Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of arginine based nanocarriers for targeting and treatment of intracellular Salmonella†
Rajeev J.
Mudakavi‡
 abc,
Surya
Vanamali‡
abc,
Surya
Vanamali‡
 b,
Dipshikha
Chakravortty
*ac and
Ashok M.
Raichur
b,
Dipshikha
Chakravortty
*ac and
Ashok M.
Raichur
 *bcd
*bcd
aDepartment of Microbiology and Cell Biology, Indian Institute of Science, Bangalore, India
bDepartment of Materials Engineering, Indian Institute of Science, Bangalore, India. E-mail: amr@materials.iisc.ernet.in; Fax: +91 80 23600472; Tel: +91 80 22932705
cCentre for BioSystems Science and Engineering, Indian Institute of Science, Bangalore, India
dNanotechnology and Water Sustainability Unit, University of South Africa, Florida, 1710 Johannesburg, South Africa
First published on 20th January 2017
Abstract
Arginine decorated nanocarriers exhibited intravacuolar targeting capability which was utilized to deliver antibiotics into the intracellular niche of pathogens like Salmonella and Mycobacterium. The arginine based nanocarrier system (Arg-MSN) was developed on a mesoporous silica nanoparticle (MSN) template by conjugating L-arginine to protamine and pectin coated MSN by using a layer-by-layer coating approach. The synthesized nanocarriers were characterized using microscopy, FTIR spectroscopy, and zeta potential analyses. Lower cytotoxicity and hemolysis was observed for Arg-MSN nanocarrier compared to bare MSN template. Ciprofloxacin, a fluoroquinolone antibiotic was entrapped in Arg-MSN which showed gradual release of ciprofloxacin over a period of 24 h. In vitro experiments in Salmonella infected macrophages and epithelial cells exhibited two-fold higher antibacterial activity with ciprofloxacin-loaded Arg-MSN (Cip Arg-MSN) compared to free ciprofloxacin. The increased antibacterial activity of Cip Arg-MSN is believed to result from co-localization of Arg-MSN with the intravacuolar Salmonella and localized delivery of the antibiotic. We also observe an increase in reactive nitrogen species upon Arg-MSN treatment in the infected cells. In vivo bacterial burden and morbidity studies exhibited nearly ten-fold lower Salmonella burden in the infected organs such as spleen, liver and MLN (mesenteric lymph nodes). Similar survival rates were observed at a lower dosage of Cip Arg-MSN over free ciprofloxacin. The coordinated effect of improved antibiotic delivery, intracellular targeting and production of reactive nitrogen species was found to result in enhanced antibacterial activity. The developed Arg-MSN system is expected to be an attractive carrier system for delivery of antibiotics for clearing intravacuolar infections.
Introduction
An increased frequency of treatment failure and increase in severity of bacterial infection is being observed globally for Salmonella infections.1,2 Decreased exposure of the pathogen to the antibacterial agent leads to ineffective clearance, evasion of the pathogen to more inaccessible locations and development of resistance, all of which lead to failure of therapy. The emergence of multidrug resistance (MDR) to antibiotics as a consequence of ineffective clearance of pathogens is a major impetus for development of newer antibiotic therapies. One such method is to improve the activity by targeted delivery of existing antibiotics into the niche of intracellular pathogens such as M. tuberculosis, Francisella tularensis, Yersinia pestis and Salmonella which persist by adapting to an intracellular lifestyle within a host.3 This presents a unique challenge in the treatment of Salmonella infection owing to decreased antibiotic penetration. It is reported that the bactericidal concentration required for eliminating intracellular bacteria is higher than the bactericidal concentration for extracellular bacteria due to poor intracellular antibiotic penetration.4–7Salmonella has gained resistance to antibiotics such as chloramphenicol, trimethoprim–sulphamethoxazole, ampicillin etc.8 Resistance to these antibiotics is now so prevalent that treatment with these antibiotics is no longer recommended. The currently recommended therapy involves administering ciprofloxacin (Cip; fluoroquinolone antibiotic) or ceftriaxone (cephalosporin antibiotic), however, recent reports have shown decreased susceptibility to even ciprofloxacin.9,10 We have utilized the Salmonella infection model as a tool to study and develop antibiotic delivery systems to target intravacuolar infections.Salmonella, Listeria, Shigella, Staphylococcus etc., can invade and even survive in the intracellular environments of immune cells such as dendritic cells and macrophages.11 In macrophages, Salmonella forms a protective membrane called Salmonella containing vacuole (SCV) within which it replicates.12 The vacuolar Salmonella is capable of obtaining nutrients from the host for survival and replication. Other intracellular pathogens like Legionella,13,14Brucella,15Francisella16 and Shigella17 are also known to scavenge several amino acids such as arginine from the host cell and even result in severe amino acid starvation.18 Arginine is an important modulator of the cellular immune response of macrophage. Nitric oxide (NO), one of the mediators responsible for the cytotoxic activity of macrophages against intracellular pathogens is produced from arginine.19,20 Two host pathways utilize arginine, one mediated by nitric oxide synthase forming nitric oxide and the other mediated by arginase forming ornithine and urea. Mycobacterium infected cells showed increased uptake of arginine by cationic transporters (CAT1 and CAT2) and diversion of arginine into arginase pathway leading to decreased production of antibacterial nitric oxide.21,22Salmonella infection also results in upregulation of CAT1 transporters responsible for increased arginine uptake. It was observed that CAT1 transporters were recruited to the Salmonella containing vacuole (SCV).23 Hence, we have investigated whether nanoparticles functionalized with arginine could show a differential uptake in Salmonella infected cells and be exploited as a targeting strategy against intravacuolar Salmonella.
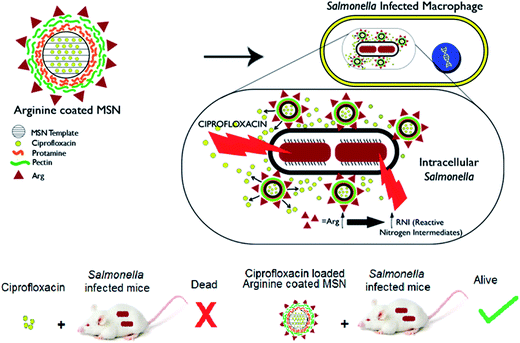
Recently halloysite clay nanotubes which are predominantly silicate based have been formulated for loading antimicrobial drugs into their porous lumen.24 The release of the pharmacological agent can be modified by employing layer-by-layer chemistry and enzyme responsive end capping.25–27 There are very few reports of therapeutic systems for targeting intracellular and especially intravacuolar pathogens. Among the available reports, a polyproline based antimicrobial peptide delivery system was shown to eliminate intracellular bacteria.28,29 Peptide-based formulation was shown to facilitate vancomycin uptake in Listeria infected macrophages.30,31 Nanoparticle formulation using PLGA or silica template was shown to be effective in Mycobacterium infections.32–34 Systems targeting intracellular Salmonella such as chitosan based nanoparticles,35–38 xerogels39 did not demonstrate any intracellular targeting mechanism. The efficacy of nanoparticle formulations can be increased by exploiting pathogen specificity and targeting the intracellular niche. We have developed a mesoporous silica based nanoparticle delivery system capable of targeting the intracellular compartment, deliver antibiotic payload and augment intracellular defenses of the host cell. We have evaluated the effectiveness of ciprofloxacin encapsulated in Arg-MSN system over free ciprofloxacin by in vitro as well as in vivo studies using Salmonella as the model intravacuolar pathogen Scheme 1.
