Open Access Article
Open Access ArticleNanostructured surfaces of biodegradable silica fibers enhance directed amoeboid cell migration in a microtubule-dependent process†
Martin Emmertab,
Patrick Witzelab,
Miranda Rothenburger-Glaubitta and
Doris Heinrich*ac
aFraunhofer Institute for Silicate Research ISC, Neunerplatz 2, 97082 Würzburg, Germany
bJulius-Maximilians-Universität Würzburg, Chemical Technology of Material Synthesis, Röntgenring 11, 97070 Würzburg, Germany
cLeiden University, LION Leiden Institute of Physics, Niels Bohrweg 2, 2333 CA Leiden, The Netherlands. E-mail: Heinrich@Physics.LeidenUniv.nl
First published on 21st November 2016
Abstract
Cell motion and in particular amoeboid migration is crucial for the survival of living organisms. To advance biomedical research, a constant need for novel materials and surface structures arises, improving immigration of cells into artificial structures used for tissue engineering and for advancing therapy in modern medicine. This work focuses on investigating amoeboid single-cell migration on intrinsically nano-structured, biodegradable silica fibers in comparison to chemically equivalent plain glass surfaces. Cell migration trajectories are classified into directed runs and quasi-random migration by a local mean squared displacement (LMSD) analysis. We find that directed movement on silica fibers is enhanced in a significant manner by the fibers' nanoscale surface-patterns. Further, cell adhesion on the silica fibers is a microtubule-mediated process. Cells lacking microtubules detach from the fibers, but adhere well to glass surfaces. Knock-out mutants of myosin II migrating on the fibers are as active as cells with active myosin II, while the migration of the knock-out mutants is hindered on plain glass. Identifying relevant cytoskeletal compounds for cell migration in 2D vs. 3D will help to optimize materials or surfaces that boost specific migration strategies to open up new opportunities for innovative diagnostic and therapeutic concepts.
Introduction
Cell migration is crucial in the human organisms, involving essential processes like embryonic morphogenesis1,2 or wound healing.3,4 The introduction of novel scaffold materials to support or enhance cell migration is a major topic in tissue engineering and modern medicine, as cell behavior in vitro and in vivo is strongly influenced by the nanostructured surface of artificial scaffolds5 or the extracellular matrix.6 Amoeboid-like cell migration is an efficient concept of cell migration,7 e.g. performed by stem cells and different types of immune cells like monocytes,8 neutrophils9 and lymphocytes.10 These cell types can travel long distances in reaction to chemical cues, exit blood vessels and crawl through tissues.9,11 As several signalling cascades and basic cytoskeletal mechanisms are similar to mammalian cells, the social amoeba Dictyostelium discoideum is a perfect model organism for the study of amoeboid migration.12 Amoeboid cells react to changes in their environment very quickly, which manifests in the amoeboid cells' ability to respond to external cues within seconds and to change their direction of movement within a few minutes.13,14 The cytoskeleton provides the highly dynamic system for the transport of regulatory proteins and cellular polymers that realize these rapid changes in migration direction or the switching from resting states to active migration: in reaction to external topographic and chemotactic stimuli, an amoeboid cell polarizes by force generation of the actomyosin cytoskeleton.15 At first, the center of mass is moved into the direction of migration by extension of protrusions upon actin polymerization and material transport by stabilized microtubules.16 Migration is then achieved by further protrusion at the front and myosin II-driven contraction and concurrent retraction of adhesion sites at the rear.17 This complex interplay between actin polymerization, motor proteins and the microtubule network is crucial for force generation between substrate and cell.18As eukaryotic cells possess sensing and adhesion features in the nanometer range, directed cell migration can be induced and supported by topographic cues on the substrate.19 In the model of contact-reinforced motility, these topographic guidance effects are explained by a stabilization of pseudopods upon contact with a topographic stimulus, increasing the probability for directed migration in the direction of the stimulus.20,21 Thus, the introduction of three-dimensional structures or surface patterns that support cytoskeletal migration mechanisms can enhance cell migration without presentation of additional chemical attractants and find wide applications in medicine and medical research. For example, scaffolds made from such smart materials could greatly increase the efficiency of cell seeding in tissue engineering or serve as a scaffold for the repair of tissue injuries.22 This work investigates (i) the influence of the intrinsic surface nanotopography of biodegradable silica fibers on amoeboid single cell migration and (ii) the role of specific cytoskeletal structures during migration on these fibers, in particular microtubules and myosin II. Our findings will contribute to the evaluation of the fiber material as a scaffold for applications in in vitro and in vivo situations.
Materials and methods
Fabrication and characterization of silica fiber samples
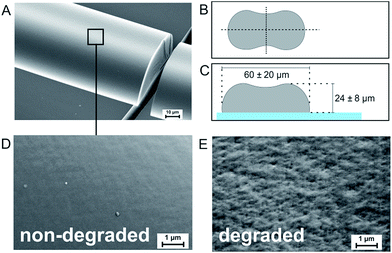
Silica fibers were spun directly onto microscopy cover glasses for cell migration experiments. The standard precursor was prepared in a three-step synthesis: a mixture of tetraethoxysilane (TEOS), ethanol (EtOH), distilled water (H2O) and nitric acid (HNO3) (all from Sigma-Aldrich Chemie GmbH) was stirred at room temperature for several hours. EtOH was removed from the mixture in a rotary evaporator, followed by sol ripening carried out at low temperature.23 During sol ripening, an abrupt increase of viscosity was detected. For fiber spinning, the gel was filled into a pressure vessel and extruded through a nozzle plate at pressures of 15–20 bar. After exiting the 150 μm nozzle diameter, the fibers were led through a laminar air flow with a drop distance of two meters onto microscopy cover glasses (borosilicate glass 3.3, 80.6% SiO2, 12.5% B2O3, 4.2% NaO, 2.2% Al2O3, 0.5% trace elements, Th. Geyer GmbH & Co. KG), placed on a programmable X–Y-table (Bosch Rexroth AG). The chemical composition of the resulting silica gel fibers was determined to be [Si(OH)0.2O1.9]n by thermal analysis in a previous work.23 Due to the high silica content, the chemical composition of the cover glasses is very similar to the fiber surface. The fiber diameter, shape and surface homogeneity were analyzed by SEM (Supra 25, Carl Zeiss Microscopy GmbH) after platinum sputtering. Fig. 1A shows a fiber fixed on a glass substrate. Due to a collapse of the circular surface upon solvent diffusion and evaporation in laminar air flow, free-standing fibers exhibit a dog-bone-like shape (Fig. 1B). The dog-bone-shape at the surface is retained upon when fibers are directly spun onto cover glasses for migration experiments (Fig. 1C). Fiber widths were measured to be 60 ± 20 μm with fiber heights of 24 ± 8 μm. The surface of freshly fabricated fibers is smooth, with no distinct surface patterns (Fig. 1D). As the fiber material is biodegrading at a rate of 10–100 nm per day,23 nano-sized surface patterns emerge upon contact to aqueous media. SEM investigations on fibers that have been stored in simulated body fluid for several days revealed pits and elevations on the fiber surface with feature sizes below 200 nm (Fig. 1E). In comparison, the surface of the cover glass around the fibers appears smooth (Fig. 1A and S3†), exhibiting an average surface roughness below 0.3 nm.24 Thus, the comparison of amoeboid cell migration on silica fibers with cover glasses of similar chemical composition – but a flat surface – is expected to reveal the impact of the fibers' nanoscale topography on amoeboid cell migration.Cell preparation and migration experiments
