Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
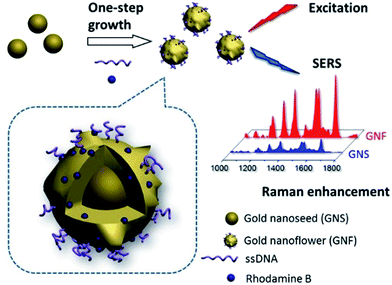
One-pot synthesis of a DNA-anchored SERS nanoprobe with simultaneous nanostructural tuning and Raman reporter encoding†
Yun Li‡
a,
Xiangjiang Liu‡ b,
Di Jianga,
Zhongzhi Yua,
Dalin Tiana,
Chang Lub,
Mingyu Lia,
Jianjun Hea and
Longhua Tang
b,
Di Jianga,
Zhongzhi Yua,
Dalin Tiana,
Chang Lub,
Mingyu Lia,
Jianjun Hea and
Longhua Tang *a
*a
aState Key Laboratory of Modern Optical Instrumentation, College of Opticall Science and Engineering, Zhejiang University, Hangzhou 310027, China. E-mail: lhtang@zju.edu.cn
bCollege of Biosystems Engineering and Food Science, Zhejiang University, Hangzhou 310058, China
First published on 17th January 2017
Abstract
We report a new simple synthesis method for a DNA-anchored SERS nanoprobe encoded with a Raman reporter by a one-pot overgrowth process, allowing the nanostructure to be controllably tuned to provide improved Raman enhancement, and simultaneously introduce Raman reporters and DNA onto nanoparticles whilst retaining their SERS activity and bio-recognition ability.
Surface-enhanced Raman scattering (SERS), as a vibrational spectroscopic technique, holds great potential for molecular diagnostics and biomedical imaging, which under optimal conditions enables highly sensitive detection and high-resolution imaging.1–8 The SERS technique has several advantageous properties, such as having efficient single photoexcitation, narrow bandwidth of <1 nm, high signal-to-noise ratio, and non-photobleaching features.9 Despite its emerging applications in sensing and bioimaging, SERS has still fallen behind other optical methods, such as fluorescence spectroscopy with organic dyes or quantum dots.10–12 The most critical reason for the lag is that the direct synthesis of robust SERS nanoprobes (also called as “SERS tags”) with bright, stable SERS signals and biofunctionality remains highly challenging.13 Ideally, SERS probes intended for analytical use should (i) provide large, uniform and stable SERS signal, (ii) be amenable for introducing Raman reporter molecules as SERS-active tags and (iii) be facilely biofunctionalized with specific molecular recognition units (e.g., antibody, functional DNA for targeting specific biomamkers).14 Thereafter, to design the robust SERS nanoprobes with combinatorial capabilities is a worthy goal to pursue for effective detection and bioimaging applications.
Typically, most of the SERS nanoprobe designs reported to now involve single plasmonic nanostrucutre uniformly fixed with Raman reporters. For instance, optically tunable SERS substrates (e.g., nanostars, nanorods and core–shell nanostrucutres, etc.) have been developed to create SERS-active hot spots and then to directly adhere Raman molecules onto the surface.15,16 However, this approach might offer enhanced SERS signal, whereas suffer from poor stability due to desorption or degradation of the Raman reporter. Coating with silica could improve stability and unfortunately result in significant weakening on SERS activity. Aiming to tackle these problems, another type of SERS nanotag was recently proposed by using interior nanogaps in core–shell nanostructures to embed Raman moloecules.17–20 The nanogap type-SERS nanoprobes can provide large uniform Raman enhancement and keep the stability of Raman reporters,21 however, they still lack the biocompatibility and biofunctionality, which need further functionalization of bioactive probes on gold shells even using poly-adenine (polyA) to mediate the synthesis probe as Fan's group reported.22 Therefore, current critical techniques still remain challenging in simultaneously forming high SERS-active nanostructure, encoding Raman tags, as well as functionalizing with bioactive molecules.
According to our previous studies,23–25 DNA can be used as the shape-controlling agent during growth process of nanoparticles, thereby influencing both the structure and function of nanomaterials. In the present work, we demonstrated that the combination of DNA and typical Raman dye (Rhodamine B, RhB) can synergistically fine tune the gold nanoparticles (GNPs) into a uniform gold flower-like nanoparticles (defined as gold nanoflowers, GNFs) in a one-pot synthesis (Scheme 1). Moreover, the RhB molecule were found to be efficiently anchored onto the as-prepared GNFs, resulting in a stable and enhanced Raman signature. More importantly, the partial DNAs can be stably embedded to the GNFs and keep their bio-recognition ability.
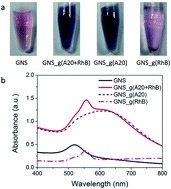
To synthesize these SERS nanoprobes, seed-mediated synthesis methods were used according to our previous reports with modification.7 Typically, DNA with a homooligomer of 20 adenine deoxyribonucleotides (called as A20, 1 μM) and model organic dye, RhB (10 μM) were firstly sequentially incubated with purified gold nanosphere seeds (GNSs, 0.5 a.u.) in a total volume of 300 μL at room temperature. After incubation for 2 hours, a mild reducing agent, hydroxylamine (NH2OH, 400 mM), and a gold precursor, hydrogen tetrachloroaurate(III) (HAuCl4, 1%), were added to initiate particle growth with fiercely shakening. These procedures resulted in different color-changed solution and UV-vis absorbance spectra (Fig. 1). In the absence of A20, the procedure led to visible precipitate at the bottom of the tube, as shown the lighter color of solution than original GNS and the appearance of a weak and blue-shifted localized surface plasmon resonance (LSPR) peak at 560 nm, which further illustrated the formation of unstable large clusters with irregular shapes (Fig. 1).
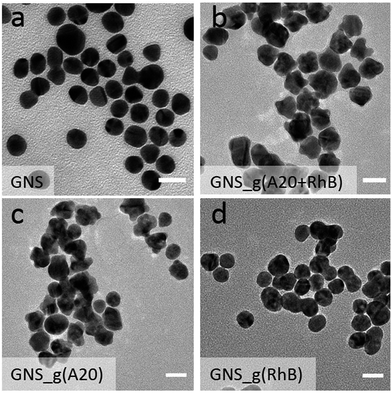
In contrast, the presence of only A20 or the mixture of A20 and RhB transformed the solution from light pink to purple, because the LSPR peak was broaden and red-shifted to 630 nm with a significantly increased absorbance intensity (Fig. 1). The latter case of resulted solution also show the appearance of the featured peak of RhB at 560 nm even with the thorough removing of the free RhB molecules by centrifugation, which will be dictated more details later. Transmission electron microscopy (TEM) image showed the formation of uniformly flower-like nanoparticles, under the incubation of only A20 (Fig. 1b), which coincided with our previous reports. Whereas, in the presence of A20 and RhB under the concentration of 1 μM and 10 μM respectively, it also showed surface-roughened and flower-like nanostructures, with much rougher surface. On the other hand, if in the absence of A20 or RhB, GNS grew into microsized gold agglomerations and had irregular shapes. These results strongly suggest that the sequential addition of A20 and RhB played a significant role in controlling the morphologies of the nanoparticles during synthesis (Fig. 2).
Along with the above observation, the combination of RhB and A20 could resulted in the relatively rough flower-like nanoparticles. Thereafter, a critical question is whether the RhB also involved in controlling the morphological evolution of GNSs or not. To understand the role of RhB during the nanoparticle overgrowth, we fixed the A20 concentration and then varied the concentration of the RhB ranging from 1 μM to 100 μM. Noted the resulted nanoparticles solution showed an increasing pink color (Fig. S1a†), which is from the intrinsic color of RhB. Whereas, after removing the free RhB, the color of resulted nanoparticle solutions were close to the case of GNS_g(A20) (Fig. S1b†), indicating their rough and flower-like nanostructures. As shown by TEM imaging (Fig. S2a†), when the RhB was relatively low (10 μM), the nanoflower shape did not change much from those grew with merely A20. At very high RB concentration (100 μM), the GNSs were overgrown and lost their regularity in shape, resulting in stick-like nanostars rougher and branching structure (Fig. S2b†). On the other hand, the synthesized nanoparticles with higher concentration of RhB were much less stable and tended to aggregate much more readily than those synthesized with low concentration, possibility due to a weaker stabilization with RhB. The UV-vis spectra also proves the argument. (Fig. S3†). Upon increasing the concentration of RhB (<100 μM), the LSPR peak gradually blue shifted to 700 nm with increasing broad peak and absorbance intensity. In contrast, for growth solution containing 100 μM RhBs, the LSPR peak was continuously blue-shifted, but the absorbance decreased, indicating the less stable of the resulted nanoparticles. These results indicated that the A20 and RhB in our system co-mediated the growth of GNS, showing a synergistic shaping effect. As we previously reported,26 the growth mode in DNA-controlled seed-mediated growth system is dependent on the binding affinity between DNA with gold. We suppose that the existence of RhB will change the DNA density on gold seeds by increasing the RhB concentration during the growth, even for the RhB molecules with weak affinity on gold surface. A more detailed study on the mechanism is still under way. In order to ensure that the morphology of Raman probes are controlled by both DNA and RhB, as well as maintains such stability, and result in more roughly growth of nanoparticles, the concentrations of RhB and A20 were used as 10 μM and 1 μM, respectively.
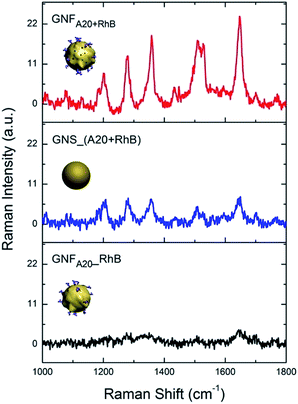
Next, we investigated the SERS activity of the obtained GNFs. Fig. 3 shows that DNA and RhBs co-mediated GNFs (GNFA20+RhB) had higher enhancement effect compared to other two probes with different synthesis methods which both only rely on the post-adhesion of Raman molecule onto gold surface, i.e. one is GNS by incubation with A20 and 1 μM RhB before growth (GNS_(A20 + RhB)) and the other is A20-controlled GNFs by direct incubation with RhB after growth (GNFA20_RhB). The calculated enhancement factor (EF) was 1.47 × 105 for GNFA20+RhB, which is about 4 times stronger than the one of GNS_(A20 + RhB) (0.32 × 105) and 10 times higher than GNFA20_RhB (0.14 × 105). As for the difference of the SERS effect, we suppose that there are two possible reasons: one is that the flower-like nanostructure should generate higher EF because of the more hot spots; the other one might be explained by more RhB molecules were attached onto the synthesized gold nanoparticles, making the Raman signal stronger. To prove the latter suppose, we calculated the number of molecules on each nanoparticle by fluorescence quantification of RhB in the supernatant after removing the GNFs with centrifugation and comparing it with the initial RhB quantity used for GNF synthesis (Fig. S4 in ESI†). The average number of attached RhBs on each GNFA20+RhB was estimated to be 2240, which is 1.14 fold than that of RhB on the GNFA20_RhB, indicating that high efficiency of loading RhB molecules onto the nanoprobes in our one-pot synthesis. To further study the stability of RhB on gold surface, we used excess mercaptopropionic acid (MPA) to treat the resulted nanoparticles. MPA has been shown to strongly bind to gold surface by effectively forming less-reactive Au(I)-MPA complex with gold ion, which can displaced the physi-chemical adsorbed RhB molecules. We found after the incubation with MPA, AuNS_(A20 + RhB) and GNFA20_RhB showed that much decrease of SERS enhancement, and as for GNFA20+RhB kept high activity, indicating that the RhB stablely adsorbed on the gold surface than the ones of post-adsorption (Fig. S5 in ESI†).
Except for the high Raman enhancement effect, the biological activity of the Raman probes is also critical for their biological sensing applications. Previously, we have demonstrated that the DNA-mediated growth of gold nanoparticle could realize DNA in situ functionalization on gold surface with high binding affinity, owing to partial DNA being buried in the GNF during the nanoparticle growth.7 Since we here also used single-stranded DNA (A20) to control the morphology of GNF in the growth, we supposed that the used DNA should have DNA hybridization activity and then mediated the self-assembly of nanoparticles.27,28 To prove the hypothesis, a complementary DNA strand, thilated-20 mer-ployT (SH-T20), were first attached onto gold nanorods (GNR) by thiol-gold chemistry and then mixed with GNFA20+RhB in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in the 10 mM Tris–HCl buffer solution with 100 mM NaCl overnight. As seen in TEM images (Fig. 4b and c), GNR was surrounded by a number of GNFA20+RhB, forming the satellite structure. Multiple satellite assembled nanostructures were also observed (Fig. 4d). As a comparison, when GNR functionalized by non-complementary DNA A20 (A20-GNR) was used to incubate with GNFA20+RhB, no any assembly was observed (Fig. S6 in ESI†). Furthermore, to testify whether any other DNA sequence can be encoded onto nanoparticle by the same synthesis method, we designed a sandwich DNA assay strategy to test the DNA hybridization activity on the Raman nanoprobe (details and Fig. S7–S9 seen in ESI†). The results confirmed that the resulted Raman nanoprobes were not only functionalized with DNA molecules in a large number but also retained their molecule recognition properties.
100 in the 10 mM Tris–HCl buffer solution with 100 mM NaCl overnight. As seen in TEM images (Fig. 4b and c), GNR was surrounded by a number of GNFA20+RhB, forming the satellite structure. Multiple satellite assembled nanostructures were also observed (Fig. 4d). As a comparison, when GNR functionalized by non-complementary DNA A20 (A20-GNR) was used to incubate with GNFA20+RhB, no any assembly was observed (Fig. S6 in ESI†). Furthermore, to testify whether any other DNA sequence can be encoded onto nanoparticle by the same synthesis method, we designed a sandwich DNA assay strategy to test the DNA hybridization activity on the Raman nanoprobe (details and Fig. S7–S9 seen in ESI†). The results confirmed that the resulted Raman nanoprobes were not only functionalized with DNA molecules in a large number but also retained their molecule recognition properties.
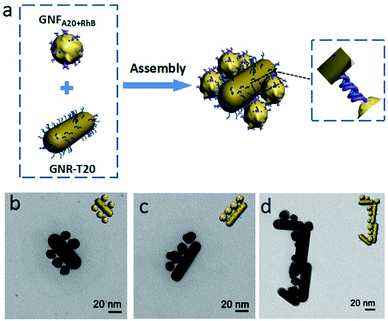 | ||
| Fig. 4 (a) Schematically illustration and (b–d) TEM imaging of DNA hybridization-mediated assembly of A20-enchored GNFs nanoprobes (GNFA20+RhB) onto T20-modified gold nanorods (T20-GNR). | ||
In summary, we reported a new simple synthesis method of SERS nanoprobe by introducing Raman dye into the one-pot DNA-mediated growth from gold nanoseeds. Under the optimal conditions, DNA and RhB effectively control the shape of nanoparticles and keep the stabilization of nanoparticles from aggregation. Since the partial RhB molecules are closely absorbed onto gold nanoparticles during the one-step synthesis, the as-resulted nanoparticles exhibit a stable and improved Raman enhancement effect. Moreover, DNA could be simultaneously introduced into the nanoparticles with high stability while retaining their biorecognition ability, allowing programmable self-assembly of new nanostructures. These SERS nanoprobes could find wide applications in fields such as nanoassembly, biological sensing, and biomedicine.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (Nos 61405176, 21305125, 61307073, 31401572), the Natural Sciences Fund of Zhejiang Province (No. LY17C130005), Zhejiang Provincial Public Welfare Technology Applied Research Project (Nos 2016C32006 and 2014C32002) and the Fundamental Research Funds for the Central Universities (No. 2016QNA5001).Notes and references
- J. H. Granger, N. E. Schlotter, A. C. Crawford and M. D. Porter, Chem. Soc. Rev., 2016, 45, 3865–3882 RSC.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- L. L. Qu, Y. T. Li, D. W. Li, J. Q. Xue, J. S. Fossey and Y. T. Long, Analyst, 2013, 138, 1523–1528 RSC.
- B. Mir-Simon, I. Reche-Perez, L. Guerrini, N. Pazos-Perez and R. A. Alvarez-Puebla, Chem. Mater., 2015, 27, 950–958 CrossRef CAS.
- Y. G. He, S. Y. Shi, N. Liu, Y. Y. Zhu, Y. S. Ding, J. Yin and Z. Q. Wu, RSC Adv., 2015, 5, 39697–39704 RSC.
- C. Qian, Q. H. Guo, M. M. Xu, Y. X. Yuan and J. L. Yao, RSC Adv., 2015, 5, 53306–53312 RSC.
- S. Saverot, X. Geng, W. Leng, P. J. Vikesland, T. Z. Grove and L. R. Bickford, RSC Adv., 2016, 6, 29669–29673 RSC.
- Y. Q. Zhang, X. J. Ye, G. X. Xu, X. L. Jin, M. M. Luan, J. T. Lou, L. Wang, C. J. Huang and J. Ye, RSC Adv., 2016, 6, 5401–5407 RSC.
- H. Kang, S. Jeong, Y. Park, J. Yim, B. H. Jun, S. Kyeong, J. K. Yang, G. Kim, S. Hong, L. P. Lee, J. H. Kim, H. Y. Lee, D. H. Jeong and Y. S. Lee, Adv. Funct. Mater., 2013, 23, 3719–3727 CrossRef CAS.
- J. F. Li, Y. F. Huang, Y. Ding, Z. L. Yang, S. B. Li, X. S. Zhou, F. R. Fan, W. Zhang, Z. Y. Zhou, D. Y. Wu, B. Ren, Z. L. Wang and Z. Q. Tian, Nature, 2010, 464, 392–395 CrossRef CAS PubMed.
- J. H. Soh, Y. Y. Lin, S. Rana, J. Y. Ying and M. M. Stevens, Anal. Chem., 2015, 87, 7644–7652 CrossRef CAS PubMed.
- H. Jans and Q. Huo, Chem. Soc. Rev., 2012, 41, 2849–2866 RSC.
- Z. D. Wang, J. Q. Zhang, J. M. Ekman, P. J. A. Kenis and Y. Lu, Nano Lett., 2010, 10, 1886–1891 CrossRef CAS PubMed.
- N. L. Rosi and C. A. Mirkin, Chem. Rev., 2005, 105, 1547–1562 CrossRef CAS PubMed.
- L. Dykman and N. Khlebtsov, Chem. Soc. Rev., 2012, 41, 2256–2282 RSC.
- X. J. Liu, M. Knauer, N. P. Ivleva, R. Niessner and C. Haisch, Anal. Chem., 2010, 82, 441–446 CrossRef CAS PubMed.
- Y. W. Cao, R. Jin and C. A. Mirkin, J. Am. Chem. Soc., 2001, 123, 7961–7962 CrossRef CAS PubMed.
- B. Zhao, J. L. Shen, S. X. Chen, D. F. Wang, F. Li, S. Mathur, S. P. Song and C. H. Fan, Chem. Sci., 2014, 5, 4460–4466 RSC.
- J. R. Anema, J. F. Li, Z. L. Yang, B. Ren and Z. Q. Tian, Annu. Rev. Anal. Chem., 2011, 4, 129–150 CrossRef CAS PubMed.
- Y. S. Wang, A. B. Serrano, K. Sentosun, S. Bals and L. M. Liz-Marzan, Small, 2015, 11, 4314–4320 CrossRef CAS PubMed.
- N. Gandra and S. Singamaneni, Adv. Mater., 2013, 25, 1022–1027 CrossRef CAS PubMed.
- H. Pei, X. Zuo, D. Zhu, Q. Huang and C. Fan, Acc. Chem. Res., 2014, 47, 550–559 CrossRef CAS PubMed.
- L. H. Tan, H. Xing and Y. Lu, Acc. Chem. Res., 2014, 47, 1881–1890 CrossRef CAS PubMed.
- Z. D. Wang, L. H. Tang, L. H. Tan, J. H. Li and Y. Lu, Angew. Chem., Int. Ed., 2012, 51, 9078–9082 CrossRef CAS PubMed.
- L. H. Tan, Y. Yue, N. S. R. Satyavolu, A. S. Ali, Z. Wang, Y. Wu and Y. Lu, J. Am. Chem. Soc., 2015, 137, 14456–14464 CrossRef CAS PubMed.
- T. J. Song, L. H. Tang, L. H. Tan, X. J. Wang, N. S. R. Satyavolu, H. Xing, Z. D. Wang, J. H. Li, H. J. Liang and Y. Lu, Angew. Chem., Int. Ed., 2015, 54, 8114–8118 CrossRef CAS PubMed.
- A. Klinkova, R. M. Choueiri and E. Kumacheva, Chem. Soc. Rev., 2014, 43, 3976–3991 RSC.
- Y. L. Wang, K. Lee and J. Irudayaraj, Chem. Commun., 2010, 46, 613–615 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section and elaboration of the results. See DOI: 10.1039/c6ra26580d |
| ‡ Y. Li and Dr X. J. Liu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |