Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Coupling the folding of a β-hairpin with chelation-enhanced luminescence of Tb(III) and Eu(III) ions for specific sensing of a viral RNA†
Cristina
Penas
,
José L.
Mascareñas
* and
M. Eugenio
Vázquez
 *
*
Centro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CIQUS), Departamento de Química Orgánica, Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Spain. E-mail: joseluis.mascarenas@usc.es; eugenio.vazquez@usc.es
First published on 11th January 2016
Abstract
Rational modification of a natural RNA-binding peptide with a lanthanide EDTA chelator, and a phenanthroline ligand yields a highly selective luminescent sensor. The sensing mechanism relies on the RNA-triggered folding of the peptide into a β-hairpin, which promotes the coordination of the phenanthroline sensitizer, and the efficient sensitization of complexed lanthanide ions.
Introduction
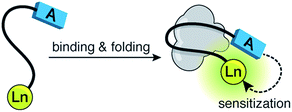
Fluorescence spectroscopy has transformed the study of biological systems by allowing the straightforward visualization of molecular events with extraordinary selectivity and sensitivity.1 The success of luminescent techniques relies on the availability of probes able to respond to specific biochemical inputs.2 In addition to organic fluorophores, lanthanide ions are privileged emissive species that display unique properties (e.g., narrow emission bands, and extremely long excited state lifetimes) that make them particularly attractive for luminescent sensing.3 In combination with peptides, lanthanides are powerful reporters that have been used to detect specific biomolecules,4,5 and monitor diverse biological processes.6,7 These luminescent sensors are typically based on the modulation of the energy transfer process from a nearby chromophore—the sensitization process—which mediates the efficient population of the excited states of the lanthanide ions.8 We have ourselves used this strategy to develop various lanthanide sensors that rely on the coupling of specific recognition events with the folding of short α-helices.4,9 Unfortunately, these sensors suffer from considerable background emission resulting from the restrictions imposed by the α-helical platform, that forces the antenna (tryptophan) and the lanthanide (terbium) ions to be placed in close proximity (relative positions i, i + 4), even in the unbound form. Besides, the folding of α-helices represents only one particular case of structural rearrangements coupled to biomolecular recognition processes, and a more general strategy applicable to the study of other conformational changes would be highly desirable.Herein, we report a new sensing strategy based on the folding of a random coil peptide into a β-hairpin, which allows the introduction of the sensitizing antenna and the lanthanide ion at distant positions in the sequence, thus ensuring low intrinsic emission in absence of the target;10 the large conformational changes experienced by the peptide upon folding give rise to a large increase in the lanthanide emission, thus ensuring high sensitivity (Fig. 1). Given that many biomolecular recognition processes involve related folding events,11 this strategy should be easily adapted to monitor different biological processes.
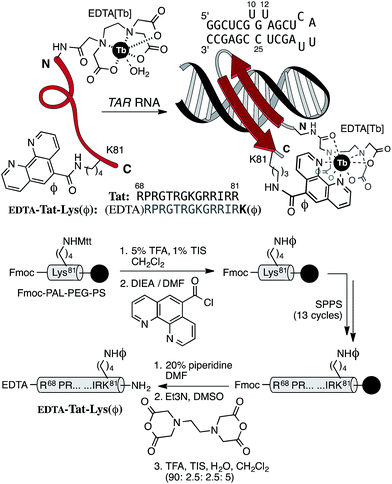
The viability of this strategy is demonstrated with the detection of the bovine immunodeficiency virus (BIV) RNA transactivation response element (TAR), a secondary RNA structure that is recognized with high affinity and high specificity by a 14-amino acid arginine-rich fragment from the Tat transcriptional activator.12Tat is a sequence-specific adaptor protein that directs the cellular transcription machinery to the viral RNA, thus promoting transcription elongation.13 Furthermore, the BIV Tat is closely related to its human immunodeficiency virus (HIV) counterpart,14 and therefore represents an attractive model to study HIV transactivation.
Results and discussion
Based on the solution structure of the peptide Tat bound to the BIV TAR hairpin RNA,15 as well as on mutational studies that identified the essential residues for high-affinity binding,16 we envisioned that extending the N-terminus of the Tat core sequence (Arg68 to Arg81) with a Trp residue, and introducing a DOTA[Tb] complex at the C-terminus should result in a metallopeptide probe that, upon binding to its target TAR RNA, would fold into a β-hairpin, placing both the DOTA[Tb] complex and the sensitizing indole ring of the Trp close to each other.17 Unfortunately, although the designed peptide was weakly emissive by itself, and displayed efficient RNA binding, addition of the target BIV TAR RNA did not induce any increase in its luminescence, likely because the excited state of the Trp is rapidly quenched by the RNA bases (most likely G11 and G19 close to the Trp in the bound state) before the sensitization takes place (see details in the ESI†).18 Straightforward modifications of that design, such as the addition of spacer residues between the Trp and the DOTA[Tb], switching their positions in the peptide chain, or using other chromophores, led in all cases to derivatives that were also spectroscopically unresponsive (see details in the ESI†). In light of these results, we reconsidered our initial design, and envisioned that forcing the direct complexation of the sensitizing chromophore to the lanthanide might favor the sensitization over the competitive quenching. Thus, we designed the peptide probe EDTA[Tb]-Tat-Lys(φ), featuring an EDTA chelator at its N-terminus (in place of the original DOTA),19 and a phenanthroline coordinating antenna (φ) attached to the side chain of a Lys residue in place of the C-terminal Arg81 (Fig. 2, top).20The target peptide EDTA-Tat-Lys(φ) was assembled following standard Fmoc/tBu solid-phase peptide synthesis procedures.21 The phenanthroline unit was connected to the side chain of an orthogonally-deprotected Lys(Mtt) residue while still attached to the solid support.22 After synthesizing the rest of the peptide sequence, the N-terminal amine (Arg68) was acylated with EDTA dianhydride (Fig. 2, bottom). The final peptide was purified by reverse-phase HPLC and identified by MS (see details in the ESI†).
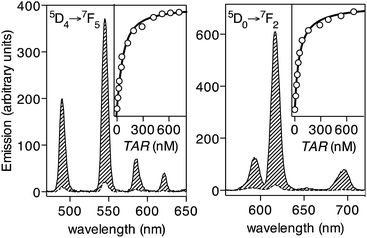
Having at hand the desired peptide probe, we studied the effect of the TAR RNA in the emission of its Tb(III) complex. As expected, the EDTA[Tb]-Tat-Lys(φ) metallopeptide, formed in situ by incubating the peptide EDTA[Tb]-Tat-Lys(φ) with 1 eq. of TbCl3, does not show significant emission from the Tb(III) ion upon irradiation of the phenanthroline antenna at 300 nm (Fig. 3, left, dashed trace, Φ[Tb] ≈ 0.010).23 Gratifyingly, incubation of this probe with increasing concentrations of the target BIV TAR RNA hairpin gives rise to a large increase in the luminescent emission bands of the Tb(III) ion (about 12-fold increase, Φ[Tb]/TAR ≈ 0.136).24 This is consistent with the folding of the Tat peptide core into a hairpin structure, and chelation of the terbium ion by the phenanthroline antenna upon binding to the RNA (Fig. 3, left, solid line). A titration experiment using the emission intensity of the hypersensitive band at 545 nm, corresponding to the 5D4 → 7F5 transition of the Tb(III) ion,25 allowed to calculate the dissociation constant of the probe KD ≈ 57 nM (Fig. 3, inset in the left graph), which is in agreement with the reported affinity of the natural Tat peptide for this hairpin RNA (KD[Tat] in the range of 50–100 nM).16 The calculated detection limit is ≈5 nM, which is well below the typical viral RNA concentration in infected cells.26 As expected, the probe is highly selective for the TAR RNA hairpin, and does not increase its emission in the presence of other non-target RNAs or DNAs; specific TAR RNA recognition by the EDTA[Tb]-Tat-Lys(φ) peptide is also demonstrated by EMSA and CD experiments (see details in the ESI†). Additionally, the phenanthroline antenna can also efficiently sensitize Eu(III) ions, and thus the analogous EDTA[Eu]-Tat-Lys(φ) probe also responds to the addition of the target TAR RNA with a large (30-fold) increase in the 5D0 → 7F2 emission band of Eu(III) at c.a. 615 nm (Fig. 3, right). The chelated EDTA[Eu]-Tat-Lys(φ) displays higher emission intensity than the bound Tb(III) probe (Φ[Eu] ≈ 0.0046; Φ[Eu]/TAR ≈ 0.165, over 30-fold increase).20,28 The lower sensitivity of the probe with Tb(III) might arise from the small energy gap between the triplet state of the phenanthroline antenna and the emitting 5D4 level in the metal ion, which favors efficient sensitization, but also allows significant energy back-transfer to the chromophore,27 thus reducing the overall sensitization efficiency.
Luminescence lifetime experiments with the EDTA[Tb]-Tat-Lys(φ) probe in D2O and H2O allowed us to calculate the number of inner-sphere water molecules bound to the metal center (q),29 which turned out to be q ≈ 1 for the probe in absence of its target RNA, and q ≈ 0 at saturating concentrations of TAR RNA. These values are in agreement with the reported number of water molecules coordinating the EDTA[Tb] complex, and support the RNA-promoted chelation of the ion upon folding of the peptide chain.30,31 Interestingly, it is known that OH oscillators in bound water molecules are efficient quenchers of lanthanide emission, and thus the displacement of the inner-sphere water of the EDTA complex upon RNA binding could also contribute to the large increase in emission observed upon folding and chelation.
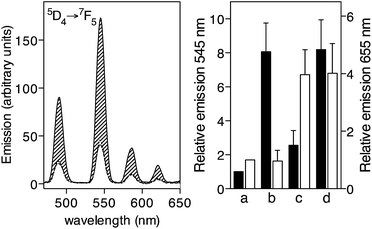
Importantly, the EDTA[Tb]-Tat-Lys(φ) probe specifically detects intracellular TAR RNA, displaying a large increase in its luminescence when added to cell lysates obtained from TAR RNA transfected HeLa cells (Fig. 4, left, solid line), while no increase in its emission was detected upon incubation with lysates from cells transfected with a non-target RNA (Fig. 4, left, dotted line), as well as with control lysates from untransfected HeLa cells (Fig. 4, left, dashed line). This experiment also demonstrates the advantages of the long luminescence lifetime of the lanthanide ions that allow time-gated conditions to isolate the emission of the lanthanide ions from the short-lived fluorescence of the biological background.32
Finally, we also demonstrated the potential of this newly developed probe, EDTA[Eu]-Tat-Lys(φ), for multiplex RNA sensing in combination with P22-NW[Tb], a previously described probe targeting the boxB RNA hairpin.9b Towards this end, we first demonstrated that the two probes, P22-NW[Tb], and EDTA[Eu]-Tat-Lys(φ), were capable of independently signaling the presence of their target hairpin RNAs, boxB and TAR, respectively. Thus, incubation of a 50 nM mixture of both sensors with boxB results in a large increase of the Tb(III) emission at 545 nm, resulting from the α-helical folding of the P22-NW[Tb] probe (Fig. 4, right, a and b),33 whereas addition of TAR RNA produced a large and selective increase of the EDTA[Eu]-Tat-Lys(φ) emission at 655 nm (Fig. 4, right, a and c). As expected, the addition of both RNA hairpins gives rise to emission increases in both probes, similar to those previously observed (Fig. 4, right, d).34
Conclusions
In summary, we have developed a new luminescence sensing strategy that takes advantage of the large conformational changes associated to recognition processes involving the folding of a β-hairpin for the effective modulation of lanthanide sensitization processes. Importantly, we found that using a lanthanide-coordinating antenna (phenanthroline) results in sensors that display both excellent dissociation constant and detection limit, with demonstrated bioanalytical applications. Finally, the modularity of this approach should allow its straightforward application for the development of sensors for monitoring other processes involving related folding events.Acknowledgements
We are thankful for the support given by the Spanish grants SAF2013-41943-R, CTQ2012-31341, and CTQ2013-49317-EXP, the Xunta de Galicia GRC2013-041, the ERDF, and the European Research Council (Advanced Grant No. 340055). Support of COST Action CM1105, and of the orfeo-cinqa network are kindly acknowledged. C. P. thanks the Spanish MCINN for her PhD fellowship. We would also like to thank Dr José R. Couceiro (USC) for his help with the RNA transfection experiments.Notes and references
- (a) J. Zhang, R. E. Campbell, A. Y. Ting and R. Y. Tsien, Nat. Rev. Mol. Cell Biol., 2002, 3, 906–918 CrossRef CAS PubMed; (b) L. D. Lavis and R. T. Raines, ACS Chem. Biol., 2008, 3, 142–155 CrossRef CAS PubMed; (c) B. N. G. Giepmans, S. R. Adams, M. H. Ellisman and R. Y. Tsien, Science, 2006, 312, 217–224 CrossRef CAS PubMed; (d) A. Mayer and S. Neuenhofer, Angew. Chem., Int. Ed. Engl., 1994, 33, 1044–1072 CrossRef.
- (a) Y. You, Curr. Opin. Chem. Biol., 2013, 17, 699–707 CrossRef CAS PubMed; (b) L. Prodi, New J. Chem., 2005, 29, 20–33 RSC; (c) K. M. Dean and A. E. Palmer, Nat. Chem. Biol., 2014, 10, 512–523 CrossRef CAS PubMed; (d) S. Wang, N. Li, W. Pan and B. Tang, Trends Anal. Chem., 2012, 39, 3–37 CrossRef CAS.
- (a) S. Shinoda and H. Tsukube, Analyst, 2011, 136, 431–435 RSC; (b) E. G. Moore, A. P. S. Samuel and K. N. Raymond, Acc. Chem. Res., 2009, 42, 542–552 CrossRef CAS PubMed; (c) A. Døssing, Eur. J. Inorg. Chem., 2005, 1425–1434 CrossRef; (d) E. Pazos and M. E. Vázquez, Biotechnol. J., 2014, 9, 241–252 CrossRef CAS PubMed.
- (a) E. Pazos, D. Torrecilla, M. Vázquez López, L. Castedo, J. L. Mascareñas, A. Vidal and M. E. Vazquez, J. Am. Chem. Soc., 2008, 130, 9652–9653 CrossRef CAS PubMed; (b) O. Vazquez, M. I. Sánchez, J. L. Mascareñas and M. E. Vazquez, Chem. Commun., 2010, 46, 5518–5520 RSC.
- L. Ancel, C. Gateau, C. Lebrun and P. Delangle, Inorg. Chem., 2013, 52, 552–554 CrossRef CAS PubMed.
- (a) L. Ancel, A. Niedźwiecka, C. Lebrun, C. Gateau and P. Delangle, C. R. Chim., 2013, 16, 515–523 CrossRef CAS; (b) E. Pazos, O. Vazquez, J. L. Mascareñas and M. E. Vazquez, Chem. Soc. Rev., 2009, 38, 3348–3359 RSC.
- (a) A. M. Lipchik and L. L. Parker, Anal. Chem., 2013, 85, 2582–2588 CrossRef CAS PubMed; (b) S. C. Zondlo, F. Gao and N. J. Zondlo, J. Am. Chem. Soc., 2010, 132, 5619–5621 CrossRef CAS PubMed.
- (a) A. J. Amoroso and S. J. Pope, Chem. Soc. Rev., 2015, 44, 4723–4742 RSC; (b) C. M. G. dos Santos, A. J. Harte, S. J. Quinn and T. Gunnlaugsson, Coord. Chem. Rev., 2008, 252, 2512–2527 CrossRef CAS.
- (a) E. Pazos, A. Jiménez, J. L. Mascareñas and M. E. Vazquez, Chem. Sci., 2011, 2, 1984–1987 RSC; (b) C. Penas, E. Pazos, J. L. Mascareñas and M. E. Vázquez, J. Am. Chem. Soc., 2013, 135, 3812–3814 CrossRef CAS PubMed.
- Key for this strategy is that lanthanide ions typically display a small Förster radius with common antennas. Thus for example, the R0 of the Tb(III)/Trp pair is between 5 and 10 Å: (a) L. S. McNemar, W. Y. Lin, C. D. Eads, W. M. Atkins, P. Dombrosky and J. J. Villafranca, Biochemistry, 1991, 30, 3417–3421 CrossRef CAS PubMed; (b) C. W. Hogue, J. P. MacManus, D. Banville and A. G. Szabo, J. Biol. Chem., 1992, 267, 13340–13347 CAS.
- (a) J. A. Robinson, Acc. Chem. Res., 2008, 41, 1278–1288 CrossRef CAS PubMed; (b) L. L. Cline and M. L. Waters, Org. Biomol. Chem., 2009, 7, 4622–4630 RSC; (c) S. M. Butterfield, W. J. Cooper and M. L. Waters, J. Am. Chem. Soc., 2005, 127, 24–25 CrossRef CAS PubMed; (d) Z. Athanassiou, R. L. A. Dias, K. Moehle, N. Dobson, G. Varani and J. A. Robinson, J. Am. Chem. Soc., 2004, 126, 6906–6913 CrossRef CAS PubMed; (e) T. Scherf, R. Kasher, M. Balass, M. Fridkin, S. Fuchs and E. Katchalski-Katzir, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 6629–6634 CrossRef CAS PubMed; (f) A. J. Maynard and M. S. Searle, Chem. Commun., 1997, 1297–1298 RSC; (g) M. Tateno, K. Yamasaki, N. Amano, J. Kakinuma, H. Koike, M. D. Allen and M. Suzuki, Biopolymers, 1997, 44, 335–359 CrossRef CAS PubMed.
- L. Chen and A. D. Frankel, Biochemistry, 1994, 33, 2708–2715 CrossRef CAS PubMed.
- M. Barboric, R. Taube, N. Nekrep, K. Fujinaga and B. M. Peterlin, J. Virol., 2000, 74, 6039–6044 CrossRef CAS PubMed.
- S. Bannwarth and A. Gatignol, Curr. HIV Res., 2005, 3, 61–71 CrossRef CAS PubMed.
- (a) J. D. Puglisi, L. Chen, S. Blanchard and A. D. Frankel, Science, 1995, 270, 1200–1203 CAS; (b) N. L. Greenbaum, Structure, 1996, 4, 5–9 CrossRef CAS PubMed.
- L. Chen and A. D. Frankel, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 507–5081 CrossRef.
- The conformational changes of small peptides upon binding to RNA targets have been exploited for the modulation of the emission of organic fluorophores: C. Matsumoto, K. Hamasaki, H. Mihara and A. Ueno, Bioorg. Med. Chem. Lett., 2000, 10, 1857–1861 CrossRef CAS PubMed.
- (a) K. E. Robinson, J. Orans, A. R. Kovach, T. M. Link and R. G. Brennan, Nucleic Acids Res., 2014, 42, 2736–2749 CrossRef CAS PubMed; (b) D. Elton, L. Medcalf, K. Bishop, D. Harrison and P. Digard, J. Virol., 1999, 73, 7357–7367 CAS.
- The EDTA[Tb] complex is very stable (log
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 17.9): J. Byegård, G. Skarnemark and M. Skålberg, J. Radioanal. Nucl. Chem., 1999, 241, 281–290 CrossRef.
K = 17.9): J. Byegård, G. Skarnemark and M. Skålberg, J. Radioanal. Nucl. Chem., 1999, 241, 281–290 CrossRef. - Y. Kitamura, S. Yamamoto, Y. Osawa, H. Matsuura and T. Ihara, Chem. Commun., 2013, 49, 285–287 RSC.
- (a) N. Sewald and H. Jakubke, Peptides: Chemistry and Biology, Wiley-VCH, Weinheim, Germany, 2002 Search PubMed; (b) I. Coin, M. Beyermann and M. Bienert, Nat. Protoc., 2007, 2, 3247–3256 CrossRef CAS PubMed.
- N. Thieriet, J. Alsina, E. Giralt, F. Guibe and F. Albericio, Tetrahedron Lett., 1997, 38, 7275–7278 CrossRef CAS.
- C. Würth, M. Grabolle, J. Pauli, M. Spieles and U. Resch-Genger, Nat. Protoc., 2013, 8, 1535–1550 CrossRef PubMed.
- This approximate quantum yield was obtained by comparing the areas under the emission curves in the time-gated experiments. This value is in the same order of those reported for related systems (see e.g., ref. 28b).
- (a) F. S. Richardson, Chem. Rev., 1982, 82, 541–552 CrossRef CAS; (b) I. Hemmilä and V. Laitala, J. Fluoresc., 2005, 15, 529–542 CrossRef PubMed.
- S. Kummer, A. Knoll, E. Socher, L. Bethge, A. Herrmann and O. Seitz, Bioconjugate Chem., 2012, 23, 2051–2060 CrossRef CAS PubMed.
- S. Quici, G. Marzanni, M. Cavazzini, P. L. Anelli, M. Botta, E. Gianolio, G. Accorsi, N. Armaroli and F. Barigelletti, Inorg. Chem., 2002, 41, 2777–2784 CrossRef CAS PubMed.
- (a) Y. Kitamura, T. Ihara, Y. Tsujimura, Y. Osawa, M. Tazaki and A. Jyo, Anal. Biochem., 2006, 359, 259–261 CrossRef CAS PubMed; (b) Y. Kitamura, T. Ihara, Y. Tsujimura, Y. Osawa, D. Sasahara, M. Yamamoto, K. Okada, M. Tazaki and A. Jyo, J. Inorg. Biochem., 2008, 102, 1921–1931 CrossRef CAS PubMed.
- (a) A. Beeby, I. M. Clarkson, R. S. Dickins, S. Faulkner, D. Parker, L. Royle, A. S. de Sousa, J. A. G. Williams and M. Woods, J. Chem. Soc., Perkin Trans. 2, 1999, 493–504 RSC; (b) E. Pazos, M. Goličnik, J. L. Mascareñas and M. E. Vázquez, Chem. Commun., 2012, 48, 9534–9536 RSC.
- Chelation-enhanced fluorescence (CHEF) of organic fluorophores has been extensively used in the development of other biosensors: (a) G. K. Walkup and B. Imperiali, J. Am. Chem. Soc., 1996, 118, 3053–3054 CrossRef CAS; (b) M. D. Shults and B. Imperiali, J. Am. Chem. Soc., 2003, 125, 14248–14249 CrossRef CAS PubMed.
- CHEL has been reported in a number of lanthanide-based sensors, although not coupled to peptide/protein folding as part of the sensing mechanism: (a) H. Akiba, J. Sumaoka, T. Hamakubo and M. Komiyama, Anal. Bioanal. Chem., 2014, 406, 2957–2964 CrossRef CAS PubMed; (b) S. Balakrishnan and N. J. Zondlo, J. Am. Chem. Soc., 2006, 128, 5590–5591 CrossRef CAS PubMed.
- (a) E. F. Dickson, A. Pollak and E. P. Diamandis, J. Photochem. Photobiol., B, 1995, 27, 3–19 CrossRef CAS; (b) W. Cui and L. L. Parker, Chem. Commun., 2015, 51, 362–365 RSC.
- Control experiments showed that the EDTA[Eu]-Tat-Lys(φ) probe has low affinity for the boxB RNA, and no increase in its luminescence emission is observed in the presence of up to 2 eq. of this RNA.
- The narrow luminescence emission bands of the lanthanide ions make them particularly appropriate for multiplex sensing: E. Pershagen and K. E. Borbas, Angew. Chem., Int. Ed., 2015, 54, 1787–1790 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Peptide and lanthanide probe synthesis, luminescence experiments, experimental description of the transfection and multiplex experiments. See DOI: 10.1039/c5sc04501k |
| This journal is © The Royal Society of Chemistry 2016 |