Different mutants of the D. discoideum strain AX2 were cultured for the experiments. For wild-type migration, cells expressing homogenously distributed Green Fluorescent Protein (GFP) in the cytoplasm (FreeGFP) were used. Additional experiments on myosin II-deficient cells were carried out with a myosin II null mutant with GFP-tagged LimEΔcc. All cell cultures were grown at 19–21 °C in Petri dishes at ambient air. 20 μg mL−1 gentamycin (G-418-Biochrom A2912, Biochrom AG) was added to the culture medium of FreeGFP cells. For myosin II null mutants, gentamycin and the additional antibiotic blasticidin (Sigma-Aldrich Chemie GmbH) were used at a concentration of 10 μg mL−1. Prior to the experiments, the cells were washed (four times) with phosphate buffered saline (PBS). Each step consisted of centrifugation at 2000 rpm for 4 minutes (Eppendorf MiniSpin® Plus, Eppendorf AG) and resuspension of the obtained cell pellet in PBS. For experiments, D. discoideum cells suspended in PBS were pipetted onto silica fibers on cover glasses at densities ranging from 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 410
000 to 410![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2, the suitable cell number for the observation of up to 10 cells in a field of view while excluding direct cell–cell interactions during cell tracking. Image acquisition was performed with a Nikon Eclipse Ti microscope (Nikon, Germany) and a 20× objective (Nikon, Germany). Fluorescence white light illumination was obtained by Intensilight (Nikon, Germany). For GFP imaging, an excitation filter (F36-525 HC, AHF, Germany) ensured excitation wavelengths of 457–487 nm and detection wavelengths of 500–540 nm. The setup is equipped with an EM-CCD camera (Hamamatsu, Japan) for detection of the fluorescence signal. A 100 W halogen lamp contained in the microscope was used for bright field illumination. Fluorescence images were acquired every 7 seconds for at least 45 minutes to exclude short-time effects. To minimize the excitation stress for the cells, exposure times were kept below 100 ms. Additionally, a bright field image was taken for every time point to ensure proper fiber localization.
000 cells per cm2, the suitable cell number for the observation of up to 10 cells in a field of view while excluding direct cell–cell interactions during cell tracking. Image acquisition was performed with a Nikon Eclipse Ti microscope (Nikon, Germany) and a 20× objective (Nikon, Germany). Fluorescence white light illumination was obtained by Intensilight (Nikon, Germany). For GFP imaging, an excitation filter (F36-525 HC, AHF, Germany) ensured excitation wavelengths of 457–487 nm and detection wavelengths of 500–540 nm. The setup is equipped with an EM-CCD camera (Hamamatsu, Japan) for detection of the fluorescence signal. A 100 W halogen lamp contained in the microscope was used for bright field illumination. Fluorescence images were acquired every 7 seconds for at least 45 minutes to exclude short-time effects. To minimize the excitation stress for the cells, exposure times were kept below 100 ms. Additionally, a bright field image was taken for every time point to ensure proper fiber localization.
For experiments with cells deprived of microtubules, the microtubule-depolymerizing agent benomyl (Sigma-Aldrich Chemie GmbH) has been used at a concentration of 50 μM on FreeGFP cells. Cell adherence after benomyl treatment was investigated by moving the microscopy table during live video-acquisition and the determination of the number of adherent and non-adherent cells.
Migration analysis
For tracking analysis of migrating cells, the fluorescence image stacks were loaded into the image processing software ImageJ (National Institutes of Health, USA), analogously edited and converted to binary images. The obtained sequences of binary images were then analysed by the ImageJ plugin “Cell Evaluator” which tracks the center of mass of each cell.25 A text file is created, containing the X–Y coordinates for each time point. The cell trajectories were analyzed by the MATLAB® algorithm “CellMovement”,21,26 distinguishing two modes of cell migration: directed cell runs and quasi-random migration phases. It utilizes a local mean squared displacement (LMSD), which is calculated for every time point ti over a rolling time window of 16 frames (corresponding to a duration T = 112 s) as a function of different lag times τ:27| 〈ΔR2(τ)〉i = 〈(R(ti + τ) − R(ti))2〉t−T/2<ti<t+T/2 | (1) |
The obtained 〈ΔR2(τ)〉 is then fitted by the power law
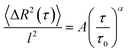 | (2) |
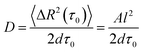 | (3) |
| 〈ΔR2(τ)〉 = vd2τ2. | (4) |
Results and discussion
Migration analysis
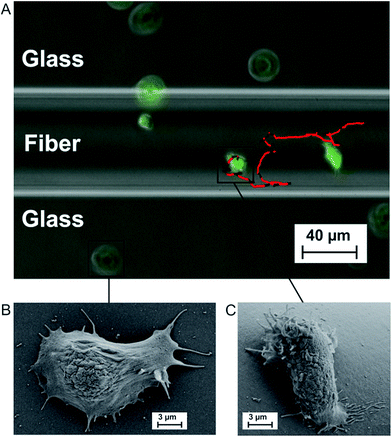
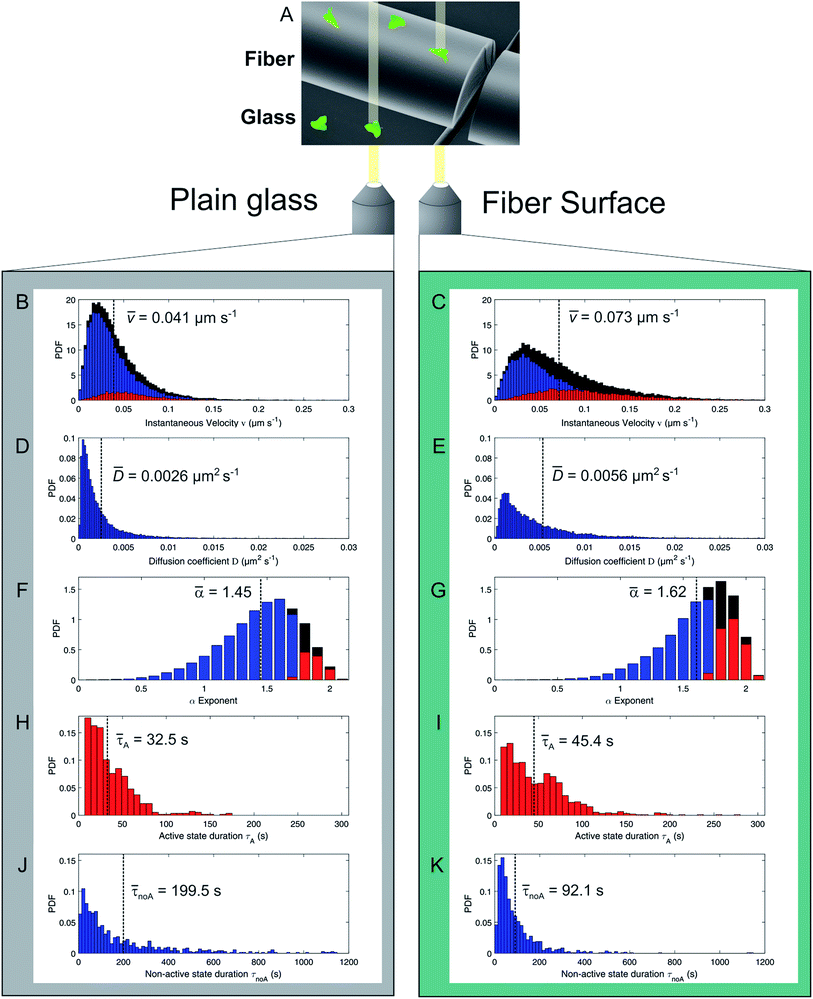
We investigate amoeboid cell migration on nanostructured silica fibers in comparison to chemically analog flat glass surfaces in order to reveal the impact of the fibers' nanoscale topography on amoeboid cell migration. Fibers exhibit a dog-bone-shape (Fig. 1A, width = 60 ± 20 μm, height = 24 ± 8 μm) and nanoscale surface patterns emerging upon biodegradation in aqueous media (Fig. 1E).Fig. 2A shows a cell migration trajectory: every 7 s, D. discoideum cells were imaged in bright field and the fluorescent channel. The fluorescent channel image was binarized and each cell's center of mass was tracked for every time point. By local mean squared displacement (LMSD) analysis, the trajectories were divided into directed cell runs (red) and quasi-random migration (black). This analysis is based on a rolling window of 16 frames, in which the LMSD is calculated as (1) (see Materials and methods) and the exponent α (2) (see Materials and methods) determines directed runs. In these directed migration states, cells are more polarized (Fig. 2C) and exhibit extended sections of directionally persistent movement. These are interrupted by short phases of quasi-random migration, where the cell is protruding into different directions without significant movement of the center of mass (Fig. 2B). We analyzed 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 438 data points (47.5 h) on flat glass and 22
438 data points (47.5 h) on flat glass and 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 413 data points (43.6 h) on silica fibers. Fig. 3 displays cell migration parameters of the LMSD analysis and their mean values.
413 data points (43.6 h) on silica fibers. Fig. 3 displays cell migration parameters of the LMSD analysis and their mean values.
The distribution of instantaneous velocities for cells migrating on silica fibers (Fig. 3B) is significantly shifted to higher values as compared to migration on plain glass (Fig. 3C). The overall mean velocity increases by 78% for cells migrating on the fibers. The velocity distribution of directed states (red) is clearly increased on silica fibers, while the number of non-directed, slow tracks is significantly decreased as compared to the 2D glass surface.
The mobility of cells in quasi-random migration states is characterized by the generalized diffusion coefficient D,27,29 displayed in Fig. 3D/E. In general, cells migrating on flat glass (Fig. 3D) exhibit lower diffusion coefficients than cells on silica fibers (Fig. 3E). The mean diffusion coefficient increases by 115% for cells on the fiber surface. Thus, quasi-randomly migrating cells are highly motile on silica fibers. For a measure of the motility mode of cells on a substrate, the dimensionless α coefficient of the LMSD is analyzed. It is the exponent of the MSD power law fit (2) (see Materials and methods) and describes the migration mode of a cell for every time point of the cell trajectory. For α values ≈ 2, a cell performs directed migration, while α < 2 characterizes diffusive, quasi-random motion. The α distributions (Fig. 3F/G) and the corresponding mean values are shifted to higher values on the silica fibers. Compared to the migration on plain glass, we find an increase by 12% from α = 1.45 to α = 1.62. Additionally, the lifetime distributions of directed and quasi-random migration states confirm this trend: on fibers, directed states are prolonged compared to on cover glass (Fig. 3H/I), with a 37% increase of the mean value from 33 s to 45 s. The distribution of the lifetimes of quasi-random states is shifted to smaller values on silica fibers (Fig. 3J/K), with the mean value decreasing from 200 s to 92 s. This result is strengthened by the significantly longer total time spent in directed migration phases on silica fibers: the portion of directed cell tracks on silica fibers (27%) is twice larger than on cover glasses (11%) (Table 1). This indicates a stabilization of directed migration phases and a nanotopography-triggered induction of directed migration for cells in the reorientation phase.
| Coverglass | Fiber | ||||
|---|---|---|---|---|---|
| FreeGFP | MyoII0 | FreeGFP | MyoII0 | ||
| Number of cells | N | 46 | 31 | 49 | 30 |
| Data points | n | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 438 438 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 212 212 |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 413 413 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 084 084 |
| Mean alpha value | ![[small alpha, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0c2.gif) |
1.45 | 1.34 | 1.62 | 1.61 |
| Mean diffusion coefficient (μm2 s−1) | ![[D with combining macron]](https://www.rsc.org/images/entities/i_char_0044_0304.gif) |
0.0026 | 0.0014 | 0.0056 | 0.0038 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Mean instantaneous velocity | |||||
| Of directed states (μm s−1) | ![[v with combining macron]](https://www.rsc.org/images/entities/i_char_0076_0304.gif) A A |
0.057 | 0.046 | 0.104 | 0.081 |
| Overall (μm s−1) | ![[v with combining macron]](https://www.rsc.org/images/entities/i_char_0076_0304.gif) all all |
0.041 | 0.029 | 0.073 | 0.059 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Mean duration of state lifetimes | |||||
| Of directed states (s) | ![[small tau, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0d4.gif) A A |
32.5 | 39.2 | 45.4 | 52.0 |
| Of quasi-random states (s) | ![[small tau, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0d4.gif) noA noA |
199.5 | 171.7 | 92.1 | 99.2 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| % of time spent in phases | |||||
| Directed | 11 | 15 | 27 | 30 | |
| Quasi-random | 85 | 81 | 64 | 61 | |
Direct impact of surface nanotopography on amoeboid migration
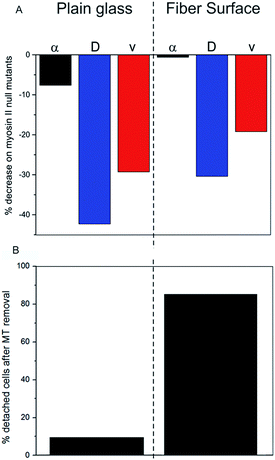
The presented results indicate a direct impact of the surface topography of the silica fibers on amoeboid cell motility. Due to biodegradation upon contact with aqueous media, nanoscaled surface features emerge at the fiber surface (Fig. 1E). On the cell level, sensing of these nanoscale features by filopodia leads to polarization with stabilized protrusions and enhanced directed migration.19,30 When analyzing cell migration on fibers, it is important to consider the effect of surface curvature. In a study on amoeboid migration of lymphocytes Song et al. fabricated sinusoidal wavy surfaces with constant amplitude but different wavelengths.31 They found an enhancement of amoeboid migration for small wavelengths but a less pronounced to no effect for higher wavelengths and flat surfaces. Transferring this curvature-dependence from this study to the fiber surfaces we investigated, none or only a very slight effect of curvature on amoeboid migration can be expected in our study. Additionally, even the most pronounced effect of surface curvature on migration velocities found by Song et al. was four-fold lower than the difference we found between flat glass and fiber surfaces. Thus, the migration data clearly display the differences in amoeboid migration with pronounced directed runs on nano-structured silica fibers and significantly decreased motility on the flat surface (Table 1). The mean generalized diffusion coefficient D is significantly higher on silica fibers than on plain glass substrates (Fig. 3D/E). This indicates that even cells in quasi-random migration states are more motile on the fiber surface than on plain glass. SEM micrographs of D. discoideum cells on the different surfaces underline these findings: more polarized, motile cells are found on the silica fibers (Fig. 2C), whereas cells on plain glass surfaces mostly lack polarization (Fig. 2B).In our experiments, migration of myosin II null mutant cells of D. discoideum on plain glass is clearly hindered compared to FreeGFP-cells. This effect is consistent throughout the whole set of migration parameters: we see a distinct decrease in the mean values of the α coefficient from 1.45 to 1.34 and decreases in the mean diffusion coefficient and the mean instantaneous velocities (Fig. 4A and S1†) (Table 1). In contrast, cell migration analysis of myosin II null mutant cells on the silica fibers reveals the same mean value for the α coefficient as for the FreeGFP cells (Fig. 4A). This indicates that the enhanced overall motility is conserved compared to FreeGFP cells, with distinct decreases in the other migration parameters due to the loss of actin cortex tension and contractibility. Therefore, we can conclude that the nanoscale topography of the silica fibers induces and supports pseudopod stabilization and facilitates cell migration even in cells with greatly decreased cortical tension.
This result points towards a migration mechanism based on actin foci as a direct functional dependency between microtubules and the actin cortex. This relation has also been found in axon growth cones: benomyl treated cells lost adhesion on microtubule deprivation due to a loss of actin foci, pointing towards an analog mechanism in mammalian cells.38
Conclusion
We investigated the influence of the intrinsically nano-patterned surface of biodegradable silica fibers on amoeboid cell migration. We found pronounced directed runs on the silica fibers compared to plain glass surfaces and further investigated underlying mechanisms. Experiments with D. discoideum deprived of microtubules revealed that the adhesion and migration mechanisms on silica fibers were greatly microtubule-dependent: adhesion to fibers was lost without microtubules, while microtubule-deprived cells on flat glass surfaces remained adherent. We found that the migration effect on fibers was less influenced by myosin II, as null mutant cells were as active as cells with active myosin II, while migration of cells lacking myosin II was greatly hindered on plain glass.Our work demonstrates the importance of nanoscale surface features and scaffold structure for the motility of cells in physiological processes and thus, in the application for medical purposes. The fact that amoeboid motion is greatly facilitated by the silica fibers used in this work opens future prospects for the use as a scaffold material in regenerative medicine. As some important cells of the innate and adaptive immune system, namely neutrophils, monocytes and some leukocytes exhibit amoeboid movement, the immune response after wounding might be greatly enhanced: a 3D silica fiber scaffold implanted into the site of injury can serve as a guiding structure, enabling immune cells to immigrate into the injury and facilitating cell movement in every spatial direction. An accelerated infiltration of immune cells could down-regulate the overall immune response which has been shown as a potential route to scarless regeneration of tissue.39,40 Knowledge of the functional connectivities and the underlying intracellular mechanisms may lead to novel strategies for cell guidance and the manipulation of cell functions. For patients, such “smart implants” for tissue regeneration would have great benefit.
Acknowledgements
D. discoideum cells were kindly provided by Dr Günter Gerisch (Max-Planck Institut für Biochemie, Martinsried). D. H. acknowledges funding from the Deutsche Forschungsgemeinschaft (grant HE5958-2-1), the Volkswagen-Foundation (grant I85100) and the Fraunhofer Attract Program.References
- C. J. Weijer, J. Cell Sci., 2009, 122(18), 3215 CrossRef CAS PubMed.
- S. Kurosaka and A. Kashina, Birth Defects Res., Part C, 2008, 84(2), 102 CrossRef CAS PubMed.
- G. C. Gurtner, S. Werner, Y. Barrandon and M. T. Longaker, Nature, 2008, 453(7193), 314 CrossRef CAS PubMed.
- J. R. Hanna and J. A. Giacopelli, J. Foot Ankle Surg., 1997, 36(1), 2 CrossRef CAS PubMed.
- S. J. Lee, J. S. Choi, K. S. Park, G. Khang, Y. M. Lee and H. B. Lee, Biomaterials, 2004, 25(19), 4699 CrossRef CAS PubMed.
- P. Friedl and E.-B. Bröcker, Cell. Mol. Life Sci., 2000, 57, 41 CrossRef CAS PubMed.
- E. Evans, Biophys. J., 1993, 64(4), 1306 CrossRef CAS.
- P. Friedl, Curr. Opin. Cell Biol., 2004, 16(1), 14 CrossRef CAS PubMed.
- P. W. Oakes, D. C. Patel, N. A. Morin, D. P. Zitterbart, B. Fabry, J. S. Reichner and J. X. Tang, Blood, 2009, 114(7), 1387 CrossRef CAS PubMed.
- K. Wolf, R. Müller, S. Borgmann, E. B. Bröcker and P. Friedl, Blood, 2003, 102(9), 3262 CrossRef CAS PubMed.
- P. Friedl, S. Borgmann and E.-B. Bröcker, J. Leukocyte Biol., 2001, 70(4), 491 CAS.
- S. J. Annesley and P. R. Fisher, Mol. Cell Biochem., 2009, 329(1–2), 73 CrossRef CAS PubMed.
- B. Meier, A. Zielinski, C. Weber, D. Arcizet, S. Youssef, T. Franosch, J. O. Rädler and D. Heinrich, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(28), 11417 CrossRef CAS PubMed.
- O. D. Weiner, Curr. Opin. Cell Biol., 2002, 14(2), 196 CrossRef CAS PubMed.
- T. Lämmermann and M. Sixt, Curr. Opin. Cell Biol., 2009, 21(5), 636 CrossRef PubMed.
- A. J. Ridley, M. A. Schwartz, K. Burridge, R. A. Firtel, M. H. Ginsberg, G. Borisy, J. T. Parsons and A. R. Horwitz, Science, 2003, 302(5651), 1704 CrossRef CAS PubMed.
- Y. Iwadate and S. Yumura, J. Cell Sci., 2008, 121(8), 1314 CrossRef CAS PubMed.
- E. Sackmann, F. Keber and D. Heinrich, Annu. Rev. Condens. Matter Phys., 2010, 1(1), 257 CrossRef CAS.
- J. Albuschies and V. Vogel, Sci. Rep., 2013, 3, 1658 CAS.
- M.K. Driscoll, X. Sun, C. Guven, J. T. Fourkas and W. Losert, ACS Nano, 2014, 8(4), 3546 CrossRef CAS PubMed.
- D. Arcizet, S. Capito, M. Gorelashvili, C. Leonhardt, M. Vollmer, S. Youssef, S. Rappl and D. Heinrich, Soft Matter, 2012, 8(5), 1473 RSC.
- M. Emmert, P. Witzel and D. Heinrich, Soft Matter, 2016, 12(19), 4287 RSC.
- M. Kursawe, W. Glaubitt and A. Thierauf, J. Sol-Gel Sci. Technol., 1998, 13(1), 267 CrossRef CAS.
- E. Preedy, S. Perni, D. Nipic, K. Bohinc and P. Prokopovich, Langmuir, 2014, 30(31), 9466 CrossRef CAS PubMed.
- S. Youssef, S. Gude and J. O. Rädler, Integr. Biol., 2011, 3(11), 1095 RSC.
- M. Gorelashvili, M. Emmert, K. F. Hodeck and D. Heinrich, New J. Phys., 2014, 16(7), 75012 CrossRef.
- D. Arcizet, B. Meier, E. Sackmann, J. O. Rädler and D. Heinrich, Phys. Rev. Lett., 2008, 101(24), 248103 CrossRef PubMed.
- A. Nandi, D. Heinrich and B. Lindner, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2012, 86(2), 21926 CrossRef PubMed.
- J. Tan and W. M. Saltzman, Biomaterials, 2002, 23(15), 3215 CrossRef CAS PubMed.
- C. A. Heckman and H. K. Plummer, Cell. Signalling, 2013, 25(11), 2298 CrossRef CAS PubMed.
- K. H. Song, S. J. Park, D. S. Kim and J. Doh, Biomaterials, 2015, 51, 151 CrossRef CAS PubMed.
- P. Y. Jay, P. A. Pham, S. A. Wong and E. L. Elson, J. Cell Sci., 1995, 108, 387 CAS.
- Y. Fukui, A. de Lozanne and J. A. Spudich, J. Cell Biol., 1990, 110(2), 367 CrossRef CAS PubMed.
- S. Yumura, J. Plant Res., 1997, 110(4), 501 CrossRef CAS.
- C. Pasternak, J. A. Spudich and E. L. Elson, Nature, 1989, 341(6242), 549 CrossRef CAS PubMed.
- E. Shelden and D. A. Knecht, Cell Motil. Cytoskeleton, 1996, 35(1), 59 CrossRef CAS PubMed.
- K. S. K. Uchida and S. Yumura, J. Cell Sci., 2004, 117(8), 1443 CrossRef CAS PubMed.
- P. W. Grabham, B. Reznik and D. J. Goldberg, J. Cell Sci., 2003, 116(18), 3739 CrossRef CAS PubMed.
- T. Wilgus, Pharmacol. Res., 2008, 58(2), 112 CrossRef CAS PubMed.
- S. R. Zins, M. F. Amare, K. Anam, E. A. Elster and T. A. Davis, J. Inflammation, 2010, 7(1), 25 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra25739a |
| This journal is © The Royal Society of Chemistry 2017 |