Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Antibacterial and catalytic activities of biosynthesized silver nanoparticles prepared by using an aqueous extract of green coffee bean as a reducing agent
Mengmeng Wanga,
Wenjing Zhanga,
Xuesong Zheng*b and
Peizhi Zhu *a
*a
aSchool of Chemistry and Chemical Engineering, Yangzhou University, Jiangsu 225002, PR China
bSchool of Perfume and Aroma Technology, Shanghai Institute of Technology, China. E-mail: xuesongzheng10@gmail.com
First published on 21st February 2017
Abstract
Spherical biogenic silver nanoparticles (AgNPs) were synthesized using aqueous green coffee bean extract as a reducing agent. The green coffee bean extract was used for the time to reduce silver ions via a one-step, green, and rapid approach. The prepared AgNPs were characterized via transmission electron microscopy (TEM), atomic force microscopy (AFM), and UV-visible spectroscopy. Dynamic light scattering (DLS) was performed to measure the particle size. These AgNPs exhibit strong antibacterial activity against both Gram-negative and Gram-positive bacteria and the minimum inhibitory concentration (MIC) of AgNPs for Escherichia coli and Staphylococcus aureus are 0.05 μmol L−1 and 0.1 μmol L−1, respectively. Furthermore, AgNPs could serve as an effective catalyst for the reduction of 4-nitrophenol to 4-aminophenol in the presence of NaBH4.
Introduction
In recent years, nanoparticles (NPs) have attracted extensive attention due to their size effect, surface effect, quantum size effect, macroscopic quantum tunnelling effect, and dielectric confinement effect. NPs have been widely used as magnetic materials,1 ceramic materials,2 sensing elements,3 semiconductors,4 catalysts,5 and even in biomedicine.6 As a noble metal with excellent performance, silver nanoparticles (AgNPs) exhibit broad applications in catalysis,7 biomedicine,8 as an antimicrobial agent,9 in chemical and biological sensings,10 and cancer therapy.11 The conventional method for preparing silver nanoparticles, which is based on the chemical reduction of a silver salt, is simple and effective, but the removal of the agents used is both a costly and time-consuming process. Furthermore, the residual agents show biological toxicity.12,13Thus, there is an increased interest in using biodegradable and biocompatible reducing agents to prepare AgNPs. Over the last decade, polysaccharide-based biopolymers such as alginate, starch, and chitosan14–16 have been used as green alternatives17–20 to conventional agents due to their low-cost, non-toxicity, and environment-friendly properties. In most cases, the chemical dispersants used for controlling the growth of metallic nanoparticles were replaced by these biopolymers due to their low-toxicity to animal cells.21 Nano-sized metal catalysts play a key role in the chemical industries; among these, silver nanoparticles have gained significant attention owing to their unique electronic properties and high surface to volume ratio.22 In addition, silver nanocatalysts are cost-effective, efficient, and environmentally friendly. In particular, silver nanoparticles present highly efficient catalytic activity in the oxidation of methanol and ethylene,23,24 as well as in the reduction of nitric oxides (NOx).25
For the synthesis of silver nanoparticles, a variety of methods have been employed such as chemical reduction,26 electrochemical process,27 microemulsion method,28 laser ablation,29 and hydrothermal method.30 Chemical reduction is still the most common method. Recently, inspired by the concept of green chemistry, several studies have been focussed on the development of novel eco-friendly reducing agents for synthesizing AgNPs. Many interesting biomolecules including those obtained from plant extract,31–33 bacteria,34 viruses,35 DNA,36 and peptides37–39 have been used to successfully synthesize AgNPs.40
In this study, we report the synthesis of silver nanoparticles using green coffee bean extract as a reducing agent for the first time. Green coffee bean extracts were obtained from the seeds of Coffea arabica, Coffea canephora, and Coffea liberica. Green coffee bean extracts have been used as a healthy food supplement for long time because the main ingredient chlorogenic acid is a native antioxidant and can reduce blood pressure. We used TEM, AFM, and DLS to characterize the morphology and particle size of the silver nanoparticles synthesized using green coffee bean extracts. The formation of silver nanoparticles was studied by UV-visible spectroscopy. In antibacterial experiments, Escherichia coli and Staphylococcus aureus were used to evaluate the antimicrobial activity of AgNPs against Gram-negative and Gram-positive bacteria, respectively. Moreover, the catalytic activity of the as-prepared silver nanoparticles for the reduction of 4-nitrophenol in the presence of NaBH4 was also studied.
Materials and methods
Chemicals and materials
Silver nitrate (AgNO3, 99.8%), 4-nitrophenol (4-NP), sodium borohydride (NaBH4, 98%) were purchased from Sinopharm Chemical Reagent Co. Ltd (Shanghai, China). Green coffee bean extracts were purchased from Xi'an Zebang Biological Technology Co. Ltd. Chlorogenic acid (CGA, 98%) was purchased from Bioduly Biochemical Reagent (Nanjing, China). All the chemicals were of analytical reagent grade and were used without further purification. All the solutions were freshly prepared using deionized double-distilled water.Preparation of silver nanoparticles
For the biosynthesized silver nanoparticles, 0.2175 g silver nitrate was dissolved in 15 mL deionized double-distilled water. Then, 25 mL of a 1 mM aqueous green coffee bean extract solution was added dropwise to a silver nitrate solution under stirring at 30 °C. To ensure a complete mixing, stirring was maintained for 5 minutes. The color of the as-prepared silver nanoparticles appeared similar to that of the green coffee bean extracts solution. The synthesis of CGA–AgNPs was performed in a water-based system; 2.8 mL of 0.5 mM AgNO3 was placed in a glass vial with a cap and heated to 80 °C on a hot plate under stirring for 3 min. A 1.2 mL of 0.05 mM CGA was added dropwise under stirring on a hot plate within 2 min. Then, the reaction mixture was further incubated in an oven for 10 h.TEM analysis of the silver nanoparticles
Transmission electron microscopy (TEM, Tecnai C2 F30 S-Twin, FEI, USA) was used to observe the morphologies and particle sizes of AgNPs, and selected-area electron diffraction (SAED) patterns were obtained via high-resolution transmission electron microscopy (HRTEM). Transmission electron microscopy was performed by fixation on a 200-mesh carbon-coated copper grid.UV-visible spectra analysis
The absorbance spectra of AgNPs and CaP-coated AgNPs were obtained by UV-Vis spectroscopy (Varian Cary 500, USA).Dynamic light scattering measurement
The particle size distribution of the biosynthesized AgNPs was analyzed using a Zetasizer Nanoseries apparatus (Malvern) with laser He–Ne (4 mW) at 632.8 nm.Atomic force microscope analysis
AFM (Bruker, Santa Barbara, CA) images were obtained using Shimadzu nano search microscope SFT-3500 in QNM (quantitative nanomechanical mapping) mode. The typical resonance frequency of the cantilever with an integrated silicon tip (spring constant 40 N m−1) was between 283.8 and 360 kHz.Antibacterial test
The antibacterial activity of the synthesized AgNPs was tested by growth inhibition curve method, and the minimum inhibitory concentration (MIC) was also determined. The assay was performed on Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 25923), which acted as the representatives for Gram-negative and Gram-positive bacteria, respectively.First, the suspension solutions of E. coli and S. aureus were prepared by shaking in an incubator at 37 °C for 8 h. The AgNPs solutions of various concentrations were prepared by diluting the nanosilver solution of 50 μmol L−1 with double-distilled H2O. Then, 100 μL of AgNPs solutions and 900 μL of LB were completely mixed in 1.5 mL microcentrifuge tubes. The tube of ddH2O in place of the AgNPs solution in the mixture was used as the control. Therefore, the final concentration of AgNPs in the media were 0.5 μmol L−1, 0.2 μmol L−1, 0.1 μmol L−1, 0.05 μmol L−1, 0.04 μmol L−1, 0.03 μmol L−1, 0.02 μmol L−1, 0.01 μmol L−1, and 0.005 μmol L−1. Two hundred microliters of the mixture was transferred with pipette into each well of a Corning 96-well microtiter plate and 2 μL of a bacteria suspension solution was inoculated in each well. Immediately, this plate was incubated at 37 °C for 180 min in Multimode Plate-Reader (Tecan infinite 200PRO). The OD600 values were measured at the intervals of 5 min, following 20 s plate-shaking every time.
Catalytic reduction of 4-nitrophenol
In a typical run for the reduction of 4-NP by NaBH4, 0.2 mL of biosynthesized silver nanoparticles (1 mM) was added to 5 mL reaction solution containing 0.05 mmol fresh solution of 4-NP and 0.2 mmol NaBH4 solution. To set a blank group, the reaction system was kept same as the abovementioned system except that the prepared silver nanoparticles were replaced by ultra-pure water. The reaction in the presence of NaBH4 was immediately monitored by UV-visible spectrophotometric monitoring instrument over a 210–600 nm wavelength.Results and discussion
Characterization of the biosynthesized silver nanoparticles
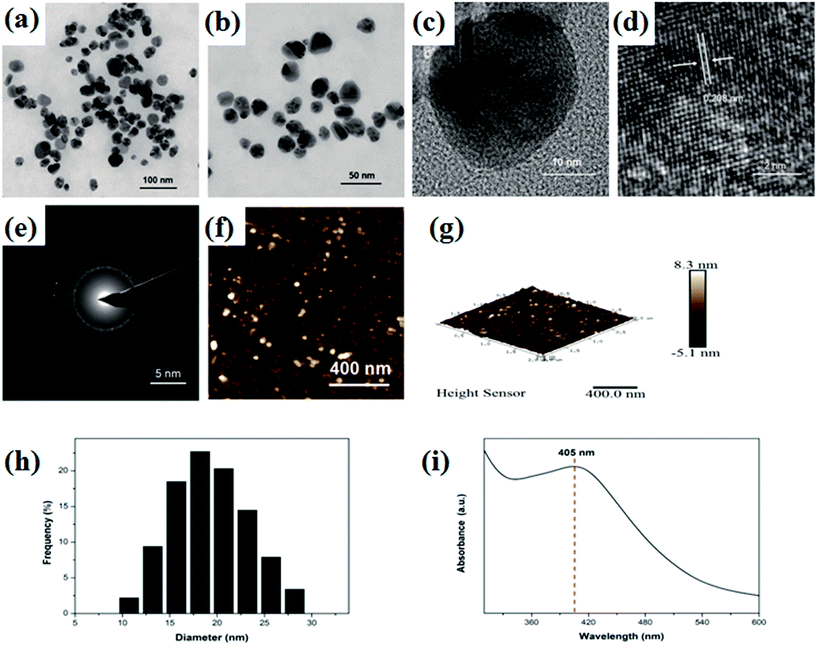
TEM was carried out to investigate the morphology and size of the obtained AgNPs. Fig. 1a, d and e shows typical TEM images of the AgNPs and most AgNPs nanoparticles were well-dispersed in the solution of green coffee bean extract. Fig. 1a and b shows that the nanoparticles were spherical in shape. The high-resolution TEM image, as shown in Fig. 1c, shows polycrystalline structure of the particles. From Fig. 1d, clear lattice fringes can be easily observed, and the interplanar spacing of these was measured to be 0.209 nm that corresponds to the Ag (200) plane.31 The selected-area electron diffraction (SAED) pattern, as shown in Fig. 1e, proves the polycrystalline structure of the synthesized AgNPs, which is consistent with the TEM result. The AFM topography image (Fig. 1f and g) shows that the size of AgNPs ranges from 10 to 30 nm in diameter, which is consistent with the DLS histogram (Fig. 1h). Fig. 1i displays the UV-vis absorption spectra of the as-prepared AgNPs. An obvious absorption peak at 405 nm, as shown in Fig. 1i, confirmed the formation of AgNPs since the characteristic silver SPR (surface plasmon resonance) bands are around 400–450 nm.41Antibacterial properties of the biosynthesized AgNPs
The 180 min growth curve for E. coli and S. aureus in LB media containing various concentrations of AgNPs are displayed in Fig. 2. Based on these curves, the excellent antibacterial activity of the AgNPs against both Gram-negative and Gram-positive bacteria can be concluded, and the minimum inhibitory concentration (MIC) (MIC = 0.04 μmol L−1 for E. coli and 0.2 μmol L−1 for S. aureus) can be determined. For E. coli (Fig. 2a), the concentration of 0.02 μmol L−1 AgNPs exhibited a weak growth inhibition. During 180 min of the assay, 0.05 μmol L−1 AgNPs absolutely repressed the bacterial growth. However, for S. aureus (Fig. 2b), 0.05 μmol L−1 AgNPs just slightly repressed the growth and the absolute inhibition concentration was approximately 0.1 μmol L−1.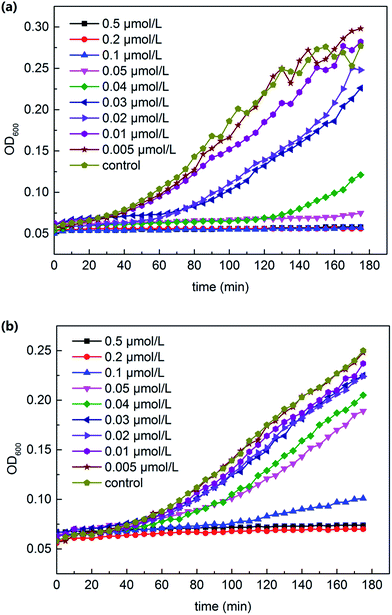 | ||
| Fig. 2 180 min growth curve for E. coli (a) and S. aureus (b) in LB media containing various concentrations of AgNPs. | ||
This difference may be due to the difference in the bacterial cell wall structure. Shrivastava et al. elucidated in their study that the different cell wall structure leads to variety in the penetration of AgNPs.42 The cell wall of the Gram-positive bacteria is composed of a thick layer of peptidoglycan that constitutes linear polysaccharides chains cross-linked by short peptides, thus forming a rigid structure. This leads to difficulty in the penetration of AgNPs compared to that for Gram-negative bacteria, where the cell wall possesses a much thinner layer of peptidoglycan.
Catalytic reduction of 4-nitrophenol
The reduction of 4-nitrophenol by NaBH4 was taken as a model reaction to examine the catalytic activity of the AgNPs. The absorption peak of 4-NP with a light yellow color shifted from 317 nm to 400 nm after the addition of freshly prepared NaBH4 solution. This is due to the presence of NaBH4 that changed the pH of the solution.43 Moreover, the light yellow color turned to intense yellow, indicating the formation of 4-nitrophenolate ion.The catalytic process of this reaction was monitored by UV-vis spectroscopy; the absorption intensity at 400 nm for 4-nitrophenolate ion remained unvaried without the silver nanocatalyst. As shown in Fig. 3a, a new band at around 300 nm emerged after adding 2 mL AgNPs as a catalyst, indicating the reduction of 4-NP to 4-AP by NaBH4. Until the end of the reaction, the intensity of the absorption peak at 400 nm gradually decreased with time, whereas the absorption peak at 300 nm simultaneously increased. The kinetic process of the reduction was monitored by measuring the extinction of the solution at 400 nm as a function of time. A good linear correlation is shown in Fig. 3b and the apparent rate constant (K) of this catalytic reaction was 0.0456 S−1, suggesting that the biosynthesized AgNPs possessing a larger surface area exhibit high-effective catalytic activity for the reduction of 4-NP.
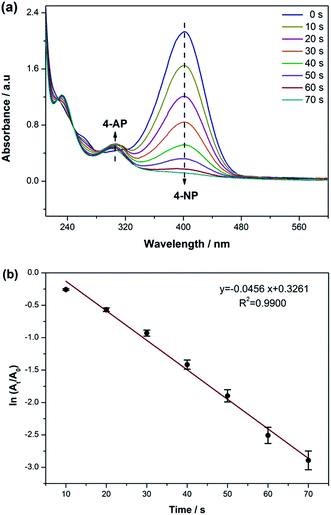 | ||
| Fig. 3 (a) UV-vis spectra of the 4-NP reduction in the presence of NaBH4 and 0.2 mL biosynthesized AgNPs and (b) catalytic degradation curves of 4-NP over the biosynthesized AgNPs. | ||
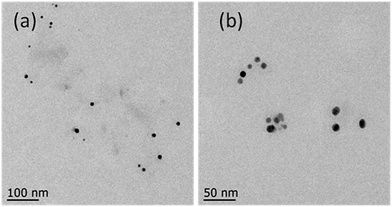
The main content of the green coffee bean is chlorogenic acid (CGA). However, we could not synthesize silver nanoparticles using only chlorogenic acid as a reducing agent at 30 °C. Then, reaction temperature was increased by a step of every 10 °C. The CGA–AgNPs were synthesized at 80 °C, which is consistent with the results of previous report.44 TEM images showed that the CGA–AgNPs were well-dispersed nanoparticles. The average particle sizes of the CGA–AgNPs were smaller than those of the AgNPs synthesized using aqueous green coffee bean extracts (Fig. 4). The synthesis of CGA–AgNPs showed that the reducing agent in the aqueous green coffee bean extracts was mainly chlorogenic acid. Green coffee bean extracts contain a variety of components such as caffeine, sucrose, citric acid, and trigonelline, many isomers of chlorogenic acid, amino acids, etc.45 Chlorogenic acid is the main component in the green coffee bean extracts, which is accountable for the reduction of Ag+ to Ag0 for the green synthesis of AgNPs. We hypothesized that other components, such as citric acid, can also act as reducing agents and the synergistic effect of several reducing agents lowered the energy barrier for the reduction of silver ions. These chemicals also act as capping agents to stabilize the synthesized silver nanoparticles. Since green coffee bean extract is much cheaper than chlorogenic acid, silver nanoparticles can be synthesized using a green coffee bean extract as a reducing agent at 30 °C, which a much lower temperature than that required by chlorogenic acid only. This environmentally friendly approach is suitable for the large-scale biosynthesis of silver nanoparticles due to its operational simplicity, low-cost, and green conditions. These well-caped silver nanoparticles produced by the green coffee bean extract are more stable and can strongly inhibit the growth of both Gram-negative and Gram-positive bacteria. In addition, these silver nanoparticles have potential in the simple and safe reduction of phenol compounds in water.
Conclusions
In this study, biogenic spherical AgNPs were successfully synthesized using aqueous green coffee bean extracts for the first time. The approach was reproducible, cost-effective, and eco-friendly. TEM and DLS analysis showed that the particle size of the as-prepared silver nanoparticles ranged from 12 nm to 31 nm. These small AgNPs exhibit strong antibacterial activity against E. coli and S. aureus and its MICs were evaluated to be 0.05 μmol L−1 and 0.1 μmol L−1, respectively. Moreover, biosynthesized silver nanoparticles could be an effective catalyst for the reduction of 4-nitrophenol to 4-aminophenol in the presence of NaBH4.Acknowledgements
We would like to thank Jiangsu Province for specially appointed professorship to Professor Peizhi Zhu and Testing Center of Yangzhou University for the support. This work was also supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions.Notes and references
- H. Lin, R. R. Zhang and Q. W. Chen, Nanoscale, 2014, 6, 14064–14105 RSC.
- F. Hua, T. H. Cui and Y. M. Lvov, Nano Lett., 2004, 4, 823–825 CrossRef CAS.
- P. Ginzburg, N. Berkovitch, A. Nevet, I. Shor and M. Orenstein, Nano Lett., 2011, 11, 2329–2333 CrossRef CAS PubMed.
- X. L. Hu, G. S. Li and J. C. Yu, Langmuir, 2010, 26, 3031–3039 CrossRef CAS PubMed.
- W. Wang, T. Y. Chiang, D. Velego and T. E. Mallouk, J. Am. Chem. Soc., 2013, 135, 10557–10565 CrossRef CAS PubMed.
- B. H. Dong, C. Y. Li, G. C. Chen, Y. J. Zhang, Y. Zhang, M. J. Deng and Q. B. Wang, Chem. Mater., 2013, 25, 2503–2509 CrossRef CAS.
- M. X. Zhang, Y. H. Cao, Y. Chong, Y. F. Ma, H. L. Zhang, Z. W. Deng, C. H. Hu and Z. J. Zhang, ACS Appl. Mater. Interfaces, 2013, 5, 13325–13332 CAS.
- S. Dasa and B. B. Dhar, RSC Adv., 2014, 4, 46285–46292 RSC.
- M. S. Noh, B. H. Jun, S. Kim, H. Kang, M. A. Woo, A. Minai-Tehrani, J. E. Kima, J. Kim, J. Y. Park, H. T. Lim, S. C. Park, T. Hyeon, Y. K. Kim, D. H. Jeong, Y. S. Lee and M. H. Cho, Biomaterials, 2009, 30, 3915–3925 CrossRef CAS PubMed.
- M. R. Reithofer, A. Lakshmanan, A. T. K. Ping and J. M. Chin, Biomaterials, 2014, 35, 7535–7542 CrossRef CAS PubMed.
- D. Kagan, P. Calvo-Marzal, S. Balasubramanian, S. Sattayasamitsathit, K. M. Manesh, G. U. Flechsig and J. Wang, J. Am. Chem. Soc., 2009, 131, 12082–12083 CrossRef CAS PubMed.
- Y. Yang, T. Liu, L. Cheng, G. S. Song, Z. Liu and M. W. Chen, ACS Appl. Mater. Interfaces, 2015, 7, 7526–7533 CAS.
- P. Sanpui, A. Chattopadhyay and S. S. Ghosh, ACS Appl. Mater. Interfaces, 2011, 3, 218–228 CAS.
- E. Y. Furuya and F. D. Lowy, Nat. Rev. Microbiol., 2006, 4, 36–45 CrossRef CAS PubMed.
- S. A. Jones, P. G. Bowler, M. Walker and D. Parsons, Wound Repair Regen., 2004, 12, 288–294 CrossRef PubMed.
- V. Prabu, P. Karthick, A. Rajendran, D. Natarajan, M. S. Kiran and D. K. Pattanayak, RSC Adv., 2015, 5, 50767–50777 RSC.
- Z. Abdali, H. Yeganeh, A. Solouk, R. Gharibi and M. Sorayya, RSC Adv., 2015, 5, 66024–66036 RSC.
- R. S. Chen, H. W. Ni, H. S. Zhang, G. Yue, W. T. Zhan and P. Y. Xiong, Vacuum, 2013, 89, 249–253 CrossRef CAS.
- A. M. Mittelman, D. S. Lantagne, J. Rayner and K. D. Pennell, Environ. Sci. Technol., 2015, 49, 8515–8522 CrossRef CAS PubMed.
- H. J. Klasen, Burns, 2000, 33, 117–138 CrossRef.
- W. Lee, K. J. Kim and D. G. Lee, BioMetals, 2014, 27, 1191–1201 CrossRef CAS PubMed.
- L. Sintubin, W. D. Windt, J. Dick, J. Mast, D. vanderHa, W. Verstraete and N. Boon, Appl. Microbiol. Biotechnol., 2009, 84, 741–749 CrossRef CAS PubMed.
- E. Qayyum, V. A. Castillo, K. Warrington, M. A. Barakat and J. N. Kuhn, Catal. Commun., 2012, 28, 128–133 CrossRef CAS.
- M. J. Lippits and B. E. Nieuwenhuys, Catal. Today, 2010, 154, 127–132 CrossRef CAS.
- S. Wunder, F. Polzer, Y. Lu, Y. Mei and M. Ballauff, J. Phys. Chem. C, 2010, 114, 8814–8820 CAS.
- Y. Sun and Y. Xia, Science, 2002, 298, 2176–2179 CrossRef CAS PubMed.
- C. D. Gu and T. Y. Zhang, Langmuir, 2008, 24, 12010–12016 CrossRef CAS PubMed.
- S. Pal, E. J. Yoon, Y. K. Tak, E. C. Choi and J. M. Song, J. Am. Chem. Soc., 2009, 131, 16147–16155 CrossRef CAS PubMed.
- H. S. Shin, H. J. Yang, S. B. Kim and M. S. Lee, J. Colloid Interface Sci., 2004, 274, 89–94 CrossRef CAS PubMed.
- M. Feng, M. Zhang, J. M. Song, X. G. Li and S. H. Yu, ACS Nano, 2011, 5, 6726–6735 CrossRef CAS PubMed.
- A. A. Kajani, A. K. Bordbar, S. H. Z. Esfahani, A. R. Khosropour and A. Razmjou, RSC Adv., 2014, 4, 61394 RSC.
- S. Iravani, Green Chem., 2011, 13, 2638–2650 RSC.
- S. K. Li, Y. H. Shen, A. J. Xie, X. R. Yu, L. G. Qiu, L. Zhang and Q. F. Zhang, Green Chem., 2007, 9, 852–858 RSC.
- H. Zhang, Q. Li, Y. Lu, D. Sun, X. Lin, X. Deng, N. He and S. Zheng, J. Chem. Technol. Biotechnol., 2005, 80, 285–290 CrossRef CAS.
- C. X. Yang, S. Jung and H. Yi, Biochem. Eng. J., 2014, 89, 10–20 CrossRef CAS.
- S. Kundu, Phys. Chem. Chem. Phys., 2013, 15, 1107–14119 RSC.
- K. T. Nam, Y. J. Lee, E. M. Krauland, S. T. Kottmann and A. M. Belcher, ACS Nano, 2008, 2, 1480–1486 CrossRef CAS PubMed.
- G. Upert, F. Bouillere and H. Wennemers, Angew. Chem., Int. Ed., 2012, 51, 4231–4234 CrossRef CAS PubMed.
- H. Wennemers, J. Pept. Sci., 2012, 18, 437–441 CrossRef CAS PubMed.
- P. R. Rathi Sre, M. Reka, R. Poovazhagi, M. Arul Kumar and K. Murugesan, Spectrochim. Acta, Part A, 2015, 135, 1137–1144 CrossRef CAS PubMed.
- E. C. Njagi, H. Huang, L. Stafford, H. Genuino, H. M. Galindo, J. B. Collins, G. E. Hoag and S. L. Suib, Langmuir, 2011, 27, 264–271 CrossRef CAS PubMed.
- S. Shrivastava, T. Bera, A. Roy, G. Singh, P. Ramachandrarao and D. Dash, Nanotechnol., 2007, 18, 225103–225122 CrossRef.
- B. Baruah, G. J. Gabriel, M. J. Akbashev and M. E. Booher, Langmuir, 2013, 29, 4225–4234 CrossRef CAS PubMed.
- H. J. Noh, H. S. Kim, S. H. Jun, Y. H. Kang, S. Cho and Y. Park, J. Nanosci. Nanotechnol., 2013, 13, 5787–5793 CrossRef CAS PubMed.
- F. Wei, K. Furihata, F. Hu, T. Miyakawa and M. Tanokura, Magn. Reson. Chem., 2010, 48, 857–865 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |