Open Access Article
Open Access ArticleFabrication of HA/PEI-functionalized carbon dots for tumor targeting, intracellular imaging and gene delivery
M. Zhang†
a,
X. Zhao†a,
Z. Fanga,
Y. Niua,
J. Loua,
Y. Wua,
S. Zoub,
S. Xiaa,
M. Sun*c and
F. Du *ab
*ab
aSchool of Medicine, Jiangsu University, Zhenjiang, 212013, P. R. China. E-mail: xiasheng1519@163.com; biodfy@ujs.edu.cn
bDepartment of Hepatosis, The Third Hospital of Zhenjiang Affiliated to Jiangsu University, Zhenjiang, Jiangsu, P. R. China
cDepartment of Clinical Laboratory, Affiliated Yancheng Hospital, School of Medicine, Southeast University, Yancheng, Jiangsu 224001, P. R. China
First published on 13th January 2017
Abstract
Carbon quantum dots (CDs) as emerging carbon nano-materials have attracted tremendous attention in biomedical fields due to unique properties. In this study, hyaluronate (HA) and polyethylenimine (PEI) functionalized carbon dots (HP-CDs) were synthesized by a facile bottom-up method for tumor targeting and gene delivery. After HA modification, the HP-CDs exhibited superior dispersibility in water and good biocompatibility and were internalized readily into the cytoplasm of cancer via HA-receptor mediated endocytosis. Meanwhile, the HP-CDs with PEI functionalization were shown to have excellent gene condensation capability via electrostatic attraction and protective capacity by preventing nuclease degradation. By virtue of good photoluminescence properties, the HP-CDs as a gene carrier were successfully applied to intracellular imaging and gene delivery. Taken together, the resultant HP-CDs displayed great potential in tumor targeting, intracellular imaging and gene delivery.
1. Introduction
Recently, fluorescent carbon dots (CDs),1,2 a new member of the nano carbon materials, firstly produced by Sun and co-workers3 have drawn extensive attention in the nanotechnology field.1 Similar to metallic quantum dots, CDs can also emit multicolour bright fluorescence by adjusting the particle size.4,5 However, CDs show significant advantages due to their water dispensability, chemical inertness, good photostability, low cytotoxicity, and excellent biocompatibility compared with organic dyes and other semiconductor nanodots with heavy metal elements. The above-mentioned capabilities of CDs endow them a wide range of technology applications in gene/drug delivery,6,7 bio-imaging,8 photocatalysis,9 bio-sensing,10 optoelectronic devices,11 energy conversion/storage devices12 and other bio-applications.13Up to now, the ways methods of synthesizing CDs are classified into two groups, including the top-down and bottom-up methods.1,14 Top-down methods generally have to break down the large carbonaceous materials and polymerize into the nanoparticles by using physical or chemical methods, but the fluorescence of CDs was relatively low. On the contrary, the bottom-up methods involves the carbonization of small molecular precursors, which are carbon-containing materials and passivation agents into the CDs, including hydrothermal carbonization,15,16 micro-wave hydrolysis17 and combustion methods.18 The passivation agents always contain terminal NH2 groups, use of which illustrated that nitrogen-containing groups acting as auxochromes could dramatically improve the photoluminescence of CDs.19 It is more conducive to control the chemical compositions and physical characters by bottom-up compared with top-down methods. Bottom-up methods were therefore mostly adopted for the synthesis of various CDs.
Hyaluronic acid (HA), a natural formation high molecular weight anionic mucopolysaccharide, has the advantages of good biocompatibility, biodegradability, non-immunogenicity and nontoxic, which endow HA grant applications in biomedicine.20,21 Most importantly, HA can bind with targeting ligand CD44, which are overexpressed in many kinds of tumor cells.22–25 In recent years, HA–drug conjugate, HA-amphiphilic derivatives, HA surface modified and HA gene carriers targeting drug systems have been widely studied and applied to active tumor targeting.26–30 Moreover, polyethyleneimine (PEI) was first reported as gene delivery vector by Boussif and his co-workers in 1995,31 which is rated as the fastest growing, the most well researched and the most potential non-viral gene delivery vector at present. For transfection efficiency, the desired PEI molecular weight range is from 11.9 × 103 to 70.0 × 103 and branched 25k PEI had the highest transfection efficiency. The unique proton sponge mechanism of PEI is beneficial for endosomal escape in cells.32–34 In view of their transfection capability, PEI has already been applied for preparation of multifunctional fluorescent CDs using distinct carbonization method.35,36 Liu et al. has already constructed CDs based on PEI, but lacked of targeting modification and hardly been used as tumour-targeting imaging.37 In this study, multifunctional CDS were synthesized by facile bottom-up method for tumour targeted and gene delivery via hydrothermal reaction of HA used as targeting ligand and PEI used as transfection motif.
2. Experimental
2.1. Materials
Branched polyethyleneimine with Mw of 25 kDa (water-free) and cell counting kit-8 (cck-8) were purchased from Sigma-Aldrich (New York, USA). Hyaluronic acid sodium salt (Mw, 200–400 kDa) from Streptococcus equi was obtained from Aladdin (Shanghai, China). RT-PCR kit were purchased from Promega (Madison, WI, USA). Lyso Tracker Red was purchased from Life technology (New York, USA). PCR green master mix was obtained from Thermo (New York, UAS). PCR primer and Plasmid DNA extract kit was obtained from Shanghai Sangon Company (Shanghai, China). Fetal bovine serum was purchased from Excell (Shanghai, China). Dulbecco's Minimum Essential Medium (DMEM) was purchased from Invitrogen China Limited. All chemicals were of analytical grade and were used without further purification.2.2. Synthesis of HP-CDs
0.14 g HA, 0.8 mL commercial branched 25k PEI (1.030 g mL−1) and ddH2O up to 20 mL were mixed under vigorous stirring to form a homogeneous solution in a common 100 mL beaker. Then the transparent solution was transferred into a 50 mL Teflon-lined stainless steel autoclave and heated at 180 °C for 4 h. When cooled down to room temperature, the colour-changed solution was centrifuged at 2000 rpm for 15 min to remove the black precipitates. The supernatant was transferred into a dialysis membrane (MWCO of 1000) and was dialyzed against pure water for 4 days to remove residues. Finally, the brown-yellow solution was collected and freeze-dried using a vacuum freeze dryer at −50 °C for 72 hours. The HP-CDs powder were obtained and saved for further characterization.2.3. Instrumentation and characterizations
The morphologies of the HP-CDs were examined by high resolution transmission electron microscopy (HRTEM) on a JEM-2100 microscope (Jeol, Japan) with an accelerating voltage of 200 kV. UV-Vis absorption was characterized by UV-2450 UV-Vis Spectrophotometer (Shimadzu, Japan). PL emission measurements were performed using Cary Eclipse Fluorometer (Varian, America). The chemical structure of HP-CDs was identified by Fourier Transform Infrared (FT-IR) spectra (Nicolet Nexus 470, America). The crystal structure of the HP-CDs was characterized via X-ray diffraction (XRD) patterns on a Rigaku-D/MAX2500 diffractometer (Rigaku, Japan) equipped with Cu Kα (λ = 0.15405 nm) radiation at a scanning speed of 4° min−1 in the range from 5° to 80°. The surface compositions of HP-CDs were identified by X-ray photoelectron spectroscopy (XPS) on Escalab 250Xi (Thermo Scientific, America).2.4. Cell viability assay
The influence of HP-CDs on cell viability was studied via Cell Counting Kit 8 assay (CCK-8 assay). MGC-803 cells (gastric cancer cells, ATCC number: TCP-1008) were seeded in a 96-well plate at a density of 3 × 103 cells per well and cultured at 37 °C in 5% CO2 incubator for 24 hours, then using DMEM containing different concentrations of HP-CDs (0 μg mL−1, 25 μg mL−1, 50 μg mL−1, 100 μg mL−1 and 200 μg mL−1) replacing the growth medium. After incubation for another 4 hours, adding 10 μL CCK-8 solution to each well, cells were incubated for 4 hours in a dark place. The absorbance was measured at 490 nm using Synergy HT Multi-Mode Microplate Reader (Bio Tek, Winooski, VT, USA). Non-treated cells (in DMEM) were used as control, and the relative cell viability (mean SD, n = 5) was expressed as Abssample/Abscontrol × 100%.2.5. Hemolytic test
Briefly, 19–21 g, ages of 6 weeks female BALB/c mice were kindly prepared by Animal Management Rules of the Ministry of Health of the People's Republic of China, then taking about 3 mL blood by removing eyeball, stabilized with heparin sodium, centrifuged (1, 200 rpm, 15 minutes) to remove the supernatant according to the literature,40 washed with PBS for five times to obtain the mouse red blood cells (MRBCs). Diluting ten times with PBS, 0.1 mL MRBCs were transferred into 1.5 mL tubes prefilled with 0.9 mL PBS containing different particle concentrations (50–400 μg mL−1) of HP-CDs, 0.9 mL ddH2O (as positive control), and 0.9 mL PBS (as negative control), respectively. The mixture was incubated for 2 hours at room temperature after a gentle shaking, then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 minute. Finally, the photographs of all the samples were taken and the absorbance of the supernatant (hemoglobin) was measured by a UV-2450 UV-Vis spectrophotometer. The hemolysis percentages of different samples were calculated by dividing the difference in absorbance among the sample, the positive and negative control at 541 nm.
000 rpm for 1 minute. Finally, the photographs of all the samples were taken and the absorbance of the supernatant (hemoglobin) was measured by a UV-2450 UV-Vis spectrophotometer. The hemolysis percentages of different samples were calculated by dividing the difference in absorbance among the sample, the positive and negative control at 541 nm.
2.6. Cell imaging and labeling
Hela cells (Henrietta Lacks strain of cancer cells, ATCC number: CCL-2) were chosen to demonstrate the feasibility of HP-CDs for cell imaging. Hela cells were seeded at a density of 0.8 × 105 per well in a 24-well plate which has already covered 14 mm diameter glass coverslips. The cells were cultured in DMEM containing 10% FBS and incubated at 37 °C and 5% CO2 overnight. The original supernate was discard and HP-CDs mixed with culture medium at 100 μg mL−1 were join in the holes. Cells were incubated at 37 °C under 5% CO2 for 6 h. After that, the supernate was discard and cells were incubated with pre-diluted Lyso Tracker (60 nM) for 90 minutes. Three times after washing, cells were fixed by 4% paraformaldehyde for 30 minutes at room temperature. After washing with PBS for 3 times, pre-diluted Hoechst 33342 (5 μg mL−1) was added to each hole for half an hour at room temperature to staining the nucleus. Three times after washing, the coverslips was taken out and mounted by resin. Then confocal lasers scanning fluorescence microscopy (Zeiss LSM-710) was used to observe the fluorescence in the cells with 496 nm laser to excite the HP-CDs.2.7. Measurement of CD44 expressed by PCR
MGC-803 cells were seeded in six-well plates at 4 × 105 per well and incubated at 37 °C and 5% CO2 for 24 h. Then 0 μg mL−1, 100 μg mL−1, 200 μg mL−1, 300 μg mL−1 and 400 μg mL−1 HP-CDs solution were added into each well respectively. Total RNA of the cells in each well were extracted after incubated for 6 h. Briefly, 0.1 μg of the total RNA of each well was first reverse transcribed into cDNA using the reverse transcription system. Subsequently, 2 μL of the resultant cDNA was amplified by Thermo® Green Master Mix. PCR was performed in PTC-200 Peltier thermal cycler with the following program: an initial denaturation at 94 °C for 3 min followed by 30 cycles (for CD44 gene and GAPDH gene) at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s. After the last cycle, the program was ended by a final extension at 72 °C for 10 min. The sequences of the PCR primers used as follows:CD44 forward primer: 5′-CTACAGCATCTCTCGGAC-3′
CD44 reverse primer: 5′-GTCTCAAATCCGATGCT C-3′;
GAPDH forward primer: 5′-TGGGTGTGAACCATGAGAAG-3′
GAPDH reverse primer: 5′-GAGTCCTTCCACGATACCAAAG-3′.
The electrophoretic showed the difference of each PCR product by 3% agarose gel, which run in a 40 mM TAE and stained using ethidium bromide (EB, 0.5 mg mL−1).
2.8. Flow cytometry
Hela cells were seeded in three wells of six-well plates at 3 × 104 per well and incubated at 37 °C and 5% CO2 for 24 hours. In the three wells, the first well was negative control, the second well was HA participating group, the third well was HP-CDs group. HA dilution solution (2 mL, 0.1 g HA dissolved in 10 mL DMEM with 10% FBS) was added in the second well. After 1 h later, the HA dilution solution was discard and 300 μg mL−1 HP-CDs solution was added to the second and third well. All of three wells of cells were digested for flow cytometry after 6 h incubation to detecting the change of fluorescence intensity.2.9. Gel retardation assay
The gene condensation capability of HP-CDs was evaluated by agarose gel electrophoresis. Plasmid at concentration of 640 ng μL−1 was chosen as the DNA carried by as-prepared HP-CDs. HP-CDs solution was pre-diluted at concentration of 400 ng μL−1. The DNA and HP-CDs were complexes at different weight ratios from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 to 1
0 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 for half an hour while the weight of DNA is invariably 640 ng. However with the increasing weight of HP-CDs, the volume of HP-CDs solution is also increased from 0, 1.6, 3.2, 4.8, 6.3, 7.9, 9.5 μL. Then the DNA/HP-CDs mixture was added to each well loaded with loading buffer in 1% agarose gel. After gel electrophoresis for 30 minutes in a 40 mM Tris–acetate–EDTA buffer, pH 8.0 (TAE), the HP-CDs bands were visualized under a UV transilluminator at a wavelength of 365 nm while DNA bands were visualized under the same condition after immersing in EB at about 10 minutes.
6 for half an hour while the weight of DNA is invariably 640 ng. However with the increasing weight of HP-CDs, the volume of HP-CDs solution is also increased from 0, 1.6, 3.2, 4.8, 6.3, 7.9, 9.5 μL. Then the DNA/HP-CDs mixture was added to each well loaded with loading buffer in 1% agarose gel. After gel electrophoresis for 30 minutes in a 40 mM Tris–acetate–EDTA buffer, pH 8.0 (TAE), the HP-CDs bands were visualized under a UV transilluminator at a wavelength of 365 nm while DNA bands were visualized under the same condition after immersing in EB at about 10 minutes.
2.10. Nucleic acid protective assay
5 μL pre-diluted HP-CDs solution (2265 ng μL−1), 5 μL RNA (3963 ng μL−1) and 5 μL RNaseA (10 mg mL−1) were mixed (HP-CDs–RNA–RNaseA) respectively as positive control for four groups, while 5 μL ddH2O (double distilled water), 5 μL RNA (3963 ng μL−1) and 5 μL RNaseA (10 mg mL−1) were complexed (H2O–RNA–RNaseA) as negative control as well as for four groups. Each group of HP-CDs–RNA–RnaseA and H2O–RNA–RnaseA was incubated successively at 37 °C in 60 min, 40 min, 20 min, 0 min. The trends were analyzed by electrophoresis for 20 minutes through a 2% agarose gel. The gel was run in a 40 mM TAE and trends were stained using EB.2.11. Transfection experiment
HEK-293T cells (human embryonic kidney 293T cells, ATCC number: CRL-11268) were used to identify transfection capability of HP-CDs. Plasmid PLV (red fluorescent protein) was chosen as reporter gene. HEK-293T cells were seeded at 1 × 105 per well in a 24-well plate which has already covered 14 mm diameter glass coverslips and incubated at 37 °C and 5% CO2 overnight. 20 μL pre-diluted HP-CDs (100 μg mL−1) were mixed with 2000 ng PLV and hold for 25 min at room temperature. After that, the complex was added in the hole blending with 2 mL culture medium. 24 h later, discarding the supernate and washing with PBS for 3 times. Then the cells were fixed with 1 mL 4% paraformaldehyde for 30 minutes at room temperature. After washing three times, Hoechst 33342 (5 μg mL−1) was used to stain nucleus for 30 min. After washing with PBS, the coverslips was taken out and mounted by resin. Then confocal lasers scanning fluorescence microscopy was used to observe the transfection efficiency of HP-CDs.3. Results and discussion
3.1. The synthesis of HP-CDs
The synthetic process of HP-CDs was given in Scheme 1. The mixture of HA and PEI experienced the process of dehydration, polymerization, aromatization and aromatization by hydrothermal process of 180 °C for 4 h. Finally, light yellow solution was dialyzed against distilled water to obtain highly fluorescent HP-CDs. In this experiment, a certain amount of PEI in the raw solution was necessary to enhance the HP-CDs formation which was critical for carbonization degree and carried positive charge combined with nucleic acid. HA endowed HP-CDs with target-specific capability via interaction with CD44 molecules, which was the receptor of HA overexpressing in most tumour cells.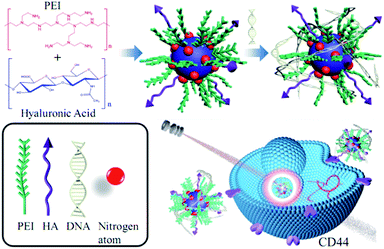 | ||
| Scheme 1 Schematic synthesis of HP-CDs from hydrothermal carbonization method and following biomedical application. | ||
The formation process of carbon dots by carbonization between carboxyl and amino groups is extremely complicated, and a clear mechanism is still unknown. Based on previously reports37,39 and our research,38 carbon dots may undergo dehydration, polymerization, aromatization and carbonization via hydrothermal reaction. These findings verified that the function of these molecules on the surface of resultant PEI-based carbon dots was remained. In this study, dehydration and polymerization between amino groups of PEI and carboxyl groups of hyaluronic acid may form special construction structure similar to amphiphilic polymer structure, while the function of each other was not affected.
3.2. The morphology and characterization of HP-CDs
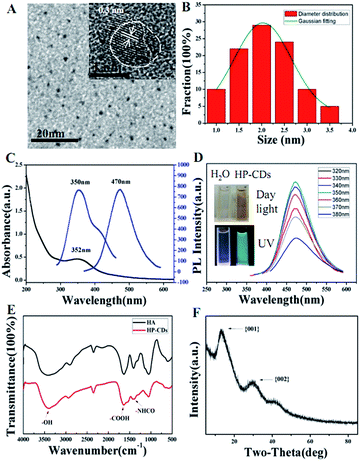
The morphology was identified by high-resolution transmission electron microscopy (HRTEM). As shown in Fig. 1A and B, HP-CDs exhibited uniform dispersion and discrete quasi-spherical shape without apparent aggregation. HP-CDs have a well-defined structure of about 2.25 nm diameter, and such ultra-small size could be conducive to cross the physiological barrier and passive target to tumour tissue. The HRTEM (Fig. 1 inset) showed that HP-CDs were mainly composed of a carbon crystalline core with a lattice spacing of approximately 0.3 nm, which reflected of the {002} facet of graphite. In addition, a small amount of amorphous structure appeared at the particle edge locations. The results of TEM showed the HP-CDs possessed heterogeneous crystal phase structures.The optical properties of HP-CDs were characterized by UV/Vis spectrum (Fig. 1C) and photoluminescence (PL) spectrum (Fig. 1D). The HP-CDs aqueous solution exhibited a long-term homogeneous phase without any noticeable precipitation under ambient conditions at room temperature. The absorption spectrum (Fig. 1C) showed that just an absorption peak was observed at 352 nm, which may give rise to a series of emission traps between n and π* states of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C. The HP-CDs aqueous solution was pale yellow and transparent in daylight but changed to bright blue fluorescence under UV excitation with quantum yield of 12.4%, which could be easily observed with naked eyes (Fig. 1D, inset). As shown in Fig. 1D, the HP-CDs displayed broad range of emission wavelengths and the strongest emission peak shifts from 450 nm to 500 nm and the intensity decrease gradually as the excitation from 360 nm to 380 nm. The strongest fluorescence emission was observed centered at approximately 470 nm under 350 nm excitation wavelength. This phenomenon has been widely observed in the luminescent carbon nanomaterials, which may be due to the optical selection of different surface defect states near the Fermi level of CDs.1 FT-IR spectra results indicated that there were a large number of the active groups on the surface of HP-CDs (Fig. 1E), such as the hydroxyl, carboxyl and acylamino groups, which endow HP-CDs with superior hydrophility and dispersibility in water. The crystal structure of HP-CDs was characterized by X-ray diffraction (XRD). As shown in Fig. 1F, the two broad main peaks centered at X = 13.42 (d = 0.74 nm) and X = 29.45 (d = 0.3 nm) indicated the amorphous nature of phase structure, which similar to the characteristic lattice spacing of graphene {002 plane} and graphene oxide {001 plane} respectively.
C. The HP-CDs aqueous solution was pale yellow and transparent in daylight but changed to bright blue fluorescence under UV excitation with quantum yield of 12.4%, which could be easily observed with naked eyes (Fig. 1D, inset). As shown in Fig. 1D, the HP-CDs displayed broad range of emission wavelengths and the strongest emission peak shifts from 450 nm to 500 nm and the intensity decrease gradually as the excitation from 360 nm to 380 nm. The strongest fluorescence emission was observed centered at approximately 470 nm under 350 nm excitation wavelength. This phenomenon has been widely observed in the luminescent carbon nanomaterials, which may be due to the optical selection of different surface defect states near the Fermi level of CDs.1 FT-IR spectra results indicated that there were a large number of the active groups on the surface of HP-CDs (Fig. 1E), such as the hydroxyl, carboxyl and acylamino groups, which endow HP-CDs with superior hydrophility and dispersibility in water. The crystal structure of HP-CDs was characterized by X-ray diffraction (XRD). As shown in Fig. 1F, the two broad main peaks centered at X = 13.42 (d = 0.74 nm) and X = 29.45 (d = 0.3 nm) indicated the amorphous nature of phase structure, which similar to the characteristic lattice spacing of graphene {002 plane} and graphene oxide {001 plane} respectively.
3.3. The surface chemical structure of HP-CDs
The surface chemical structure of the HP-CDs was characterized by X-ray photoelectron spectroscopy (XPS). As shown in Fig. 2A, there were three peaks separately at 285.0 eV, 400.0 eV and 530.0 eV, which illustrated that our as-prepared HP-CDs were truly composed of three elements including carbon, nitrogen and oxygen. In the high-resolution spectrum of C1s (Fig. 2B), the three main peak at 285.0 and 288.0 eV represented C–C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N/C
N/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O respectively. The binding energy peak at 285.0 eV demonstrated the HP-CDs had the structure of graphite-like, which was consistent with the results of XRD. As shown in Fig. 2C, the spectra of O1s showed two conspicuous peak at 530.8 eV (C
O respectively. The binding energy peak at 285.0 eV demonstrated the HP-CDs had the structure of graphite-like, which was consistent with the results of XRD. As shown in Fig. 2C, the spectra of O1s showed two conspicuous peak at 530.8 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 532.2 eV (C–O), which was in agreement with the spectrum of C1s. The remarkable N–H peaks at 399.5 eV and N–C peak at 400.6 eV of N1s (Fig. 2D) were corroborate the FTIR spectra of HP-CDs, which also confirmed the successful incorporation of N atoms into the HP-CDs by the hydrothermal treatment.
O) and 532.2 eV (C–O), which was in agreement with the spectrum of C1s. The remarkable N–H peaks at 399.5 eV and N–C peak at 400.6 eV of N1s (Fig. 2D) were corroborate the FTIR spectra of HP-CDs, which also confirmed the successful incorporation of N atoms into the HP-CDs by the hydrothermal treatment.
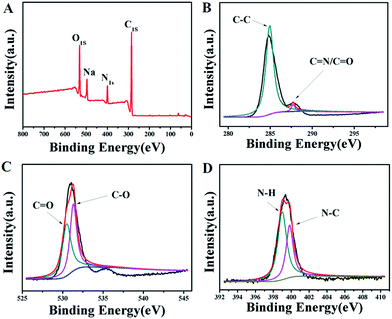 | ||
| Fig. 2 XPS spectra of the HP-CDs. (A) Survey spectrum. (B) C1s spectrum. (C) O1s spectrum. (D) N1s spectrum. | ||
3.4. Biomedical applications of HP-CDs
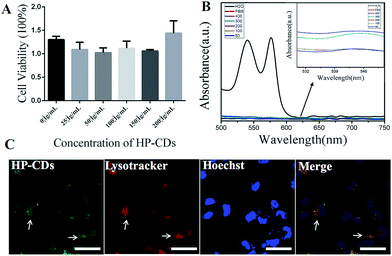
The biocompatibility of HP-CDs, as potential biomedical agents, is critical for their biomedical application. The cytotoxicity of HP-CDs was investigated using CCK-8 in Hela cells. Compared with control group, no obvious toxicity was revealed with the increase of HP-CDs concentration from 50 μg mL−1 to 200 μg mL−1 (Fig. 3A). Meanwhile, PBS treatment showed no influence on blood cells in Fig. 3B, but ddH2O treatment gave rise to severe hemolysis phenomenon, which caused obvious increase of absorbance value at 541 and 575 nm. On the contrary, each group absorbance of HP-CDs aqueous solution from different concentration with blood cells showed no obvious change by absorbance value compared with ddH2O treatment group. The insert images in Fig. 3B showed that HP-CDs resulted in no solution colour change caused by hemolysis after centrifugation. These results clearly suggested that the as-synthesized HP-CDs had very low cytotoxicity and good hemocompatibility.3.5. The internalization and tumour cell imaging of HP-CDs
The potential application of the as-prepared HP-CDs was evaluated by laser scanning confocal microscopy. After incubation with HP-CDs for 6 h, the Hela cells were washed to remove extracellular HP-CDs and incubated with lysotracker for another 90 min. It was found that the labelled cells were brightly illuminated due to strong fluorescence emitting from HP-CDs which distributed in cytosol (Fig. 3C). Consistent with optical results, the HP-CDs exhibited green fluorescence upon excitation 496 nm. It was noted that the HP-CDs were clearly observed in cytoplasm regions, especially exhibited a co-location in the lysosomes with red fluorescence. In view of this result, we supposed that large amount of HP-CDs could be readily internalized into the cells across the cell membrane and further transferred to the lysosome. It might be explained by the fact that HP-CDs uptaken by cells could be internalized into the lysosomes via HA receptor-mediated endocytosis. These results gave us the conclusion that HP-CDs readily entered into the cytoplasm, which agreed well with the results of previous investigation on the interaction of living cells with other CDs.19,41,42 Thus, it can be seen that HP-CDs can be effectively applied in tumour cell imaging and biological labelling.3.6. The endocytosis of HP-CDs mediated by HA receptors
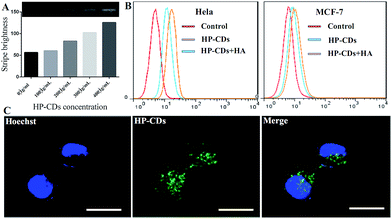
In order to verify the HA receptor-mediated endocytosis, competitive inhibition test were employed via adding free HA molecules between HA-receptor high expression and low expression tumour cell lines. Natural HA macromolecule, a kind of main ingredient of extracellular matrix (ECM) and intercellular substance, could specific bind to CD44 which is a surface molecule over-expressed in cancer cells. Firstly, PCR assay was conducted to verify whether the HA functionalized HP-CDs could induce CD44 gene expression via interact with CD44 molecules. As shown in Fig. 4A, the increased CD44 signal intensity with the increased HP-CDs concentration demonstrated as-prepared HP-CDs could induce the gene expression of CD44 at RNA level.Secondly, to prove the tumor-specific uptake behaviour of HP-CDs by the HA receptor-mediated endocytosis, Hela cells with CD44-high as positive group expression and MCF-7 cells with CD44-low expression as negative group were chosen as the model cell lines. From flow cytometry analysis (Fig. 4B), it was found that Hela cells effectively took up a large number of HP-CDs, but the endocytosis behaviour was markedly inhibited after pre-treated free HA group. That may be because free HA molecules competitive bounded with CD44 and blocked the HP-CDs binding site, resulting in lower intake. However, the amount of HP-CDs by MCF-7 cells took up was lower and not significantly changed between free HA pre-treatment and not, which suggested that this competitive inhibition was disappeared. According to the confocal microscopy images (Fig. 4C), it was further confirmed that HP-CDs could really distribute in cytosols of Hela cells. All mentioned-above suggested that HP-CDs could be effectively taken up by the HA receptor-mediated endocytosis and serve as cell-specific targeting probe for imaging and labeling of CD44-high expression tumor cells.
3.7. Nucleic acid condensation and transfections of HP-CDs
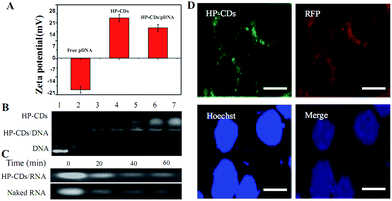
As depicted in Scheme 1, we speculated that the PEI functionalized HP-CDs may be capable of condensing nucleic acid molecules by electrostatic interactions. Thus, the surface charge of HP-CDs/pDNA complexes was measured to determine the ability of cationic vectors to condense DNA. The zeta potential data revealed (in Fig. 5A) that of HP-CDs displayed highest potential, free DNA showed lowest potential. However, the potential was obviously neutralized but still a positive charge, when HP-CDs bounded with pDNA complexes. Next, gel retardation assay was further used to illustrate whether HP-CDs possessed the capacity of condensing DNA (Fig. 5B). The HP-CDs were mixed with DNA at different weight ratios (DNA/HP-CDs) to form DNA/HP-CDs complexes. DNA with the negative charge run toward positive pole but HP-CDs with positive charge run toward negative pole in the electrophoresis process. As can be seen from Fig. 5B, with the increasing weight of HP-CDs, they showed increased fluorescence while the intensity of DNA migration band decreased significantly and finally disappeared at the DNA/HP-CDs weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. This result confirmed that HP-CDs indeed have the capability of condensing and loading nucleic acid substances.
3. This result confirmed that HP-CDs indeed have the capability of condensing and loading nucleic acid substances.
We further investigated whether HP-CDs could prevent nucleic acid from the RnaseA degradation. 5 μL pre-diluted HP-CDs solution (2265 ng μL−1) mixed with 5 μL RNA (3963 ng μL−1) and 5 μL RnaseA (10 μg mL−1) were prepared as the experimental group, while 5 mL ddH2O mixed with above-mentioned 5 μL RNA (3963 ng μL−1) and 5 μL RnaseA (10 μg mL−1) were prepared as control group. The four samples of each group were respectively incubated in 37 °C from 0 to 60 min. As shown in Fig. 5C, with the incubation time prolonging, the degradation of RNA were relieved by the protection of HP-CDs compared with the control group especially until 60 min. These data demonstrated that HP-CDs showed favourable nucleic acid protective function.
To illustrate the transfection efficiency of HP-CDs, gene transfection experiment in HEK-293T cells were performed by using PLV which enhancing red fluorescence as the reporter gene in order to differentiate with green fluorescence of HP-CDs. As can be seen from Fig. 5D, the nucleus was dyed blue with Hoechst 33342. Intrinsic green fluorescence endowed HP-CDs with advantageous real-time tracking as living cells probe. The present of green fluorescence showed the HP-CDs have been readily internalized into cytoplasm, which may be regulated by HA receptor-mediated endocytosis. Bright red fluorescence of PLV could be observed from the cytoplasm, which indicated the successful delivery and expression of PLV. These images indicated that the HP-CDs not only served as fluorescence probe could real-time monitor the internalization by receptor-mediated endocytosis, but also as efficient gene vector could carry exogenous gene into the cell. More importantly, these transfected genes could escape from lysosome and effectively express to produce gene product, which may be caused by the protonation of polycation on the surface of HP-CDs.
4. Conclusions
In summary, we synthesized a kind of novel multifunctional HP-CDs through a facile and simple bottom-up route. The as-prepared HP-CDs possessed stable photoluminescence, superior disperbility in water and surface biocompatibility. The application of HA modification in HP-CDs interacting with CD44 was applied to targeting to tumour cells. Furthermore, in this efficient nano-carrier system, PEI played two crucial roles: endow the HP-CDs with intense photoluminescence by surface passivation and DNA condensation for gene transfection. Furthermore, we further demonstrated that HP-CDs with good biocompatibility were easily and fast internalized by cancer cells and particularly suitable for real-time cellular imaging and labelling.Acknowledgements
The research was supported by the National Natural Science Foundation of China (No. 81301316, 31200676), China Postdoctoral Science Foundation (2013M540425, 2014T70487, 2015M571705), Health and family planning commission of Jiangsu province scientific research subject (H201557), Natural Science Foundation of Jiangsu Province (BK20161317), Senior Talents Scientific Research Foundation of Jiangsu University (No. 13JDG022, 11JDG113).Notes and references
- S. N. Baker and G. A. Baker, Angew. Chem., Int. Ed. Engl., 2010, 49, 6726–6744 CrossRef CAS PubMed.
- A. L. Himaja, P. S. Karthik and S. P. Sigh, Chem. Rec., 2015, 15, 595–615 CrossRef CAS PubMed.
- Y. P. Sun, B. Zhou, W. Wang, K. A. Shiral Fenando, P. Pathak and M. J. Meziani, J. Am. Chem. Soc., 2006, 128, 7756–7757 CrossRef CAS PubMed.
- D. Chen, N. A. Monteiro-Riviere and L. W. Zhang, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2016 DOI:10.1002/wnan.1419.
- F. Zhang, X. Feng, Y. Zhang, L. Yan, Y. Yang and X. Liu, Nanoscale, 2016, 8, 8618–8632 RSC.
- Q. L. Wang, X. X. Huang, Y. J. Long, X. L. Wang, H. J. Zhang and R. Zhou, Carbon, 2013, 59, 192–199 CrossRef CAS.
- E. Keles, Y. Song, D. Du, W. J. Dong and Y. H. Lin, Biomater. Sci., 2016, 4, 1291–1309 RSC.
- A. W. Zhu, Q. Qu, X. L. Shao, B. Kong and Y. Tian, Angew. Chem., Int. Ed. Engl., 2012, 51, 7185–7189 CrossRef CAS PubMed.
- H. T. Li, X. D. He, Z. H. Kang, H. Huang, Y. Lin and J. L. Liu, Angew. Chem., Int. Ed. Engl., 2010, 49, 4430–4434 CrossRef CAS PubMed.
- M. Long, A. N. Hoang, J. Chung, N. Gural, C. B. Ford and C. Min, Nat. Commun., 2013, 4, 216–219 Search PubMed.
- D. Wu, Y. Jiang, S. Y. Li, F. Z. Li, J. W. Li and X. Z. Lan, Nanotechnology, 2011, 22, 3785–3793 Search PubMed.
- J. J. Huang, Z. F. Zhong, M. Z. Rong, X. Zhou, X. D. Chen and M. Q. Zhang, Carbon, 2014, 70, 190–198 CrossRef CAS.
- S. Patra, E. Roy, R. Madhuri and P. K. Sharma, Biomater. Sci., 2016, 4, 418–429 RSC.
- Y. Yang, J. C. Zhang, J. Zhuang and X. Wang, Nanoscale, 2015, 7, 12284–12290 RSC.
- Q. H. Liang, W. J. Ma, Y. Shi, Z. Li and X. M. Yang, Carbon, 2013, 60, 421–428 CrossRef CAS.
- C. Z. Zhu, J. F. Zhai and S. J. Dong, Chem. Commun., 2012, 48, 9367–9369 RSC.
- S. N. Qu, X. Y. Wang, Q. P. Lu, X. Y. Liu and L. Y. Wang, Angew. Chem., Int. Ed. Engl., 2012, 51, 12215–12218 CrossRef CAS PubMed.
- R. Zhang, Y. B. Liu, L. B. Yu, Z. Li and S. Q. Sun, Nanotechnology, 2013, 24, 225601 CrossRef PubMed.
- F. Y. Du, J. Yuan, M. M. Zhang, J. N. Li, Z. Zhou and Z. Li, RSC Adv., 2014, 4, 37536–37541 RSC.
- W. C. Yen and W. W. Lamph, Prostate, 2006, 66, 305–316 CrossRef CAS PubMed.
- A. K. Yadav, P. Mishra, S. Jain, P. Mishra, A. K. Mishra and G. P. Agrawal, J. Drug Targeting, 2008, 16, 464–478 CrossRef CAS PubMed.
- S. Ganesh, A. K. lyer, F. Gattacceca, D. V. Morrissey and M. M. Amiji, J. Controlled Release, 2013, 172, 699–706 CrossRef CAS PubMed.
- K. K. Vangara, J. L. Liu and S. Palakurthi, Anticancer Res., 2013, 33, 2425–2434 CAS.
- K. Cohen, R. Emmanuel, E. Kisinfinfer, D. Shabat and D. Peer, ACS Nano, 2014, 8, 2183–2195 CrossRef CAS PubMed.
- S. C. Ghosh, A. S. Nesihan and J. Klostergaard, Expert Opin. Ther. Targets, 2012, 16, 635–650 CrossRef CAS PubMed.
- C. E. Galer, D. Sano, S. C. Ghosh, J. H. Hah, E. Auzenne and A. N. Hamir, Oral Oncol., 2011, 47, 1039–1047 CrossRef CAS PubMed.
- H. J. Cho, H. Y. Yoon, H. Koo, S. H. Ko, J. S. Shim and J. H. Lee, Biomaterials, 2011, 32, 7187–7190 Search PubMed.
- R. E. Eliaz and F. C. Szoka Jr, Cancer Res., 2001, 61, 2592–2601 CAS.
- W. Hyung, H. Ko, J. Park, E. Lim, S. B. Park and Y. J. Park, Biotechnol. Bioeng., 2008, 99, 442–454 CrossRef CAS PubMed.
- J. Yao, Y. Fan, R. H. Du, J. P. Zhou, Y. Lu and W. Wang, Biomaterials, 2010, 31, 9357–9365 CrossRef CAS PubMed.
- B. O. Boussif, F. Lezoualc’H and M. A. Zanta, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 7297–7301 CrossRef.
- A. E. Nel, L. Madler, D. Velegol, X. Tian, E. M. V. Hoek and P. Somasundaran, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- R. Z. Ur, D. Hoekstra and I. S. Zuhorn, ACS Nano, 2013, 7, 3767–3777 CrossRef PubMed.
- M. Breunig, C. Hozsa, U. Lungwitz, K. Watanabe, I. Umeda and H. Kato, J. Controlled Release, 2008, 130, 57–63 CrossRef CAS PubMed.
- L. Hu, Y. Sun, S. Li, X. Wang, K. Hu, L. Wang, X. Liang and Y. Wu, Carbon, 2014, 67, 508–513 CrossRef CAS.
- Q. Wang, C. Zhang, G. Shen, H. Liu, H. Fu and D. Cui, J. Nanobiotechnol., 2014, 12, 58 CrossRef PubMed.
- C. Liu, P. Zhang, X. Zhai, F. Tian, W. Li, J. Yang, Y. Liu, H. Wang, W. Wang and W. Liu, Biomaterials, 2012, 33, 3604–3613 CrossRef CAS PubMed.
- S. K. Bhunia, A. Saha, A. R. Maity, S. C. Ray and N. R. Jana, Sci. Rep., 2013, 3, 1473 Search PubMed.
- L. Hu, Y. Sun, S. Li, X. Wang, K. Hu, L. Wang, X. Liang and Y. Wu, Carbon, 2014, 67, 508–513 CrossRef CAS.
- F. Du, J. Yuan, M. Zhang, J. Li, Z. Zhou and Z. Li, RSC Adv, 2014, 4, 37536–37541 RSC.
- F. Du, X. Jin, J. Chen, Y. Hua, M. L. Gao and L. Zhang, J. Nano Res., 2014, 16, 1–10 CAS.
- F. Du, J. Li, Y. Hua, M. Zhang, Z. Zhou and Y. Jing, J. Biomed. Nanotechnol., 2015, 11, 780–789 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |