIGN-R1 on the cell surface triggers cellular signaling and induces ROS production.12 Thus, the cellular response to glycans is directly associated with glycan binding to cell-surface SIGN-R1. In the screening system, the differential response of cells to different glycans was quantitatively determined by using the boronate-based fluorescent H2O2 probe PF1, which is more selective and sensitive than the widely used DCFH-DA (2′,7′-dichlorofluorescin diacetate).16,17
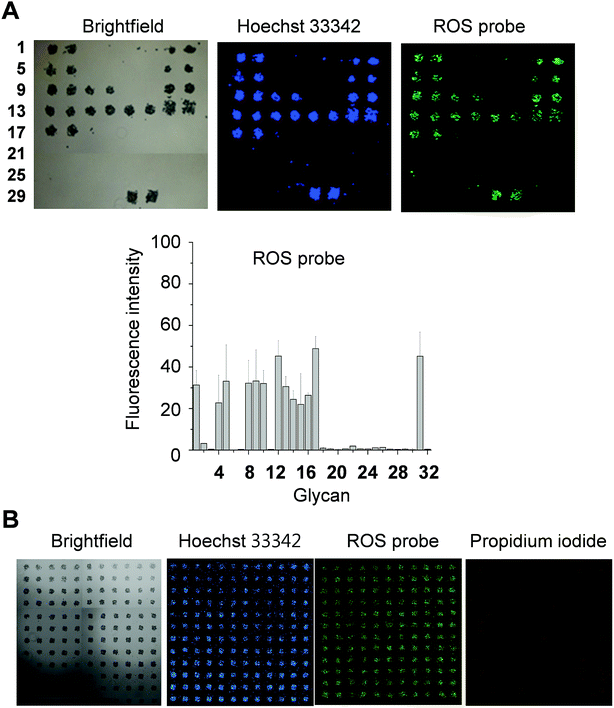
DCEK-SIGN-R1 cells cultured under non-adherent conditions were incubated with a mixture of PF1 and Hoechst 33342 (a nucleus staining dye) for 1 h and then applied to carbohydrate microarrays. After incubation for 1 h, the bound cells were analyzed by using a microscope and/or a microarray scanner. The results reveal that cells bound to glycans on the microarrays produce ROS, as inferred from the appearance of the PF1 derived fluorescence (Fig. 4A and S5†). Specifically, ROS are generated in cells bound to α-mannose, fucose and GlcNAc containing glycans. The binding tendency of free SIGN-R1 to glycans (Fig. 3) is relatively well correlated with the level of ROS produced in cells after adherence to glycans. Importantly, ROS produced in even small numbers of cells (less than 50 cells) after their binding to glycans immobilized on the surface could be detected. We also examined whether cells bound to glycans on the microarrays are alive or dead. In this study, microarrays containing 132 spots of mannan were incubated with cells treated with propidium iodide (PI), which is normally employed to detect dead cells. As shown in Fig. 4B, cells binding to immobilized glycans were seldom stained by PI, indicating that the bound cells are alive. The results of the immobilization concentration-dependent study show that as immobilization concentrations of glycans increase, more cells bind to the glycans to generate ROS (Fig. S6†).
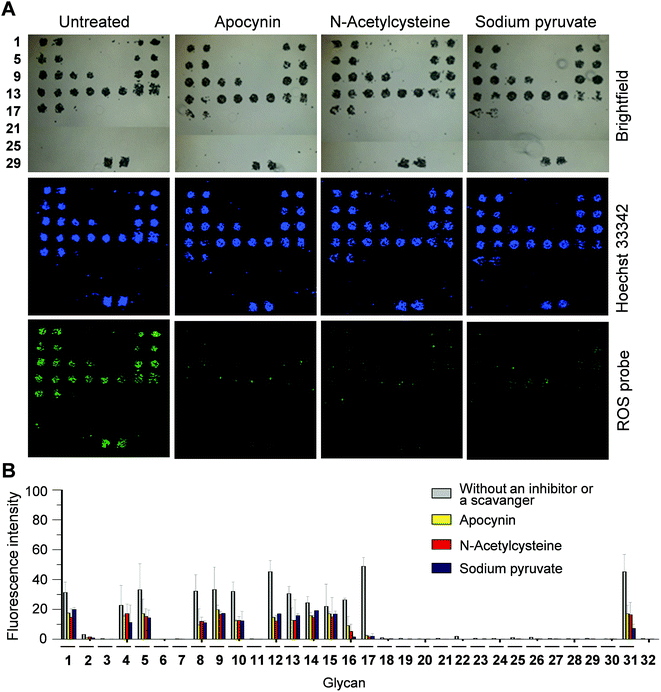
Next, we tested if PF1 derived fluorescence signals result from the response of PF1 to ROS produced in cells. For this purpose, the effect of a ROS scavenger or an NADPH oxidase inhibitor, which destroys ROS or suppresses ROS production, on PF1 fluorescence was investigated. DCEK-SIGN-R1 cells were incubated for 1 h with a mixture of Hoechst 33342 and PF1 in the absence or presence of apocynin (an inhibitor of NADPH oxidase that produces superoxide anion radicals), N-acetylcysteine (a ROS scavenger) or sodium pyruvate (a ROS scavenger),18 and then applied to carbohydrate microarrays. Analysis of microarray data after 1 h incubation shows that when either an ROS scavenger or a NADPH oxidase inhibitor is present, the fluorescence intensities of PF1 in cells bound to glycans are significantly reduced but the ability of cells to bind glycans is not significantly affected (Fig. 5). Collectively, the results indicate that the binding of cell-surface SIGN-R1 to glycans on the microarrays promotes ROS generation by eliciting the lectin-associated cellular response.
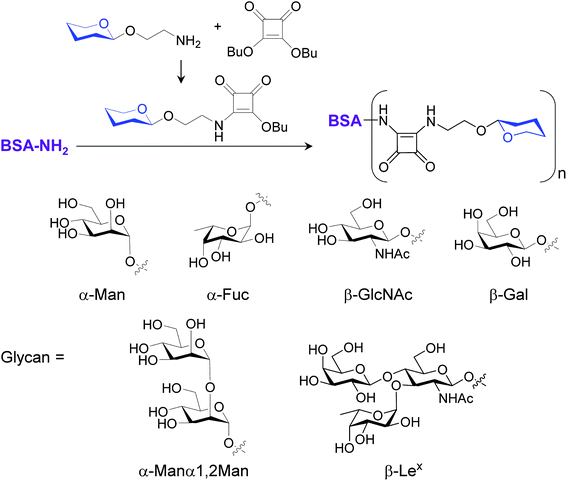
To gain confirmatory evidence supporting the results obtained from the microarray experiments, we conducted a traditional cell assay to probe six glycan–bovine serum albumin (BSA) conjugates (BSA–Man, BSA–Fuc, BSA–GlcNAc, BSA–Gal, BSA–Manα1,2Man and BSA–Lex) and S. cerevisiae mannan for their abilities to stimulate ROS generation. The multivalent BSA–glycan conjugates used in these experiments were prepared by reacting lysine amine groups of BSA with squarate-activated glycans, which were obtained from reactions of dibutyl squarate with aminoethylated α-Man, α-Fuc, β-GlcNAc, β-Gal, α-Manα1,2Man and β-Lex (Fig. 6).9a,19 MS analysis of BSA–glycan conjugates shows that 8–10 glycans are linked to one BSA molecule (see ESI†).
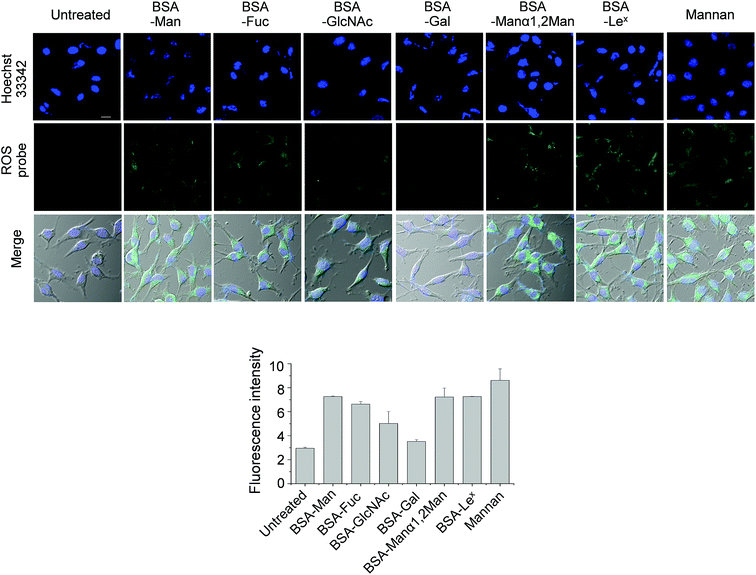
DCEK-SIGN-R1 cells in culture media were incubated with various concentrations (0–100 μg mL−1) of mannan or a BSA–glycan conjugate for 3 h. Observations made in time and concentration dependent studies indicate that 50 μg mL−1 of BSA–glycan conjugates and 1–2 h incubation time are sufficient for ROS production in treated cells (Fig. S7 and S8†). However, the addition of unconjugated BSA (0–200 μg mL−1) does not induce ROS generation in cells (Fig. S9†). Next, DCEK-SIGN-R1 cells were separately incubated with six different BSA–glycan conjugates (50 μg mL−1) and S. cerevisiae mannan (50 μg mL−1). The results show that incubation with mannan and all of the glycoconjugates except BSA–Gal leads to increases in the intensities of fluorescence signals from cells compared to the untreated control (Fig. 7). However, when N-acetylcysteine was added to the cells 1 h prior to stimulation with BSA–glycan conjugates or mannan, ROS production was markedly attenuated (Fig. S10†). The pattern of ROS production inside cells after treatment with BSA–glycan conjugates is similar to that in cells after binding to glycans on the microarrays. It is worth mentioning that because the glycans are attached to the glass surface on the microarrays and BSA via the different linkage (N-linkage vs. O-linkage) and glycans are conjugated to BSA with slightly different conjugation ratios (8 to 10), the pattern of ROS production in carbohydrate microarrays and traditional cell assays is similar but not the same.
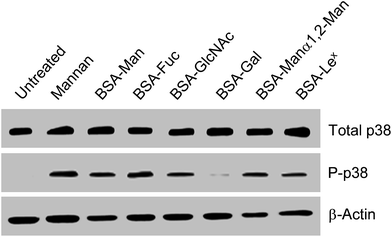
Because glycan binding to SIGN-R1 stimulates cellular signaling, the effect of BSA–glycan conjugates on signaling was finally evaluated. DCEK-SIGN-R1 cells were separately treated with six conjugates (50 μg mL−1, BSA–Man, BSA–Fuc, BSA–GlcNAc, BSA–Gal, BSA–Manα1,2Man and BSA–Lex) and S. cerevisiae mannan (10 μg mL−1). The expression level of a phosphorylated form of p38 MAPK was then determined by using western blot analysis. The results show that treatment with mannan and all of the glycoconjugates except for BSA–Gal leads to significantly increased levels of phosphorylated p38 MAPK in cells compared to that of the untreated control (Fig. 8). These results suggest that binding of cell-surface SIGN-R1 to glycans induces ROS generation, thereby activating p38 MAPK signaling.20
Conclusion
Carbohydrate microarrays have been applied for the rapid evaluation of glycan–protein interactions, the quantitative determination of binding of glycans to proteins, identification of disease-related anti-glycan antibodies for diagnosis, the rapid analysis of acceptor specificities of glycosyltransferases and detection of mammalian cells and pathogens. Although glycan arrays with low density spotting of a few glycan samples were used to evaluate live cell responses,11a glycan microarrays with high density spotting of various glycan samples have rarely been employed for this purpose. In the effort described above we have demonstrated that carbohydrate microarrays with high density spotting of a number of glycans by using an automatic microarrayer can be employed for the rapid screening of functional glycans that enhance the cell-surface lectin-associated cellular response. Because most animal lectins function as signal transducers after binding to glycans,2 carbohydrate microarray technology can be used to identify glycans that stimulate the cell-surface lectin-mediated cellular response in a high-throughput manner. To the best of our knowledge, this is the first application of ‘carbohydrate microarrays with the high density spotting of various glycan samples’ to screen glycans whose binding to the cell-surface lectin promotes the cellular response. Observations made in this investigation show that the carbohydrate microarray based technology will be useful for the simultaneous screening of glycans in efforts aimed at the development of novel chemical probes for studies of functional glycomics.Experimental section
Preparation of hydrazide-derivatized glass slide
Amine-coated glass slides (K-MAC, Korea) were kept in a staining jar containing a solution of poly(ethylene glycol)diglycidyl ether (3%, average Mn ∼ 526) in 10 mM NaHCO3 with gentle shaking for 1 h. After washing with water, the slides were immersed in a solution of 4,7,10-trioxa-1,13-tridecanediamine (3%) in 10 mM NaHCO3 with gentle shaking for 1 h. They were then washed with water and soaked in a solution of succinic anhydride (3%) in DMF with gentle shaking for 3 h. After washing with DMF, the slides were treated with a solution of diisopropyl carbodiimide (DIC, 3%) and N-hydroxysuccinimide (NHS, 3%) in DMF with gentle shaking for 3 h. They were then washed with DMF and immersed into a solution of hydrazine monohydrate (3%) in DMF with gentle shaking for 3 h. The slides were washed with water and dried by purging with argon gas. The derivatized slides can be stored at room temperature in a desiccator for several weeks.Construction of carbohydrate microarrays
Unmodified glycans were dissolved in sodium phosphate buffer (pH 5.0) containing 30% glycerol. A solution of glycan (2.6 nL, 30 mM) and polysaccharides (2.6 nL, 0.5 mg mL−1) from a 384-well plate was printed in a predetermined place on a hydrazide coated glass slide with a distance of 350 μm between centers of adjacent spots by using an automatic pin-type microarrayer (MicroSys 5100 from Cartesian Technologies). After the completion of printing, the slide was placed in a humidity chamber at 50 °C for 12 h and then a compartmentalized plastic film, coated by adhesive on one side (thickness: 0.1–0.2 mm), was attached to the glass slide. The slide was washed with PBS containing 0.1% Tween 20 and a solution of 0.1% Tween 20 possessing 2.5% BSA was dropped in the block. After incubation for 0.5 h, the slide was washed with the same buffer without BSA.Detection of carbohydrate–lectin interactions using carbohydrate microarrays
Carbohydrate microarrays were probed with Cy3-labeled AA (2 μg mL−1), WGA (5 μg mL−1), RCA120 (50 μg mL−1), ConA (50 μg mL−1) and SIGN-R1 (5 μg mL−1) in 20 mM Tris–HCl (pH 7.4) containing 0.1% Tween 20, 150 mM NaCl, 2 mM MgCl2 and 2 mM CaCl2 for 1 h at room temperature. The unbound proteins were removed by washing with the same buffer and rinsing with water. After drying by purging with argon gas, the slide was scanned using an ArrayWoRx scanner (Applied Precision). Fluorescence intensity was analyzed using ImaGene 6.1 software (BioDiscovery).Binding of cells cultured under adherent conditions to carbohydrate microarrays
Wild-type DCEK and DCEK-SIGN-R1 cells in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), 50 units per mL penicillin and 50 μg mL−1 streptomycin were grown in 100 cm2 tissue culture dishes at 37 °C in a cell culture incubator (adherent culture conditions). After 48 h, the cells were detached from the dishes by treatment with EDTA–trypsin for 5 min. After centrifugation, the resulting pellet was washed with culture media twice and re-suspended in culture media. The cells (107 cells per mL, the number of cells was counted by using a hemocytometer) in culture media were applied to carbohydrate microarrays washed with 70% ethanol for sterilization. After incubation for 1 h at 37 °C in a cell culture incubator, the carbohydrate microarrays were immersed into 10 mL culture media in 100 cm2 tissue culture dishes. The culture dishes were gently shaken by hand for 10 s and culture media containing unbound cells were discarded. This washing process was repeated three times to remove unbound cells. The carbohydrate microarrays were imaged using microscopy (Nikon Eclipse Ti-U). Cells did not bind to glycans on carbohydrate microarrays under these conditions.Detection of ROS production in cells bound to glycans on carbohydrate microarray
Wild-type DCEK and DCEK-SIGN-R1 cells were grown on uncoated plastic petri dishes in culture media (non-adherent culture conditions). Cell aggregates in suspension culture were disassembled by cell shearing with pipetting. After centrifugation, the cells (107 cells per mL) were incubated with 50 μM PF1 and 1 μM Hoechst 33342 in culture media lacking phenol red for 1 h at 37 °C in a cell culture incubator. The treated cells were applied to carbohydrate microarrays washed with 70% ethanol and incubated for 1 h at 37 °C in a cell culture incubator. The carbohydrate microarrays were immersed into 10 mL culture media lacking phenol red in 100 cm2 tissue culture dishes. The culture dishes were gently shaken with hand for 10 s and culture media containing unbound cells were discarded. This washing process was repeated three times to remove unbound cells. The carbohydrate microarrays were imaged using microscopy (Nikon Eclipse Ti-U and LSM 700 META, Carl Zeiss) or an ArrayWoRx microarray scanner (Applied Precision). Excitation of PF1 loaded cells was carried out with a 488 nm laser, and emission was collected using a confocal FL detector between 495 and 581 nm. Excitation of Hoechst 33342 was carried out using a 405 nm laser and emission was collected between 452 and 537 nm. Image analysis was performed using ZEN 2011.To test the effect of a ROS scavenger or an inhibitor of NADPH oxidase on ROS production on the carbohydrate microarrays, the cells (107 cells per mL) cultured under non-adherent conditions were incubated with 50 μM PF1 and 1 μM Hoechst 33342 in the presence of 2 mM apocynin, 30 mM N-acetylcysteine or 10 mM sodium pyruvate in culture media lacking phenol red for 1 h at 37 °C in a cell culture incubator. After centrifugation and washing with culture media lacking phenol red, the treated cells were applied to carbohydrate microarrays washed with 70% ethanol and incubated for 1 h at 37 °C in a cell culture incubator. The slide was washed using the same procedure described above. The bound cells were imaged using microscopy.
Detection of ROS production by cells in culture dishes
DCEK-SIGN-R1 cells in culture media were seeded in a Lac-Tek glass chamber slide (Nunc). After 24 h, the cells were incubated with 50 μM PF1 and 1 μM Hoechst 33342 in culture media lacking phenol red for 1 h at 37 °C in a cell culture incubator. The treated cells were incubated with BSA–glycan conjugates and S. cerevisiae mannan at various concentrations (0–100 μg mL−1) for 2 h or during different time periods at 50 μg mL−1 BSA–glycan conjugates and S. cerevisiae mannan at 37 °C in a cell culture incubator. After washing the cells with PBS twice to remove the remaining PF1 and glycans, bright field and fluorescence images of cells were taken by using confocal microscopy.To test the effect of a ROS scavenger on ROS production by cells in culture dishes, DCEK-SIGN-R1 cells were incubated with 50 μM PF1 and 1 μM Hoechst 33342 in the presence or absence of 30 mM N-acetylcysteine in culture media lacking phenol red for 1 h at 37 °C. The treated cells were incubated with 50 μg mL−1 BSA–glycan conjugates and S. cerevisiae mannan for 2 h. After washing the cells with PBS twice, they were imaged using confocal microscopy.
Acknowledgements
This work was supported by the National Creative Research Initiative (2010-0018272) program in Korea.Notes and references
- (a) A. Varki, Glycobiology, 1993, 3, 97–130 CrossRef CAS PubMed; (b) C. R. Bertozzi and L. L. Kiessling, Science, 2001, 291, 2357–2364 CrossRef CAS PubMed; (c) S. Park, M. R. Lee and I. Shin, Chem. Soc. Rev., 2008, 37, 1579–1591 RSC.
- (a) S. I. Gringhuis, J. den Dunnen, M. Litjens, M. van der Vlist and T. B. H. Geijtenbeek, Nat. Immunol., 2009, 10, 1081–1088 CrossRef CAS PubMed; (b) S. E. Hardison and G. D. Brown, Nat. Immunol., 2012, 13, 817–822 CrossRef CAS PubMed; (c) L. M. Kingeter and X. Lin, Cell. Mol. Immunol., 2012, 9, 105–112 CrossRef CAS PubMed; (d) E. Hebert, Biosci. Rep., 2000, 20, 213–237 CrossRef CAS PubMed; (e) I. M. Dambuza and G. D. Brown, Curr. Opin. Immunol., 2015, 32, 21–27 CrossRef CAS PubMed; (f) H. S. Goodridge, C. N. Reyes, C. A. Becker, T. R. Katsumoto, J. Ma, A. J. Wolf, N. Bose, A. S. H. Chan, A. S. Magee, M. E. Danielson, A. Weiss, J. P. Vasilakos and D. M. Underhill, Nature, 2011, 472, 471–475 CrossRef CAS PubMed.
- P. R. Crocker, J. C. Paulson and A. Varki, Nat. Rev. Immunol., 2007, 7, 255–266 CrossRef CAS PubMed.
- Y. S. Kang, S. Yamazaki, T. Iyoda, M. Pack, S. A. Bruening, J. Y. Kim, K. Takahara, K. Inaba, R. M. Steinman and C. G. Park, Int. Immunol., 2003, 15, 177–186 CrossRef CAS PubMed.
- C. Galustian, C. G. Park, W. G. Chai, M. Kiso, S. A. Bruening, Y. S. Kang, R. M. Steinman and T. Feizi, Int. Immunol., 2004, 16, 853–866 CrossRef CAS PubMed.
- (a) S. Fukui, T. Feizi, C. Galustian, A. M. Lawson and W. G. Chai, Nat. Biotechnol., 2002, 20, 1011–1017 CrossRef CAS PubMed; (b) D. N. Wang, S. Y. Liu, B. J. Trummer, C. Deng and A. L. Wang, Nat. Biotechnol., 2002, 20, 275–280 CrossRef CAS PubMed; (c) S. Park and I. Shin, Angew. Chem., Int. Ed., 2002, 41, 3180–3182 CrossRef CAS; (d) W. G. T. Willats, S. E. Rasmussen, T. Kristensen, J. D. Mikkelsen and J. P. Knox, Proteomics, 2002, 2, 1666–1671 CrossRef CAS PubMed.
- (a) C. D. Rillahan and J. C. Paulson, Annu. Rev. Biochem., 2011, 80, 797–823 CrossRef CAS PubMed; (b) S. Park, J. C. Gildersleeve, O. Blixt and I. Shin, Chem. Soc. Rev., 2013, 42, 4310–4326 RSC; (c) T. Feizi and W. G. Chai, Nat. Rev. Mol. Cell Biol., 2004, 5, 582–588 CrossRef CAS PubMed; (d) T. Horlacher and P. H. Seeberger, Chem. Soc. Rev., 2008, 37, 1414–1422 RSC; (e) P. H. Liang, C. Y. Wu, W. A. Greenberg and C. H. Wong, Curr. Opin. Chem. Biol., 2008, 12, 86–92 CrossRef CAS PubMed.
- (a) O. Blixt, S. Head, T. Mondala, C. Scanlan, M. E. Huflejt, R. Alvarez, M. C. Bryan, F. Fazio, D. Calarese, J. Stevens, N. Razi, D. J. Stevens, J. J. Skehel, I. van Die, D. R. Burton, I. A. Wilson, R. Cummings, N. Bovin, C. H. Wong and J. C. Paulson, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17033–17038 CrossRef CAS PubMed; (b) S. Park, M.-R. Lee, S.-J. Pyo and I. Shin, J. Am. Chem. Soc., 2004, 126, 4812–4819 CrossRef CAS PubMed; (c) B. Y. Xia, Z. S. Kawar, T. Z. Ju, R. A. Alvarez, G. P. Sachdev and R. D. Cummings, Nat. Methods, 2005, 2, 845–850 CrossRef CAS PubMed; (d) M.-R. Lee and I. Shin, Angew. Chem., Int. Ed., 2005, 44, 2881–2884 CrossRef CAS PubMed; (e) J. C. Manimala, T. A. Roach, Z. T. Li and J. C. Gildersleeve, Angew. Chem., Int. Ed., 2006, 45, 3607–3610 CrossRef CAS PubMed; (f) J. L. de Paz, C. Noti and P. H. Seeberger, J. Am. Chem. Soc., 2006, 128, 2766–2767 CrossRef CAS PubMed; (g) A. R. de Boer, C. H. Hokke, A. M. Deelder and M. Wuhrer, Anal. Chem., 2007, 79, 8107–8113 CrossRef CAS PubMed; (h) S. E. Tully, M. Rawat and L. C. Hsieh-Wilson, J. Am. Chem. Soc., 2006, 128, 7740–7741 CrossRef CAS PubMed; (i) K. S. Ko, F. A. Jaipuri and N. L. Pohl, J. Am. Chem. Soc., 2005, 127, 13162–13163 CrossRef CAS PubMed; (j) H. Y. Hsiao, M. L. Chen, H. T. Wu, L. D. Huang, W. T. Chien, C. C. Yu, F. D. Jan, S. Sahabuddin, T. C. Chang and C. C. Lin, Chem. Commun., 2011, 47, 1187–1189 RSC.
- (a) S. Park and I. Shin, Org. Lett., 2007, 9, 1675–1678 CrossRef CAS PubMed; (b) S. Park, M.-R. Lee and I. Shin, Nat. Protoc., 2007, 2, 2747–2758 CrossRef CAS PubMed; (c) P. H. Liang, S. K. Wang and C. H. Wong, J. Am. Chem. Soc., 2007, 128, 13668–13669 Search PubMed; (d) L. Ban, N. Pettit, L. Li, A. D. Stuparu, L. Cai, W. L. Chen, W. Y. Guan, W. Q. Han, P. G. Wang and M. Mrksich, Nat. Chem. Biol., 2012, 8, 769–773 CrossRef CAS PubMed; (e) A. Beloqui, J. Calvo, S. Serna, S. Yan, I. B. H. Wilson, M. Martin-Lomas and N. C. Reichardt, Angew. Chem., Int. Ed., 2013, 52, 7477–7481 CrossRef CAS PubMed.
- (a) L. Nimrichter, A. Gargir, M. Gortler, R. T. Altstock, A. Shtevi, O. Weisshaus, E. Fire, N. Dotan and R. L. Schnaar, Glycobiology, 2004, 14, 197–203 CrossRef CAS PubMed; (b) R. Sardzik, R. Sharma, S. Kaloo, J. Voglmeir, P. R. Crocker and S. L. Flitsch, Chem. Commun., 2011, 47, 5425–5427 RSC; (c) X. Z. Song, H. Yu, X. Chen, Y. Lasanajak, M. M. Tappert, G. M. Air, V. K. Tiwari, H. Z. Cao, H. A. Chokhawala, H. J. Zheng, R. D. Cummings and D. F. Smith, J. Biol. Chem., 2011, 286, 31610–31622 CrossRef CAS PubMed.
- (a) T. M. Puvirajesinghe, Y. A. Ahmed, A. K. Powell, D. G. Fernig, S. E. Guimond and J. E. Turnbull, Chem. Biol., 2012, 19, 553–558 CrossRef CAS PubMed; (b) L. E. Dickinson, C. C. Ho, G. M. Wang, K. J. Stebe and S. Gerecht, Biomaterials, 2010, 31, 5472–5478 CrossRef CAS PubMed; (c) N. X. Arndt, J. Tiralongo, P. D. Madge, M. von Itzstein and C. J. Day, J. Cell. Biochem., 2011, 112, 2230–2240 CrossRef CAS PubMed.
- (a) H. J. Choi, W. S. Choi, J. Y. Park, K. H. Kang, M. G. Prabagar, C. Y. Shin and Y. S. Kang, Biomol. Ther., 2010, 18, 271–279 CrossRef CAS; (b) L. Rizzetto, M. Kuka, C. De Filippo, A. Cambi, M. G. Netea, L. Beltrame, G. Napoitani, M. G. Torcia, U. D'Oro and D. Cavalieri, J. Immunol., 2010, 184, 4258–4268 CrossRef CAS PubMed.
- (a) M.-R. Lee and I. Shin, Org. Lett., 2005, 7, 4269–4272 CrossRef CAS PubMed; (b) S. Park, M.-R. Lee and I. Shin, Bioconjugate Chem., 2009, 20, 155–162 CrossRef CAS PubMed.
- K. Takahara, T. Arita, S. Tokieda, N. Shibata, Y. Okawa, H. Tateno, J. Hirabayashi and K. Inaba, Infect. Immun., 2012, 80, 1699–1706 CrossRef CAS PubMed.
- K. Takahara, Y. Yashima, Y. Omatsu, H. Yoshida, Y. Kimura, Y. S. Kang, R. M. Steinman, C. G. Park and K. Inaba, Int. Immunol., 2004, 16, 819–829 CrossRef CAS PubMed.
- S. Park, G.-H. Kim, S.-H. Park, J. Pai, D. Rathwell, J.-Y. Park, Y.-S. Kang and I. Shin, J. Am. Chem. Soc., 2015, 137, 5961–5968 CrossRef CAS PubMed.
- E. W. Miller, A. E. Albers, A. Pralle, E. Y. Isacoff and C. J. Chang, J. Am. Chem. Soc., 2005, 127, 16652–16659 CrossRef CAS PubMed.
- (a) M. Petrônio, M. Zeraik, L. Fonseca and V. Ximenes, Molecules, 2013, 18, 2821–2839 CrossRef PubMed; (b) M. Zafarullah, W. Q. Li, J. Sylvester and M. Ahmad, Cell. Mol. Life Sci., 2003, 60, 6–20 CrossRef CAS PubMed; (c) J. O'Donnell-Tormey, C. F. Nathan, K. Lanks, C. J. DeBoer and J. de la Harpe, J. Exp. Med., 1987, 165, 500–514 CrossRef.
- (a) J. Angulo, I. Diaz, J. J. Reina, G. Tabarani, F. Fieschi, J. Rojo and P. M. Nieto, ChemBioChem, 2008, 9, 2225–2227 CrossRef CAS PubMed; (b) A. Chernyak, S. Oscarson and D. Turek, Carbohydr. Res., 2000, 329, 309–316 CrossRef CAS PubMed; (c) Y. Hongbin, L. A. Aimé and Z. Yuyan, Bioorg. Med. Chem. Lett., 2007, 17, 6535–6538 CrossRef PubMed.
- K. Ito, A. Hirao, F. Arai, K. Takubo, S. Matsuoka, K. Miyamoto, M. Ohmura, K. Naka, K. Hosokawa, Y. Ikeda and T. Suda, Nat. Med., 2006, 12, 446–451 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sc03789a |
| ‡ These two authors equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2016 |