Open Access Article
Open Access ArticleScreening for the bioactive constituents of traditional Chinese medicines—progress and challenges
Xing-Xin Yang a,
Wen Gua,
Li Lianga,
Hong-Li Yana,
Yan-Fang Wanga,
Qian Bia,
Ting Zhanga,
Jie Yu*ab and
Gao-Xiong Rao*ab
a,
Wen Gua,
Li Lianga,
Hong-Li Yana,
Yan-Fang Wanga,
Qian Bia,
Ting Zhanga,
Jie Yu*ab and
Gao-Xiong Rao*ab
aCollege of Pharmaceutical Science, Yunnan University of Traditional Chinese Medicine, 1076 Yuhua Road, Kunming 650500, Yunnan Province, P. R. China. E-mail: cz.yujie@gmail.com; rao13987124569@qq.com; Fax: +86-871-65933303; Fax: +86-871-65918232; Tel: +86-871-65933303 Tel: +86-871-65918232
bEngineering Laboratory for National Healthcare Theories and Products of Yunnan Province, 1076 Yuhua Road, Kunming 650500, Yunnan Province, P. R. China
First published on 13th January 2017
Abstract
The search for lead compounds from traditional Chinese medicines (TCMs) may be promising for new drug development. Screening efficiency and efficacy is chiefly determined by the validity of the selected screening tools. Recently, many experimental screening tools have been developed to identify lead compounds. This review provides a toolbox of the experimental screening methods and models that have been applied to discover new lead compounds and drugs from TCMs. The merits and limitations of these tools are summarized, assisting readers in the rapid retrieval of useful information and providing reference values for selecting appropriate screening tools for different research.
1. Introduction
Traditional Chinese Medicines (TCMs) count among the world's most ancient herbal medicines and have been used extensively by TCM practitioners for thousands of years.1 TCM has played an indispensable role in the prevention and treatment of human disease,1–3 especially in complicated and chronic diseases such as rheumatism and arthritis. As a complementary therapeutic technique with fewer side effects than western medicines, TCM is attracting increasing attention. The chemical synthesis of new drugs has rapidly developed in recent years with the advancement of combinatorial chemistry and computer-aided drug design technology. However, the exploration of drugs and lead compounds from TCMs remains a significant avenue for drug development,4–6 because of the novel structures, therapeutic abilities, and certain unique pharmacological effects of the chemical substances in TCMs. The screening of bioactive constituents from TCMs is of great significance in innovative drug discovery with medicinal herbs as lead compounds.The clarification of the active constituents of TCMs is a precondition to understanding their action mechanism, and provides the foundation and core of TCM quality control. It also guards the safety, effectiveness, stability, and controllability of TCMs. However, multiple substances in TCMs and their likely complex mechanisms of action complicate screening and analysis for bioactive constituents, even from well-documented formulations for specific diseases. This acutely restrains the development and modernization of TCM. Thus, the development of screening methods and models that efficiently search for bioactive constituents within the complex matrices of TCMs is important for elucidating the pharmacodynamic essence (including effective substances and action mechanisms) of TCMs and in the development of new drugs from them.
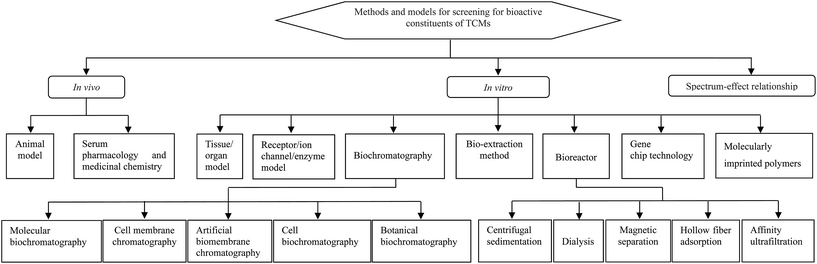
A variety of screening methods and models (Fig. 1) have been developed and used to screen for the bioactive constituents of TCMs.6–8 These include animal, tissue/organ, cell, and receptor/enzyme models; serum pharmacology; biochromatography; gene chip technology; molecularly imprinted polymers and spectrum-effect relationships. Although widely applied, these screening tools may cause confusion, with difficulty in selecting suitable ones, due to their considerable diversity. In this review, these screening tools are identified, and their merits and limitations are summarized simply yet comprehensively. This may assist readers in the rapid retrieval of useful information for the selection and application of suitable screening tools for different research objectives.
2. In vivo screening methods and models
2.1. Animal models
The extraction of a single compound or single distilled fraction from natural products and the evaluation of its bioactivity in classic pathological animal models is a routinely used screening process. Rat, mouse, rabbit, beagle dog, monkey, and zebra fish are commonly used model animals. The animal model is the most classic pharmacological screening model, and is important in medicine evaluation as it can mimic the physiopathological and clinical features of patients, demonstrating the efficacy, side effects, and toxicity of medicines. Although the use of animal models is laborious and time-consuming, and their screening throughput is low, it remains the primary indispensable method of drug discovery and evaluation.6,82.2. Serum pharmacology and serum medicinal chemistry
Although TCMs contain numerous complex chemical substances, detection of distinct compounds in TCMs after they enter the bloodstream are extremely limited.8 TCM ingredients that enter the bloodstream comprise the “crude extract” of serum, which includes constituents present in TCMs and formed in the process of decoction, as well as metabolites and endogenous bioactive substances in the body generated by stimulation from TCMs.9 The effectiveness of serum containing crude extract is evaluated using serum pharmacology, whereas serum medicinal chemistry is the process of isolating and identifying the active compounds in serum. The analytical process begins with the intragastric administration of TCM extracts. The serum is then separated and its bioactivity evaluated by pharmacological means, in vitro and in vivo. Finally, the bioactive constituents in the serum are separated and identified.8 As such, the chemical substances of TCMs that exert certain pharmacologic actions may be revealed. Substance effectiveness of many types of TCMs has been researched using this method, with valuable results obtained.8–11This method has the following advantages: (1) interference from the physical and chemical properties of crude TCM preparations is excluded, such as pH, osmotic pressure, and the presence of electrolytes or tannins; (2) TCM ingredients in the gastrointestinal tract that cannot enter the bloodstream are excluded, which simplifies exploration; (3) serum from the administered animal will contain the authentic effective TCM substances; (4) the influence of many factors that affect in vitro experiments is avoided, potentially reflecting the effectiveness of TCMs more accurately and authentically.
However, there are also some deficiencies in this method: (1) the separation of bioactive constituents from complex biological samples (serum) is more difficult than direct separation from crude TCM extracts; (2) the inference of information about the structure of bioactive compounds in TCMs from their metabolites is difficult; (3) some compounds detected in the serum may be inactive ingredients in TCMs; (4) the efficacy of purified compounds that appear in serum is only evident when the blood concentration of the compound exceeds the original concentration; (5) operational protocols require standardization for dose schedule, time and method of blood collection, serum pretreatment, differences in drug absorption from individual animals, and differences in the composition of serum and the concentration of compounds from dosage and sampling time. Therefore, although the application of this method has excellent prospects, problems remain that need to be addressed further.
3. In vitro screening methods and models
3.1. Tissue/organ models
With the development of modern medicine and pharmacology, a growing number of animal tissue/organ models have been established for evaluating the activity of drugs, such as the isolated vascular ring, heart/liver perfusion and tissue culture models. By investigating the effects of a drug on specific tissues or organs, possible pharmacological activity and mechanism of action may be explored. This model has more recently become an important means for activity screening. It overcomes some of the deficiencies of the animal model in the following ways:8 (1) the amount of screening sample required is less. Generally, a sample of between 1–5 g or more is required for use in an animal model (depending on the dosage and the size of the animal used), while the sample amount required in a tissue/organ model is as low as or less than one-tenth of that used in an animal model; (2) labor intensity and animal consumption are less, and the screening scale may be expanded; in some models only a small amount of tissue or organ (such as the isolated vascular ring model) is required for a complete screening. Multi-sample screening can be performed simultaneously, improving efficiency and decreasing costs; (3) the interference from other in vivo factors is reduced, allowing more precise evaluation.Despite the advantages of screening using tissue/organ models, limitations exist, such as a small screening scale, low efficiency, limited pharmacological indices and difficulty of realizing high-throughput and high-content screening. The more recent application of tissue microarray technology12,13 is improving screening efficiency and drug target identification at the tissue/organ level, with advantages of high throughput, low experimental error and abundant information capacity. Tissue microarray technology is also time-saving, and small sample volumes can be handled.
3.2. Cell model
The cell model is the most widely-used drug screening model, and not only more closely mimics the pathophysiological state of various diseases, but also provides useful information about the predictability of bioactive compounds in vivo, as well as on permeability and toxicity. It shows considerable advantage over whole-animal and tissue/organ models: (1) screening efficiency is greatly improved, due to the simple and fast operation of cell experiments, the short time period for cell growth, and the applicability of large-scale cultures; (2) factors interfering with efficacy are reduced and easier to control, making screening results more stable and reliable; (3) high throughput screening can be achieved as many compounds can be screened simultaneously.14,15 This makes it possible to find drugs with excellent activity solely by expanding the screening objects and scope; (4) due to the low-consumption sample amounts used in a cell model, a small amount of isolated compound (fraction) can be used with multiple pathological models, improving the probability of discovering new drugs and lead compounds; (5) high content screening can be performed. Because the action of a drug on cells may be demonstrated in a variety of ways, its efficacy or toxicity is more thoroughly revealed with a cell model, by expanding the detection range and concurrently monitoring multiple pharmacological indices.16,17Nevertheless, in vitro cell models cannot mimic the dynamic procedures of drugs in the human body, and do not exactly reflect pharmacological behavior of drugs in vivo. The efficacy of some active compounds identified by cell models may be reduced or absent once in the circulation. Thus, cell model screening processes should simulate the body's internal environment as far as possible. Additionally, activity screening using cell models remains a process in which the effect of samples on cells is observed; this cannot reveal specific pathways and targets of drug action, and only shows comprehensive effects of the drug on cell growth processes.
3.3. Receptor/ion channel/enzyme model
With the development of enzymology and receptor theory, drug targets have been revealed and new drug targets discovered consecutively. So far, more than 600 types of drug target have been found. The use of receptors, ion channels or enzymes as targets to search for bioactive constituents is an important method in new drug discovery.8,18,19 Within this process, the receptor, enzyme or channel is stimulated by the tested compound, and a variety of pharmacological indices are then detected and binding intensity between the compound and target is evaluated. This model has a number of characteristics: (1) the screening process is cut short, with high-detection sensitivity and potent specificity, enabling high-throughput and high content screenings; (2) screening occurs at the binding site, and not only is binding information between drug and target obtained, but the activity of the drug can be understood and functional screening performed; (3) some limitations of an animal model are overcome. For example, diseases can be screened for if an appropriate animal model is not available. Moreover, some drugs are metabolized in the animal gut or liver and may not arrive at their targets, diminishing the ability to assess their activity in a whole animal model.Major TCM compounds are not highly bioactive; compounds do not bind tightly with targets in this model resulting in low screening efficiency. In addition, screening results do not completely reflect multi-target and multi-pathway characteristics of effective TCM substances. Active compounds that bind with targets in this model may not provide the expected pharmacological effect after entering the body and, therefore, may not be developed as a drug.
3.4. Biochromatography
A variety of bioactive materials (enzymes, carrier proteins and cell membranes) or biomimetic membrane materials (liposomes or artificial cell membranes) are used in novel affinity chromatography method known as biochromatography, when a stationary phase is used. Because biological activity occurs with a stationary phase and various bioactive substances are specifically bonded, biochromatography has been recently applied to the screening, separation, and preparation of active substances from mixtures.20,21 Biomacromolecules, cell membrane (or biomimetic membrane), living cells and plant cells (or cell walls) have been fixed on a chromatographic carrier (e.g. silica and gel) to form different biochromatography stationary phases. Biochromatography may be subdivided into molecular biochromatography, cell membrane chromatography (or artificial biomembrane chromatography), cell biochromatography, and botanical biochromatography, according to the chromatographic stationary phase used.These methods have the characteristics of both chromatographic separation and biological activity, and separation and recognition of biological activity can be achieved simultaneously while screening for bioactive constituents. Therefore, active constituents not only bind and are selectively recognized during the stationary phase, but interference from inactive ingredients in a complex matrix such as that found in TCMs is directly eliminated, contributing to an efficient screening when used with TCMs.20,22 Two-dimensional “recognition–separation–identification” chromatographic systems have been developed by combining biochromatography with liquid chromatography/mass spectrometry (LC/MS) or gas chromatography/mass spectrometry (GC/MS), enabling highly efficient screening for bioactive constituents in complex TCM samples.23–29
Biochromatography technology has been widely used for TCM research, but disadvantages remain: (1) the bioactivity of a stationary phase can only be preserved for a short time, which may lead to failure of commercialization of the chromatographic column; (2) the pressure and temperature requirements of columns, and the properties and flow rate of a mobile phase, are highly specific. A chromatographic environment with high pressure and anoxia is not amenable to the maintenance of bioactivity in a stationary phase in the analytical process; (3) the preparation procedure of a stationary phase is complicated. A stationary phase with bioactivity is easy to inactivate allosterically during preparation and use. Furthermore, stationary phases prepared in different laboratories and used in different environments produce different results, challenging the reproducibility and applicability of biochromatography; (4) the amount of active constituents specifically retained by the stationary phase is usually small, making it difficult to obtain sufficient sample quantities for the identification of molecular structure or pharmacological verification trials. In summary, further improvements to the performance and dependability of biochromatography technology are required before widespread application of this technique to screening of TCMs.
This technology has been applied widely and Wang et al.31 used a chromatographic column filled with stationary phase of silica-bonded human serum albumin to screen Angelica sinensis extract, confirming a finding of the two most effective compounds previously reported in the literature. Additionally, Wang et al.26 used a stationary phase of silica-bonded human serum albumin with LC/MS to screen and analyze an extract of Longdan Xiegan decoction. They found hundreds of compounds interacting with human serum albumin, of which 19 were identified using LC/MS. Su et al.32 applied a stationary phase of silica-bonded DNA to screen for TCM extracts. The chromatographic retention time of the screened compounds correlated well with their affinity for DNA, and multiple active compounds that could bind to DNA were identified from these complex TCM samples. Min et al.33 coated acetylcholinesterase (AChE) on a capillary wall to prepare a bioactive stationary phase of capillary electrophoresis. The prepared phase was then used to screen for a variety of TCM extracts, including 30 extracts that interact with AChE, so this method may be used to screen for AChE inhibitors.
A variety of CMC models have been developed using cell membranes from different tissues (vascular smooth muscle, myocardium, and cerebral vessel) or cultured cell lines (red blood cells, macrophages, and cells with high expression of CD40, EGFR, and α1AAR). By combining online (or offline) with LC/MS or GC/MS, these CMC models have been widely applied to the screening of bioactive constituents from TCM extracts, and data show that CMC can be used to screen for active constituents of TCMs.22,34,35
3.5. Bio-extraction method
With bio-extraction, the analyzed sample is incubated with bioactive material (such as enzymes, receptors, carrier proteins, cell membranes, DNA, live cells, and liposomes), to bind active constituents with active material. The bound active constituents are then separated from complex matrices using specific separation techniques. As with biochromatography, bio-extraction can be used to identify biological activity and for chromatographic separation. The active constituents of complex samples such as TCMs can be recognized and separated, with direct elimination of interference from inactive ingredients.Unlike biochromatography, the screening target for bio-extraction does not require immobilization on a chromatography carrier, thus avoiding the complicated stationary phase preparation and packing process of a column, which is prone to maintaining the stereoisomers and biological activity of the screening target. Combined with online or offline LC/MS or GC/MS, bio-extraction can be performed for the convenient identification and analysis of active constituents in TCMs and has recently received widespread attention and application in TCM research. Several separation techniques may be used, including centrifugal sedimentation, dialysis, magnetic separation, hollow fiber adsorption, and affinity ultrafiltration, with a suitable separation technique selected according to screening target and sample characteristics.
Li et al.41 isolated 6 compounds from an extract of Danggui Buxue decoction that selectively combined with endothelial cells. Of these, Angelica sinensis Radix contributed 2, and Astragali Radix 4. Four of the 6 potentially bioactive compounds were elucidated using HPLC/MS and identified as ononoside, calycosin, 3-butylphthalide, and ligustilide. Fan et al.42 found 8 compounds in a sample of Mailuoning injection that may bind with platelet membrane, using centrifugal sedimentation. Zhang et al.43 found 14 active compounds from a glucose injection of zedoary turmeric oil that bound with living hepatocytes, using centrifugal sedimentation with GC/MS analysis. Of these, curzerene and β-elemene, which were of the highest content, were the main active ingredients. Zhang et al.44 identified 9, 7 and 13 active compounds that could bind with HL-7702, RAW 264.7 and Caco-2 cells, respectively, from an extract of Danggui Buxue decoction, using centrifugal sedimentation with LC/MS analysis. Using the same method, many active constituents binding different cells or cell membranes have been isolated from TCM extracts such as Scrophulariae Radix, Notoginseng Radix, Astragali Radix, Angelicae sinensis Radix, Zingiberis Rhizoma, Cordyceps, Artemisiae scopariae Herba, Epimedii Folium and Yu-ping-feng powder.20
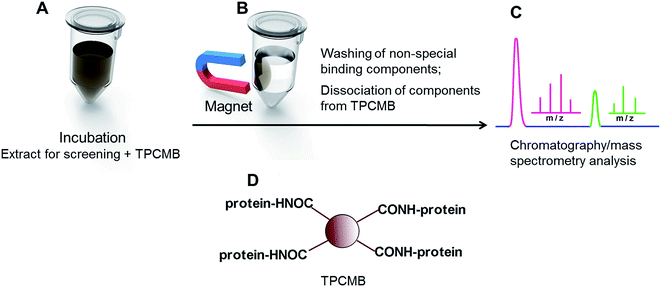
A variety of TPCMBs for different target proteins have been fabricated to screen for active constituents in complex matrices such as TCMs, and several active compounds have been found. Target proteins used have included human serum albumin,48–51 bovine serum albumin,52 hVDR protein,53 anti-streptavidin,54 protein tyrosine phosphatase 1B,55 α-glucosidase,55 maltase,56 sucrase,56 lipases,56 estrogen receptor,57 SIRT6 protein,58 acetylcholinesterase,59 heat shock protein,60 and calmodulin.61 Qing et al. identified 3 and 4 active compounds from extracts of Dioscorea nipponica48 and Dioscorea panthaica,49 respectively, using human serum albumin-coupled magnetic beads. Zhang et al.50 used human serum albumin-coupled magnetic beads to isolate 47 active substances from Astragali Radix. Liu et al.52 prepared bovine serum albumin-coupled magnetic beads to extract 13 active compounds from Puerariae lobatae Radix. Arai et al.53 prepared hVDR protein-coupled magnetic beads and found 2 active substances (ent-12(E)-labda-8(17),12,14-trien-18-ol and ent-12(Z)-labda-8(17),12,14-trien-18-ol) in the ethanol extract of Eleutherine palmifolia. Qing et al.55 prepared protein tyrosine phosphatase 1B- and α-glucosidase-coupled magnetic beads, respectively extracting 4 and 2 active compounds from the bark of Punica granatum L. Tao et al.56 developed a multi-target screening method connecting maltose-, sucrose- and lipase-coupled magnetic beads, and identified 5 active compounds inhibiting the 3 enzymes immobilized on magnetic beads from the TCM formula of Sanye Tangzhiqing. Yasuda et al.58 isolated 2 active compounds from an extract of common Fenugreek seed using prepared SIRT6 protein-coupled magnetic beads.
Although these results suggest that screening active constituents from complex matrices of TCMs can be achieved using TPCMB with magnetic field separation, the target protein may be denatured or its three-dimensional configuration may be changed when the target protein is coupled to magnetic beads. This results in mild binding with active compounds, or even unbinding. Additionally, coupling conditions should be optimized to maximally immobilize protein, which improves screening sensitivity.
Hollow fiber adsorption has been used to screen living cell, cell membranes, organelles, and enzyme targets. Xue et al.62 identified a diverse active compounds from extracts of Schisandrae chinensis Fructus, Cnidii Fructus and Psoraleae Fructus using hollow fiber adsorbed with living cells. Liu et al.63 isolated several active compounds from Coptidis Rhizoma using 3 types of hollow fiber, respectively adsorbed with 3 different targets (MCF-7 cells, cell membranes and organelles). Yan et al.64 found several anthraquinone compounds that could penetrate cells from extracts of Polygoni cuspidati Rhizoma, Cassiae obtusifoliae Semen and Polygoni multiflori Radix. Tao et al.65 isolated 3 lipase-bound ligands (quercetin-3-O-β-D-arabinopyranosyl-(1→2)-β-D-galactopyranoside, quercetin-3-O-β-D-glucuronide and kaempferol-3-O-β-D-glucuronide) from extracts of lotus leaf, using hollow fiber adsorbed with lipases from porcine pancreas. Despite these successes, limitations persist. The target adsorbed on the inner wall of the hollow fiber has a short survival time and only few targets are adsorbed, restricting the sensitivity of this method.
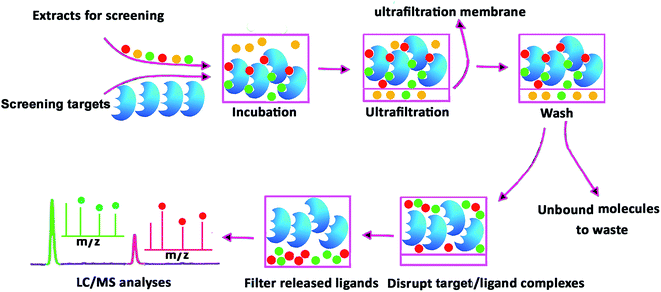 | ||
| Fig. 3 Analytical procedure for the identification of bioactive constituents from complex mixtures, using centrifugal ultrafiltration/liquid chromatography/mass spectrometry method. | ||
Compared with other active molecular screening methods such as biochromatography, magnetic separation, and hollow fiber adsorption, affinity ultrafiltration has significant advantages:70 (1) it is not necessary to immobilize the screening targets on a chromatographic carrier. The target is directly incubated with the samples, which maintain their native conformation and avoid a reduction in activity, deactivation, or conformational change originating from the target immobilization; (2) pulse ultrafiltration can be conveniently coupled with online MS (or with a fraction trap, followed by online LC/MS), for online analysis of active constituents; (3) affinity ultrafiltration is convenient to operate, low-cost, and has a high hit ratio.
Many active molecules have been found in a variety of natural products and combinatorial libraries using affinity ultrafiltration combined with MS or LC/MS (Table 1), suggesting that affinity ultrafiltration could be used for screening active constituents from products with complex matrices, such as TCMs.
| Screened sample | Target | Screening results | |
|---|---|---|---|
| Combinatorial libraries | 20–30 closely related molecules71 | Benzodiazepine antibodies | Benzodiazepines recognized |
Combinatorial library comprising 123![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 405 compounds72 405 compounds72 |
Streptococcus pneumoniae cell wall biosynthesis enzyme (MurF) | 2 specific ligands | |
Combinatorial library comprising 237![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 797 compounds73 797 compounds73 |
DNA damage checkpoint kinase Chk1 | 58 specific ligands | |
Combinatorial library comprising 263![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 382 compounds74 382 compounds74 |
Anti-apoptosis protein Bcl-xL | 12 specific ligands | |
| Combinatorial library comprising 5 compounds75 | Topoisomerase I | 3 specific ligands | |
| Combinatorial library comprising 133 compounds76 | Plasmodium falciparum thioredoxin reductase (PfTrxR) | 9 specific ligands | |
| Series of analogues of the adenosine deaminase inhibitor77 | Adenosine deaminase | 3 enzymic inhibitors | |
| Combinatorial library comprising 22 compounds78 | Dihydrofolate reductase | 6 enzymic inhibitors | |
| 22 chymostatin derivatives79 | Chymotrypsin | 10 enzymic inhibitors | |
| Mixtures of estradiol and 6 equine estrogens80 | Estrogen receptor (ER) | Method can be used for evaluation of affinity with ER and affinity discrepancy between ER-β and ER-α | |
| Natural products | Different cyclo-oxygenase (COX-2) inhibitors dissolved in DMSO, plant extract and bacterial fermentation broth extract81 | COX-2 | 2 or 3 ligands for COX-2 were identified in each sample |
| Methanolic fermentation broth extract spiked with known inhibitor to carbonic anhydrase82 | Carbonic anhydrase and serum albumin | Non-covalent ligand binding to both targets observed | |
| TCM Huo-Luo-Xiao-Ling Dan83 | Cyclo-oxygenase | 17 specific ligands | |
| Botanical extracts and their metabolites obtained by hepatic cytochrome P450 (ref. 84) | Glutathione (GSH) | Several compounds that formed GSH conjugates | |
| Lonicerae japonicae Flos extract85 | Liposome | 8 permeable compounds | |
| Extract of a marine sediment bacterial culture86 | Human retinoid X receptors (RXRs) | 2 specific ligands | |
| Cells grown in culture64 | Topoisomerase I | 1 enzymic inhibitor | |
| Mulberry leaf extracts87 | Tyrosinase | 2 enzymic inhibitors | |
| Scutellaria Radix extract88 | Neuraminidase | 6 enzymic inhibitors | |
| Salviae miltiorrhizae Radix extract89 | Xanthine oxidase | 11 enzymic inhibitors | |
| Water extract of Eucommia ulmoides bark90 | Phosphodiesterase | 7 enzymic inhibitors | |
| Extract of marine sediment bacteria91 | Quinine reductase-2 | 1 enzymic inhibitor | |
| Extract of Humulus lupulus L.91 | Quinine reductase-2 | 2 enzymic inhibitors | |
| Water extract of Eucommia ulmodies leaves92 | Bovine serum albumin | 6 special ligands | |
| Lonicerae japonicae Flos extract93 | Bovine serum albumin | 11 special ligands | |
| Extract of Lonicera japonica Thunb.94 | DNA | 4 DNA binders | |
| TCM red clover isoflavone extracts95 | DNA | 5 DNA binders | |
| Extracts of Macleaya cordata, Chelidonium majus and Coptis chinensis96 | DNA | 9 DNA binders | |
| Hawthorn leaf flavonoids extract97 | α-Glucosidase | 4 enzymic inhibitors | |
| Extract of leaves of Acanthopanax senticosus harms98 | α-Glucosidase | 8 enzymic inhibitors | |
| Total saponins of ginseng stems and leaves99 | α-Glucosidase | 12 enzymic inhibitors | |
| Extract of Polygonatum odoratum100 | α-Glucosidase | 9 enzymic inhibitors | |
| Astragali Radix extract and its human microsomal metabolites101 | α-Glucosidase | 13 prototype isoflavonoids and 1 monohydroxylated metabolic isoflavonoid with enzymic binding activity | |
| Extracts of Polygoni cuspidati Rhizoma et Radix and Scutellariae Radix102 | Mitochondria | 19 active compounds detected and identified | |
3.6. Bioreactor
In a bioreactor, analytical samples are incubated with screening targets, and reaction substrates are added to initiate a biochemical reaction under enzymatic catalysis. The concentrations (or contents) of active compounds change during this process and reactions can be divided into pre-, in- and after-column according to the installation site of the bioreactor.With a pre-column reaction, reacted samples are separated and measured with chromatography (or electrophoresis), which is convenient and sensitive. Mou et al.103 cultured LoVo cells on a porous PHC scaffold to develop a three-dimensional cell bioreactor. Combined with HPLC/MS, this bioreactor was used to screen for active constituents in Polygonum cillinerve (Nakai) Ohwi extract, resulting in the identification of 2 active compounds (aristolochic acid A and aristolochic acid B).
With an in-column reaction, the screening targets (enzymes) are immobilized on a support and then packed in a column. Samples are injected into the column and incubated with the enzymes and substrates. After the biochemical reaction, reactive samples are separated using electrophoresis (or chromatography). This method is highly stable and repeatable, with little enzyme consumption, but the immobilized enzymes may be denatured or inactivated. L-Glutamate dehydrogenases were immobilized on the surface of gold nanoparticles, and then packed into a capillary column to develop an online enzyme microreactor, by Zhao et al.104 Using this microreactor, 3 TCM extracts that had inhibitory activity were identified from 25 extracts. Ji et al.105 prepared a microreactor of adenosine deaminase and identified one inhibitory extract from 19 TCM extracts.
In an after-column reaction, the analytical samples are separated by chromatography (electrophoresis), and separated fractions are reacted with screening targets (enzymes). Changes in sample components are then measured. Interference within the biochemical reaction from matrices and other components of complex samples is less with this method. However, online sample detection is difficult and special equipment and techniques are required. Li et al.106 developed online microreactors for xanthine oxidase and ABTS radical cation (ABTS˙+) to screen for xanthine oxidase inhibitors and free radical scavengers, respectively, from Oroxyli Semen extract. Six free radical scavengers, a xanthine oxidase inhibitor and a compound with free radical scavenging and xanthine oxidase inhibiting activity were identified. Lin et al.107 next prepared luminol and ferricyanide microreactors to screen Aurantii Fructus for antioxidants and found 25 active compounds.
3.7. Gene chip technology
Genes carry genetic information and drugs act through different targets, directly or indirectly affecting cellular gene expression. DNA microarray gene expression profiling is the gene chip generally used for drug screening.108 By comparing genetic expression of two samples before and after drug administration, changes in expression may help to identify the target gene of the drug. Gene chips for drug screening is low with respect to labor intensity and animal consumption and can be carried out with small samples. Gene chip screening also has a short cycle, which decreases time and cost of drug screening, so it is popular for screening active constituents in TCMs.109–113 Gene chip analysis can be improved by an increase in chip specificity, simplification of sample preparation and marking, improvements in signal detection sensitivity and stability, and additional cost reduction.3.8. Molecularly imprinted polymers
Molecular imprinting is used to prepare polymers containing recognition sites of predetermined specificity. Molecularly imprinted polymers (MIPs) are complexed in a solution of template molecules with functional monomers by covalent or non-covalent bonds, followed by a polymerization reaction with an excess of cross-linkers. Removal of the templates leaves specific recognition sites that are complementary to the template in terms of its shape, size and functionality in the polymer network.114 The perfect selectivity, high binding affinity and physical robustness of MIPs allow their application for non-biological screening in lead compound discovery. Structures of affinitive compounds, which were trapped by MIPs from the mixture, were similar to the template and should have similar bioactivity according to structure–activity relationships.115Despite MIPs require ∼1 mmol of pure template molecule and have difficulty dealing with macromolecules such as proteins and polynucleotides, they have been used for screening analogues with similar bioactivity in the hope of finding new drug candidates from plant extracts or combinatorial chemistry libraries. Yang's group115 prepared oseltamivir MIPs and identified an active component against influenza virus (matrine) from chloroform extracts of a TCM formula of Yandureqing. Huang and colleagues116 prepared propyl gallate MIPs and identified an antiplatelet active ingredient (protocatechuic acid) from Salviae miltiorrhizae Radix. Furthermore, (−)-ephedrine-, (E)-piceatannol-, fisetin-, harman- and RD3-4078-imprinted polymers were prepared for screening and separating active natural compounds from TCMs.114 Steroid receptor, folate receptor and α2-adrenoreceptor mimics have been made by MIPs and used to screen bioactive compounds from synthetic libraries.114
4. Spectrum-effect relationship
Chemical constituents and pharmacological studies have been unable to clarify how TCM substances can treat particular diseases. Exploring the spectrum-effect relationship (SER) by combining chemical constituent studies with pharmacological studies may clarify overall therapeutic effects and the relationship between active constituents. SER is widely applied in research into the basis for substance effectiveness, compatibility, and the processing of TCMs117,118 as follows: (1) fingerprinting of TCM samples from different environments, manufacturers or with different preparation methods is developed to profile chemical discrepancies across different samples; (2) difference in sample efficacy is evaluated by in vivo or in vitro experiments; (3) statistical analysis, such as principal components analysis, direct-vision comparison, and gray relational analysis, is performed to establish the SER, revealing the most effective TCM substances. Xu et al.118 reviewed the current status of and future perspectives in SER for TCM research and various active constituents in single TCMs and TCM formulas have been identified. Thus, SER analysis may potentially determine the effective substances in complex mixtures and reflect the internal quality of TCMs.Although SER research has been used successfully, optimization is needed: (1) TCM fingerprinting may have poor stability and resolution; (2) modern pharmacological models that are currently established within western medicine theory may not adequately reflect the clinical efficacy of TCMs; (3) important factors affecting the stability and reliability of SER research results are not investigated systematically; (4) uniform standards for quantitative indicators of efficacy are lacking; (5) the relationships between the active constituents in TCMs are not clear. In summary, the application of SERs in TCM research needs further exploration.
5. Conclusion
Identifying lead compounds from natural products plays a vital role in the search for successful drug candidates. Furthermore, clarifying the effective substance basis of TCMs is a prerequisite for recognizing TCM efficacy and ensuring TCM quality. However, the complex chemical composition and action mechanisms of TCMs mean that effective substance research is a slow process. Various methods and techniques have been applied to the exploration of the effective substances in commonly used TCMs that treat certain disorders, with valuable results. However, research methods continue to have limitations. New research ideas, and the further development of screening techniques to improve the efficiency and effectiveness of screening for the active constituents of TCMs, are required. This paper's documentation of the techniques currently used may assist readers with the rapid retrieval of useful information, and meaningfully promote the continued search for an effective substance basis to TCM, improving quality control and modernization.Acknowledgements
Financial support from the National Natural Science Foundation of China (Grant 81660596 and 81460623), the Application and Basis Research Project of Yunnan China (Grant 2016FD050), the Natural Science Foundation of Yunnan China (Grant 2014FA035), and the Young and Middle-aged Academic and Technological Leader of Yunnan (Grant 2015HB053) is gratefully acknowledged.References
- K. Chen, Trends Pharmacol. Sci., 1995, 16, 182–187 CrossRef.
- D. Normile, Science, 2003, 299, 188–190 CrossRef CAS PubMed.
- R. Yuan and Y. Lin, Pharmacol. Ther., 2000, 86, 191–198 CrossRef CAS PubMed.
- J. F. Wang, D. Q. Wei and K. C. Chou, Curr. Top. Med. Chem., 2008, 8, 1656–1665 CrossRef CAS PubMed.
- Y. Wang, X. Fan, H. Qu, X. Gao and Y. Cheng, Curr. Top. Med. Chem., 2012, 12, 1356–1362 CrossRef CAS PubMed.
- B. C. Wang, J. Deng, Y. M. Gao, L. C. Zhu, R. He and Y. Q. Xu, Fitoterapia, 2011, 82, 1141–1151 CrossRef CAS PubMed.
- S. M. Chan and J. M. Ye, J. Pharm. Pharm. Sci., 2013, 16, 207–216 Search PubMed.
- X. Huang, X. L. Kong, X. Chen, M. Guo and H. F. Zou, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2004, 812, 71–84 CrossRef CAS.
- X. J. Wang, World J. Sci. Technol., 2002, 4, 1–4 CrossRef.
- G. L. Yan, H. Sun, A. H. Zhang, Y. Han, P. Wang, X. H. Wu, X. C. Meng and X. J. Wang, Zhongguo Zhongyao Zazhi, 2015, 40, 3406–3412 Search PubMed.
- F. Gao, Y. Hu, G. Fang, G. Yang, Z. Xu, L. Dou, Z. Chen and G. Fan, J. Pharm. Biomed. Anal., 2014, 87, 241–260 CrossRef CAS PubMed.
- O. P. Kallioniemi, J. Kononen and G. Sauter, Clin. Chem., 2012, 58, 1717–1718 CAS.
- J. Z. Pan, R. Jörnsten and R. P. Hart, Physiol. Genomics, 2004, 17, 201–214 CrossRef CAS PubMed.
- F. Xu, J. Wu and S. Wang, Biofabrication, 2011, 3, 034101 CrossRef PubMed.
- K. Tsuganezawa, Y. Nakagawa, M. Taruya, S. F. Takahashi, M. Endoh, R. Utata, M. Mori, N. Ogawa, T. Honma, S. Yokoyama, Y. Hashizume, M. Aoki, T. Kasai, T. Kigawa, H. Kojima, T. Okabe, T. Nagano and A. Tanaka, J. Biomol. Screening, 2013, 18, 191–198 CrossRef PubMed.
- M. V. Tsiper, J. Sturgis, L. V. Avramova, S. Parakh, R. Fatig, A. Juan-García, N. Li, B. Rajwa, P. Narayanan, C. W. Qualls, J. P. Robinson and V. J. Davisson, PLoS One, 2012, 7, e45226 CAS.
- L. Tolosa, M. J. Gómez-Lechón, G. Pérez-Cataldo, J. V. Castell and M. T. Donato, Arch. Toxicol., 2013, 87, 1115–1127 CrossRef CAS PubMed.
- Y. Shen, Y. Hu, B. Chen and S. Yao, Comb. Chem. High Throughput Screening, 2010, 13, 885–899 CrossRef CAS PubMed.
- Y. Zhu, Z. Zhang, M. Zhang, D. E. Mais and M. W. Wang, Comb. Chem. High Throughput Screening, 2010, 13, 837–848 CrossRef CAS PubMed.
- C. Chen, F. Q. Yang, H. L. Zuo, Y. L. Song, Z. N. Xia and W. Xiao, J. Chromatogr. Sci., 2013, 51, 780–790 CAS.
- F. Yu, L. Kong, H. Zou and X. Lei, Comb. Chem. High Throughput Screening, 2010, 13, 855–868 CrossRef CAS PubMed.
- X. X. Yang, Y. L. Zhang, X. X. Zhang and X. N. Li, J. Chin. Pharm. Sci., 2011, 20, 20–25 CAS.
- X. F. Hou, M. Z. Zhou, Q. Jiang, S. C. Wang and L. C. He, J. Chromatogr. A, 2009, 1216, 7081–7087 CrossRef CAS PubMed.
- S. C. Wang, M. Sun, Y. M. Zhang, H. Du and L. C. He, J. Chromatogr. A, 2010, 1217, 5246–5252 CrossRef CAS PubMed.
- X. X. Yang, Y. W. Wang, X. X. Zhang, R. M. Chang and X. N. Li, J. Sep. Sci., 2011, 34, 2586–2593 CrossRef CAS PubMed.
- Y. Wang, L. Kong, L. Hu, X. Lei, L. Yang, G. Chou, H. Zou, C. Wang, S. W. Annie Bligh and Z. Wang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 860, 185–194 CrossRef CAS PubMed.
- S. Wang, C. Wang, X. Zhao, S. Mao, Y. Wu and G. Fan, Anal. Chim. Acta, 2012, 713, 121–129 CrossRef CAS PubMed.
- Y. Wang, L. Kong, X. Lei, L. Hu, H. Zou, E. Welbeck, S. W. Bligh and Z. Wang, J. Chromatogr. A, 2009, 1216, 2185–2191 CrossRef CAS PubMed.
- D. Jia, X. Chen, Y. Cao, X. Wu, X. Ding, H. Zhang, C. Zhang, Y. Chai and Z. Zhu, J. Pharm. Biomed. Anal., 2016, 118, 27–33 CrossRef CAS PubMed.
- X. Su, L. Kong, X. Lei, L. Hu, M. Ye and H. Zou, Mini-Rev. Med. Chem., 2007, 7, 87–98 CrossRef CAS PubMed.
- H. Wang, L. Kong, H. Zou, J. Ni and Y. Zhang, Chromatographia, 1999, 50, 439–445 CAS.
- X. Su, L. Hu, L. Kong, X. Lei and H. Zou, J. Chromatogr. A, 2007, 1154, 132–137 CrossRef CAS PubMed.
- W. A. Min, W. P. Wang, J. R. Chen, A. Wang and Z. Hu, Anal. Bioanal. Chem., 2012, 404, 2397–2405 CrossRef CAS PubMed.
- X. Hou, S. Wang, T. Zhang, J. Ma, J. Zhang, Y. Zhang, W. Lu, H. He and L. He, J. Pharm. Biomed. Anal., 2014, 101, 141–150 CrossRef CAS PubMed.
- S. Muhammad, S. Han, X. Xie, S. Wang and M. M. Aziz, J. Sep. Sci., 2016, 1–15 CAS.
- R. L. Pignatello, T. Musumeci, L. Basile, C. Carbone and G. Puglisi, J. Pharm. BioAllied Sci., 2011, 3, 4–14 CrossRef CAS PubMed.
- C. Zhang, J. Li, L. Xu and Z. G. Shi, J. Chromatogr. A, 2012, 1233, 78–84 CrossRef CAS PubMed.
- C. Zeng, Y. Zhang, L. Lu, E. Brekkan, A. Lundqvist and P. Lundahl, Biochim. Biophys. Acta, 1997, 1325, 91–98 CrossRef CAS.
- R. Ehwald, H. Woehlecke and C. Titel, Phytochemistry, 1992, 31, 3033–3038 CrossRef CAS.
- G. Dongow ski, R. Ehwald, K. Luck and G. Stoof, Phytochemistry, 1992, 31, 3039–3042 CrossRef.
- S. L. Li, P. Li, L. H. Sheng, R. Y. Li, L. W. Qi and L. Y. Zhang, J. Pharm. Biomed. Anal., 2006, 41, 576–581 CrossRef CAS PubMed.
- H. W. Fan, Q. Zhu, M. Hong and L. Yu, Chin. Pharm. J., 2006, 41, 63–66 CrossRef CAS PubMed.
- H. Y. Zhang, C. X. Hu, C. P. Liu, H. F. Li, J. S. Wang, K. L. Yuan, J. W. Tang and G. W. Xu, J. Pharm. Biomed. Anal., 2007, 43, 151–157 CrossRef CAS PubMed.
- X. Zhang, L. W. Qi, L. Yi, P. Li, X. D. Wen and Q. T. Yu, Biomed. Chromatogr., 2008, 22, 157–163 CrossRef CAS PubMed.
- X. Lei, L. Kong, H. Zou, H. Ma and L. Yang, J. Chromatogr. A, 2009, 1216, 2179–2184 CrossRef CAS PubMed.
- L. W. Qi, P. Li and L. H. Sheng, Chin. J. Anal. Chem., 2006, 34, 196–199 CAS.
- Q. Q. Dong, R. Y. Li and P. Li, Chin. Pharm. J., 2007, 42, 1208–1211 CAS.
- L. S. Qing, Y. Xue, Y. Zheng, J. Xiong, X. Liao, L. S. Ding, B. G. Li and Y. M. Liu, J. Chromatogr. A, 2010, 1217, 4663–4668 CrossRef CAS PubMed.
- L. S. Qing, X. Q. Shan, X. M. Xu, Y. Xue, W. L. Deng, B. G. Li, X. L. Wang and X. Liao, Rapid Commun. Mass Spectrom., 2010, 24, 3335–3339 CrossRef CAS PubMed.
- Y. Zhang, M. Nie, S. Shi, Q. You, J. Guo and L. Liu, Food Chem., 2014, 146, 56–64 CrossRef CAS PubMed.
- L. S. Qing, Y. Xue, W. L. Deng, X. Liao, X. M. Xu, B. G. Li and Y. M. Liu, Anal. Bioanal. Chem., 2011, 399, 1223–1231 CrossRef CAS PubMed.
- L. Liu, Y. Ma, X. Chen, X. Xiong and S. Shi, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2012, 887–888, 55–60 CAS.
- M. A. Arai, E. Kobatake, T. Koyano, T. Kowithayakorn, S. Kato and M. Ishibashi, Chem.–Asian J., 2009, 4, 1802–1808 CrossRef CAS PubMed.
- X. L. Yang, J. Pu, H. Zhao, X. Y. Li, J. Liao, Y. L. Xie, S. Zhu, G. B. Long, Y. H. Yuan and F. Liao, Microchim. Acta, 2012, 176, 243–249 CrossRef CAS.
- L. S. Qing, N. Tang, Y. Xue, J. Liang, Y. M. Liu and X. Liao, Anal. Methods, 2012, 4, 1612–1615 RSC.
- Y. Tao, Z. Chen, Y. Zhang, Y. Wang and Y. Cheng, J. Pharm. Biomed. Anal., 2013, 78–79, 190–201 CrossRef CAS PubMed.
- Y. Choi and R. B. van Breemen, Comb. Chem. High Throughput Screening, 2008, 11, 1–6 CrossRef CAS PubMed.
- M. Yasuda, D. R. Wilson, S. D. Fugmann and R. Moaddel, Anal. Chem., 2011, 83, 7400–7407 CrossRef CAS PubMed.
- L. Pochet, F. Heus, N. Jonker, H. Lingeman, A. B. Smit, W. M. Niessen and J. Kool, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2011, 879, 1781–1788 CrossRef CAS PubMed.
- M. P. Marszall, R. Moaddel, S. Kole, M. Gandhari, M. Bernier and I. W. Wainer, Anal. Chem., 2008, 80, 7571–7575 CrossRef CAS PubMed.
- M. J. McFadden, M. S. Junop and J. D. Brennan, Anal. Chem., 2010, 82, 9850–9857 CrossRef CAS PubMed.
- X. Xue, L. Li, X. Chen, S. Hu and X. Bai, J. Chromatogr. A, 2013, 1280, 75–83 CrossRef CAS PubMed.
- X. Liu, S. Hu, X. Chen and X. Bai, J. Pharm. Biomed. Anal., 2014, 98, 463–475 CrossRef CAS PubMed.
- Y. Yan, Y. Hao, S. Hu, X. Chen and X. Bai, J. Chromatogr. A, 2013, 1322, 8–17 CrossRef CAS PubMed.
- Y. Tao, Y. Zhang, Y. Wang and Y. Cheng, Anal. Chim. Acta, 2013, 785, 75–81 CrossRef CAS PubMed.
- D. A. Annis, E. Nickbarg, X. Yang, M. R. Ziebell and C. E. Whitehurst, Curr. Opin. Chem. Biol., 2007, 11, 518–526 CrossRef CAS PubMed.
- K. M. Comess and M. E. Schurdak, Curr. Opin. Drug Discovery Dev., 2004, 7, 411–416 CAS.
- T. E. Cloutier, Library screening using ultrafiltration and mass spectrometry, Wiley-VCH, Weinheim, 2007, pp. 157–184 Search PubMed.
- D. Liu, J. Guo, Y. Luo, D. J. Broderick, M. I. Schimerlik, J. M. Pezzuto and R. B. van Breemen, Anal. Chem., 2007, 79, 9398–9402 CrossRef CAS PubMed.
- B. M. Johnson, D. Nikolic and R. B. van Breemen, Mass Spectrom. Rev., 2002, 21, 76–86 CrossRef CAS PubMed.
- R. Wieboldt, J. Zweigenbaum and J. Henion, Anal. Chem., 1997, 69, 1683–1691 CrossRef CAS PubMed.
- K. M. Comess, M. E. Schurdak, M. J. Voorbach, M. Coen, J. D. Trumbull, H. Yang, L. Gao, H. Tang, X. Cheng, C. G. Lerner, J. O. McCall, D. J. Burns and B. A. Beutel, J. Biomol. Screening, 2006, 11, 743–754 CrossRef CAS PubMed.
- K. M. Comess, J. D. Trumbull, C. Park, Z. Chen, R. A. Judge, M. J. Voorbach, M. Coen, L. Gao, H. Tang, P. Kovar, X. Cheng, M. E. Schurdak, H. Zhang, T. Sowin and D. J. Burns, J. Biomol. Screening, 2006, 11, 755–764 CrossRef CAS PubMed.
- J. Qian, M. J. Voorbach, J. R. Huth, M. L. Coen, H. Zhang, S. C. Ng, K. M. Comess, A. M. Petros, S. H. Rosenberg, U. Warrior and D. J. Burns, Anal. Biochem., 2004, 328, 131–138 CrossRef CAS PubMed.
- H. Zhang, Q. Gu, X. Liang and Y. Pan, Anal. Biochem., 2004, 329, 173–179 CrossRef CAS PubMed.
- R. Munigunti, V. Mulabagal and A. I. Calderón, J. Pharm. Biomed. Anal., 2011, 55, 265–271 CrossRef CAS PubMed.
- Y. Z. Zhao, R. B. Van Breemen, D. Nikolic, C. R. Huang, C. P. Woodbury, A. Schilling and D. L. Venton, J. Med. Chem., 1997, 40, 4006–4012 CrossRef CAS PubMed.
- D. Nikolic and R. B. Van Breemen, Comb. Chem. High Throughput Screening, 1998, 1, 47–55 CAS.
- S. Mathur, M. Hassel, F. Steiner, K. Hollemeyer and R. W. Hartmann, J. Biomol. Screening, 2003, 8, 136–148 CrossRef CAS PubMed.
- Y. Sun, C. Gu, X. Liu, W. Liang, P. Yao, J. L. Bolton and R. B. van Breemen, J. Am. Soc. Mass Spectrom., 2005, 16, 271–279 CrossRef CAS PubMed.
- D. Nikolic, S. Habibi-Goudarzi, D. G. Corley, S. Gafner, J. M. Pezzuto and R. B. van Breemen, Anal. Chem., 2000, 72, 3853–3859 CrossRef CAS PubMed.
- M. B. Beverly, P. West and R. K. Julian, Comb. Chem. High Throughput Screening, 2002, 5, 65–73 CrossRef CAS PubMed.
- H. M. Cao, R. Yu, Y. S. Choi, Z. Z. Ma, H. Zhang, W. Xiang, D. Y. Lee, B. M. Berman, K. D. Moudgil, H. H. Fong and R. B. van Breemen, Pharmacol. Res., 2010, 61, 519–524 CrossRef CAS PubMed.
- B. M. Johnson, J. L. Bolton and R. B. van Breemen, Chem. Res. Toxicol., 2001, 14, 1546–1551 CrossRef CAS PubMed.
- X. P. Chen, Y. Q. Xia, Y. Lu and J. Liang, J. Pharm. Biomed. Anal., 2011, 54, 406–410 CrossRef CAS PubMed.
- D. Liu, J. Guo, Y. Luo, D. J. Broderick, M. I. Schimerlik, J. M. Pezzuto and R. B. van Breemen, Anal. Chem., 2007, 79, 9398–9402 CrossRef CAS PubMed.
- Z. Yang, Y. Zhang, L. Sun, Y. Wang, X. Gao and Y. Cheng, Anal. Chim. Acta, 2012, 719, 87–95 CrossRef CAS PubMed.
- S. Liu, J. P. Xing, J. Yan, F. R. Song, Z. Q. Liu and S. Y. Liu, Acta Chim. Sin., 2011, 69, 1570–1574 CAS.
- Y. Liu, S. Liu and Z. Liu, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2013, 923–924, 48–53 CrossRef CAS PubMed.
- S. Y. Shi, M. J. Peng, Y. P. Zhang and S. Peng, Anal. Bioanal. Chem., 2013, 405, 4213–4223 CrossRef CAS PubMed.
- Y. Choi, K. Jermihov, S. J. Nam, M. Sturdy, K. Maloney, X. Qiu, L. R. Chadwick, M. Main, S. N. Chen, A. D. Mesecar, N. R. Farnsworth, G. F. Pauli, W. Fenical, J. M. Pezzuto and R. B. van Breemen, Anal. Chem., 2011, 83, 1048–1105 CrossRef CAS PubMed.
- Y. Zhang, M. Peng, L. Liu, S. Shi and S. Peng, J. Agric. Food Chem., 2012, 60, 3119–3125 CrossRef CAS PubMed.
- Z. M. Qian, S. J. Qin, L. Yi, H. J. Li, P. Li and X. D. Wen, Biomed. Chromatogr., 2008, 22, 202–206 CrossRef CAS PubMed.
- J. L. Zhou, Z. M. Qian, Y. D. Luo, D. Tang, H. Chen, L. Yi and P. Li, Biomed. Chromatogr., 2008, 22, 1164–1172 CrossRef CAS PubMed.
- L. Ma, Z. F. Wang, L. N. Chen, F. R. Song, Z. Q. Liu and S. Y. Liu, Chem. J. Chin. Univ., 2013, 34, 331–335 CAS.
- Z. F. Zhang, Y. Guo and X. H. Wei, Zhongguo Zhongyao Zazhi, 2011, 36, 2684–2688 CAS.
- H. L. Li, F. R. Song, J. P. Xing, R. Tsao, Z. Liu and S. Liu, J. Am. Soc. Mass Spectrom., 2009, 20, 1496–1503 CrossRef CAS PubMed.
- H. Zhou, J. Xing, S. Liu, F. Song, Z. Cai, Z. Pi, Z. Liu and S. Liu, Phytochem. Anal., 2012, 23, 315–323 CrossRef CAS PubMed.
- Z. M. He, X. H. Wang, G. F. Li, J. M. Sun, H. Yang, Y. G. Gao and L. K. Zhang, Chin. J. Anal. Chem., 2013, 41, 1694–1698 CrossRef CAS.
- X. Zhou, J. Liang, Y. Zhang, H. Zhao, Y. Guo and S. Shi, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2015, 985, 149–154 CrossRef CAS PubMed.
- H. Zhao, Y. Zhang, Y. Guo and S. Shi, J. Pharm. Biomed. Anal., 2015, 104, 31–37 CrossRef CAS PubMed.
- X. X. Yang, F. Xu, D. Wang, Z. W. Yang, H. R. Tan, M. Y. Shang, X. Wang and S. Q. Cai, J. Chromatogr. A, 2015, 1413, 33–46 CrossRef CAS PubMed.
- Z. L. Mou, X. N. Qi, R. L. Liu, J. Zhang and Z. Q. Zhang, J. Chromatogr. A, 2012, 1243, 33–38 CrossRef CAS PubMed.
- S. Zhao, X. Ji, P. Lin and Y. M. Liu, Anal. Biochem., 2011, 411, 88–93 CrossRef CAS PubMed.
- X. W. Ji, F. G. Ye, P. Li and S. L. Zhao, Talanta, 2010, 82, 1170–1174 CrossRef CAS PubMed.
- D. Q. Li, J. Zhao and S. P. Li, J. Chromatogr. A, 2014, 1345, 50–56 CrossRef CAS PubMed.
- Z. Lin, H. Wang, Y. Xu, J. Dong, Y. Hashi and S. Chen, Food Chem., 2012, 134, 1181–1191 CrossRef CAS PubMed.
- C. Debouck and P. N. Goodfellow, Nat. Genet., 1999, 21, 48–50 CrossRef CAS PubMed.
- H. M. Cheng, C. C. Li, C. Y. Chen, H. Y. Lo, W. Y. Cheng, C. H. Lee, S. Z. Yang, S. L. Wu, C. Y. Hsiang and T. Y. Ho, J. Ethnopharmacol., 2010, 132, 429–437 CrossRef PubMed.
- S. Katz, R. Harris, J. T. Y. Lau and A. Chau, J. Evidence-Based Complementary Altern. Med., 2006, 3, 65–70 CrossRef PubMed.
- M. Yan, L. Y. Zhang, L. X. Sun, Z. Z. Jiang and X. H. Xiao, J. Ethnopharmacol., 2006, 107, 308–311 CrossRef CAS PubMed.
- A. Hara, N. lizuka, Y. Hamamoto, S. Uchimura, T. Miyamoto, R. Tsunedomi, K. Miyamoto, S. Hazama, K. Okita and M. Oka, Life Sci., 2005, 77, 991–1002 CrossRef CAS PubMed.
- N. Lizuka, M. Oka, K. Yamamoto, A. Tangoku, K. Miyamoto, T. Miyamoto, S. Uchimura, Y. Hamamoto and K. Okita, Int. J. Cancer, 2003, 107, 666–672 CrossRef PubMed.
- X. J. Xu, Drug Discovery Today: Technol., 2006, 3, 247–253 CrossRef PubMed.
- Y. J. Yang, J. Y. Li, X. W. Liu, J. Y. Zhang, Y. R. Liu and B. Li, PLoS One, 2013, 8, e84458 Search PubMed.
- M. Huang, W. Pang, J. Zhang, S. Lin and J. Hu, J. Pharm. Biomed. Anal., 2012, 58, 12–18 CrossRef CAS PubMed.
- C. S. Zhu, Z. J. Lin, M. L. Xiao, H. J. Niu and B. Zhang, Chin. J. Nat. Med., 2016, 14, 177–184 Search PubMed.
- G. L. Xu, M. Xie, X. Y. Yang, Y. Song, C. Yan, Y. Yang, X. Zhang, Z. Z. Liu, Y. X. Tian, Y. Wang, R. Jiang, W. R. Liu, X. H. Wang and G. M. She, Molecules, 2014, 19, 17897–17925 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2017 |