Results and discussion
Arg-MSN nanocarrier system: synthesis and characterization
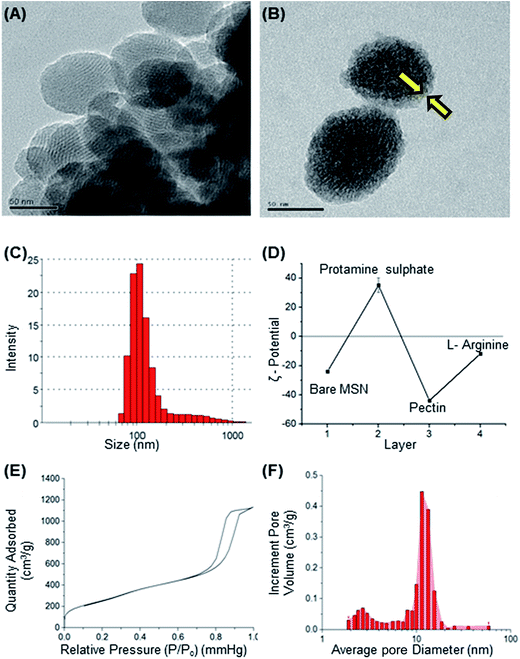
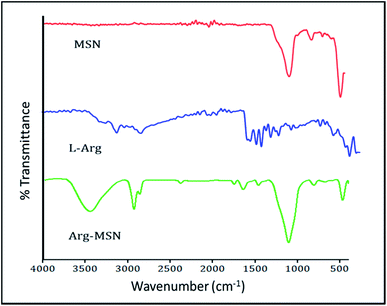
Mesoporous silica nanoparticle (MSN) and amine-MSN were prepared by modified Stöber's process.40 Mesoporous silica nanoparticles find various applications in the field of catalysis, adsorption, separation, photochemistry, nanomedicine etc. Their application in drug delivery is exceptional owing to their (i) ability to interact with large number of guest molecules owing to its high porosity and surface area, (ii) ease in surface functionalization (iii) stability in the biological media which allow for these systems to be responsive to chemical, physical or biological stimuli.41–43 Transmission electron microscopy of the synthesized bare MSN and arginine grafted MSN (Arg-MSN) showed that the particles have an average size of 75 nm and a highly ordered structure (Fig. 1A and B). Layer-by-layer coating chemistry was utilized for functionalizing the bare silica surface with polyelectrolyte coating.44,45 The outer surface of the mesoporous silica nanoparticle has a high negative zeta potential (−25 mV) due to presence of exposed silanol groups and provides an effective template for coating with positively charged polymers. First, bare MSN was coated with cationic polymer protamine and subsequently with negatively charged pectin polyelectrolyte to form a shell of oppositely charged polyelectrolyte layers. The polyelectrolyte coated MSN was decorated with L-arginine by conjugating it with the exterior pectin layer to give the arginine grafted MSN (Arg-MSN). Coating with polyelectrolytes did not show any obvious increase in size or its aggregation behaviour as observed by their narrow size distribution after coating (Fig. 1C). Coating of nanoparticles with polyelectrolytes solution was optimized using zeta-potential analyses. Protamine solution of concentration 0.5% w/v was used to coat the negatively charged bare MSN surface which develops a positive charge. The protamine-MSN particles were then coated with 0.1% w/v pectin solution to obtain a negatively charged pectin–protamine-MSN. L-Arginine was tagged to the above polyelectrolyte coated nanoparticles by EDC/NHS crosslinking chemistry. The carboxylic groups of pectin coated MSN were activated by EDC and covalently functionalized with L-arginine leading to the Arg-MSN particle. The polyelectrolyte coating and its subsequent conjugation with L-arginine was confirmed by measuring the zeta potential which showed alternating positive and negative zeta potential values characteristic of a typical layer-by-layer coating chemistry (Fig. 1D). The synthesized Arg-MSN has an isoelectric pH between 4 and 5, which develops a negative surface charge at the cytosolic pH and positive surface charge in the intralysosomal environment. This change in the charge behaviour is beneficial in preventing negatively charged ligands from binding and occluding the surface groups of the particle.To characterize the porosity and surface area of MSN, nitrogen adsorption/desorption isotherm studies was carried out. The isotherm obtained was characteristic of mesoporous materials exhibiting type IV isotherm with H1 type of hysteresis loop. The surface area of the synthesized MSN was found to be 987.7 m2 g−1 with an average pore diameter of 7.2 nm (Fig. 1E and F). The pore size is characteristic of mesoporous structures (2 nm < d > 50 nm) as defined by International Union of Pure and Applied Chemistry (IUPAC).46,47 The FTIR spectrum of bare MSN showed characteristic peak of silica at 1078 cm−1 ascribed to asymmetric vibration of Si–O groups and FTIR spectrum of Arg-MSN showed stretching vibrations at 2900–2850 cm−1 and 1730 cm−1 corresponding to alkyl groups and ester groups respectively (Fig. 2) confirming the coating of the polymers and arginine on MSN surface. The presence of high surface area, mesoporous structure and adaptability to functionalize ligand groups renders the Arg-MSN particulate system attractive for antibiotics delivery. Hence, ciprofloxacin (Cip) was loaded into the developed Arg-MSN particle system and its drug release behaviour studied. The amount of ciprofloxacin loaded into Arg-MSN was found to be 18 ± 1.2% w/w (Cip:MSN). Release profile of ciprofloxacin exhibited an initial burst release of 20% followed by release up to 60% for the next 24 h (Fig. S1†). This release pattern ensures continuous and prolonged delivery of the antibiotic reaches the intracellular niche of the pathogen present inside the macrophages and epithelial cells.
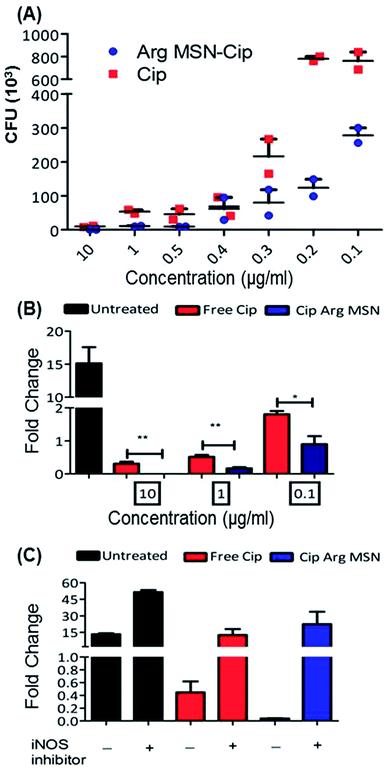
In vitro anti-bacterial activity of ciprofloxacin loaded Arg-MSN was compared with free ciprofloxacin on Salmonella typhimurium. Ciprofloxacin solutions of concentration 10 μg ml−1, 1 μg ml−1, 0.5 μg ml−1, 0.4 μg ml−1, 0.3 μg ml−1, 0.2 μg ml−1 and 0.1 μg ml−1 were incubated for 24 h and colony forming units (CFU) determined after plate count (Fig. 3A). At high antibiotic concentration (10 μg ml−1), no difference in CFU was observed. But at lower concentrations, a 3–4 fold reduction in CFU between Cip loaded Arg-MSN and equivalent concentration of free Cip was observed. Prevention of intracellular proliferation of STM in HeLa and RAW 264.7 cells after Cip Arg-MSN treatment was determined by measuring fold change in CFU of Salmonella after 16 h compared to 2 h i.e., CFU (16 h)/(2 h). The intracellular replication in RAW 264.7 cells was reduced by ten-fold, while in HeLa cells two-fold decrease was observed (Fig. S3†). To ascertain the appropriate reduction in dosage required to elicit same efficacy, different dosages of ciprofloxacin ranging from 0.1 μg ml−1 to 10 μg ml−1 was delivered via Arg-MSN carrier and compared with free ciprofloxacin (Fig. 3B). Anti-bacterial activity 0.1 μg ml−1 Cip Arg-MSN was comparable to 1 μg ml−1 of free ciprofloxacin. Thus a lower dosage of Cip Arg-MSN was found to be effective in clearing the pathogen present even in the intracellular environment.
In macrophages, nitric oxide is produced by the activity of inducible nitric oxide synthase (iNOS) enzyme. The effectors secreted by Salmonella pathogenicity island 2 (SPI 2) are reported to interfere in the iNOS pathway.48 To ascertain the role of nitric oxide radicals after delivery of Arg-MSN particles, intracellular proliferation assay was performed in the presence of iNOS inhibitor (1400w). Untreated cells, Cip treated cells and Cip Arg-MSN treated cells, all showed an increase in proliferation in the presence of iNOS inhibitor (Fig. 3C). But maximum change in the fold proliferation before and after inhibitor treatment was observed for Arg-MSN treated cells. This implicates the role of nitric oxide radicals in the observed increased activity of Cip Arg-MSN compared to free antibiotic.
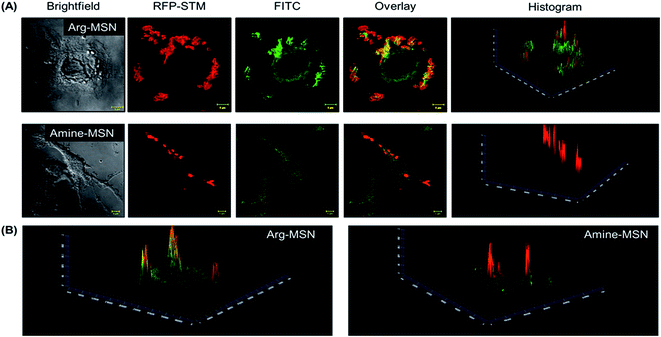
Cellular uptake and co-localization of Arg-MSN with intracellular Salmonella
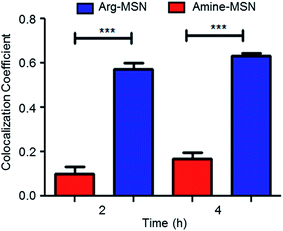
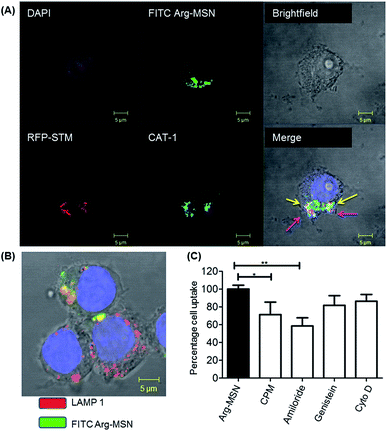
Cellular uptake and trafficking of Arg-MSN nanocarriers in Salmonella infected cells was studied using FITC tagged Arg-MSN. Co-localization of the Arg-MSN particles with intracellular STM was observed after 2 h incubation (Fig. 4A). Significant overlap was observed between intracellular STM and Arg-MSN, while no overlap was observed with amine-MSN. The cross-section of the image at the plane of intracellular STM confirmed bacterial co-localization with only Arg-MSN and not with amine-MSN (Fig. 4B). The co-localization with intracellular STM was specific to Arg-MSN nanocarrier only but not with other positively charged MSN particle (FITC tagged amine-MSN) as shown in Fig. 5.It was earlier reported that CAT1 transporters are upregulated in Salmonella infection and get trafficked to SCV presumably for delivering arginine to intracellular Salmonella.23 In line with the earlier study, we observed that CAT transporter exhibited co-localization with intracellular STM as shown in Fig. S4† and with the internalized Arg-MSN particles (Fig. 6A). We also observed that the Arg-MSN particle showed trafficking through endocytosis as the intracellular particles were found to overlap with lysosome associated membrane protein 1 (LAMP1) (Fig. 6B). We further investigated the endocytosis mechanism of nanoparticle uptake to understand the mechanism leading to co-localization of Arg-MSN with STM by using different pharmacological inhibitors of endocytosis.49,50 The pharmacological inhibitors exhibited no toxicity evaluated by FACS at the concentrations used for the endocytosis study (Fig. S5†). We found significant decrease in percentage uptake of Arg-MSN after incubating with chlorpromazine and amiloride (Fig. 6C) suggesting the internalization occurring primarily by clathrin mediated endocytosis and macropinocytosis. Hence, the presence of arginine on the nanoparticle directed the nanoparticle trafficking towards SCV and also aids in specifically targeting infected cells.
Evaluation Cip Arg-MSN nanocarriers in clearing Salmonella infection in vivo
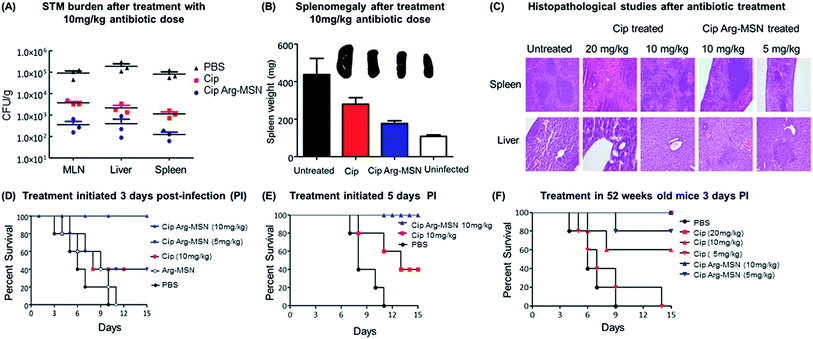
The therapeutic effectiveness of Cip Arg-MSN over free ciprofloxacin was assessed by measuring bacterial burden in different organs post Salmonella infection in BALB/c mice. Enteric pathogens such as Salmonella and Yersinia pseudotuberculosis invade the epithelial cells and disseminate to establish systemic infections. They invade the intestinal epithelial barrier into the underlying dendritic cells and macrophage cells from which they are believed to be transported by the afferent lymphatics into spleen, liver and other systemic organs.51–53 It has been recently observed that the nanominerals are present in the intestinal lumen which are predominantly phosphates particles are transported across the epithelial microfold-cells (M-cells) into the underling immune cells of the payers patches.54–56 Hence we expect the colloidal MSN particles carrying the antibiotic are similarly phagocytosed by the dendritic cells of Payers patches and transported to various organs. Salmonella CFU burden in liver, spleen and mesenteric lymph nodes (MLN) showed significant reduction after treatment with Cip Arg-MSN compared to free ciprofloxacin (Fig. 7A). Splenomegaly, a characteristic indication of bacterial infection showed marked reduction in inflammation and weight of spleen in Cip Arg-MSN treated mice compared to free drug alone, pointing towards resolving infection (Fig. 7B). Sections of liver, and spleen tissue of infected mice after treatment with 20 mg kg−1, 10 mg kg−1, and 5 mg kg−1 of Cip and Cip Arg-MSN (Fig. 7C) were studied for indication of pathology. The white pulp was significantly expanded and lymphoid hyperplasia along with infiltration of megakaryocytes was observed in the infected spleen. The cohorts treated with ciprofloxacin showed signs of resolving inflammation with congested red pulp and hyperplasia in white pulp. The inflammatory hyperplasia seen after treatment with 20 mg kg−1 and 10 mg kg−1 of free ciprofloxacin was comparable to treatment with 10 mg kg−1 Cip Arg-MSN. 5 mg kg−1 of Cip Arg-MSN showed lower lymphoid hyperplasia but fibrosis was observed. Liver sections showed moderately higher inflammation after treatment with Arg-MSN as compared to spleen tissue. Histopathological analysis reveals that ciprofloxacin delivered via Arg-MSN is more potent in arresting the spread of infection in the spleen, when compared to free ciprofloxacin.Salmonella typhimurium infection at an oral bacterial dose (108 CFU) is known to be fatal to mice and improvement in the survival of mice is considered as a measure of therapeutic efficacy. We find 100% survival of Salmonella infected mice treated with 10 mg kg−1 Cip Arg-MSN compared to 40% survival with free ciprofloxacin at same dosage (Fig. 7D). Among the mice that survived, those treated with free Cip showed higher morbidity symptoms like loss of hair and decreased movement. In another experimental setup, antibiotic treatment initiated 5 days post infection was carried out to test the effectiveness of the nanoparticle treatment in mice with advanced stage of infection. The results projected in Fig. 7E shows complete protection of infected mice even at an advanced stage of infection. To simulate Salmonella infections in elderly patients (60 years), infections in 52 week old mice were studied. 100% survival was observed at 20 mg kg−1 of ciprofloxacin dose and Cip Arg-MSN dose of 10 mg kg−1. 100% mortality was observed for ciprofloxacin dose of 5 mg kg−1 while the same concentration in the Arg-MSN carrier conferred over 50% survival (Fig. 7F).
Most of the Salmonella outbreaks and infection cases are reported from third world and developing countries where access to proper medical setup is inadequate. Hence therapeutic systems developed for intravenous delivery may find limited utility. Polyisohexylcyanoacrylate and liposomal based nanodelivery systems developed in the last decade for Salmonella treatment were limited by their intravenous delivery approach and requirement of rigorous toxicity studies.57–59 Intracellular targeting systems have been recently developed using cell penetrating peptides such as polyarginine based system60 and gold nanoparticle-conjugated to AMP (antimicrobial peptide) by Lee et al.61 It is reported that positively charged residues on the peptide conjugated particle enable efficient cellular uptake. Such highly positive charged particles exhibit increased tendencies to aggregate, which limits their application. The intracellular targeting mechanism, stability of the peptides and route of delivery remain challenging for developing peptide based particle delivery systems for Salmonella therapy.
Salmonella alters the intracellular microenvironment to prevent lysosomal degradation and persist by remodelling the endolysosomal system which can be tracked by nanoparticles.62,63 Therefore nanoparticle delivery mechanisms can be developed to utilize the intracellular microenvironment resources to target intracellular pathogens. It is known that nutrients present in the cytosol can be limiting for intracellular replication. The replication of Listeria monocytogenes was found to depend upon efficient uptake of hexose sugars of the host apart from aromatic amino acids, threonine and adenine.64–66 Similarly Shigella flexneri is known to take up guanine, thymine, p-aminobenzoic acid and diaminopimelate from the host cell.67–69 Many viruses and bacteria ensure availability of amino acids for their survival by regulating cationic amino acid receptor (CAT) present on the surface of infected host cell. We exploited arginine dependency of intracellular Salmonella as a targeting strategy for the delivery of antibiotics. Hence, for short term therapy necessitating delivery of antibiotics, the Arg-MSN formulation system developed was studied and found to be 3–4 times more efficient in clearing intracellular pathogens than free antibiotic alone. The nanoparticle targeting approach could be extended to other pathogens which show such nutritional dependencies for their intracellular survival.
Experimental
Synthesis of mesoporous silica nanoparticles (MSN) and coating of polyelectrolyte layers
MSN was prepared using a modified Stöber process described earlier.40 Coating of nanoparticles with polyelectrolytes solution was optimized using zeta-potential analyses, protamine sulphate solution and pectin solution of concentration 0.5% w/v and 0.1% w/v respectively were used. Protamine sulphate solutions (0.5% w/v) was prepared in 0.15 M NaCl of pH 5 and incubated with MSN particles for 30 minutes, washed thrice with 0.15 M NaCl and centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm to remove unabsorbed protamine, and its zeta potential (ζ) measured by Nanozetasizer (ZEN 3690, Malvern Instruments, UK). Pectin solution (0.1% w/v) in 0.15 M NaCl of pH 5 was used to coat the protamine coated MSN. The pectin coated particles 1 mg ml−1 was activated using EDC (2 mM) and NHS (5 mM) in 0.1 M MES buffer at pH 5 for 30 minutes to which 5 mg ml−1 of L-arginine solution in PBS was added. The unreacted reagents were removed by washing steps and zeta potential of the particles measured to confirm tagging of L-arginine to the LbL coated MSN particles to obtain arginine decorated MSN particles (Arg-MSN). Amine-MSN (NH2-MSN) was prepared by refluxing MSN dispersed in dry toluene with aminopropyltrimethoxysilane (APTMS) for 20 h in an inert atmosphere. Fluorescent Arg-MSN and amine MSN were prepared by incubation with 2 mg ml−1 FITC in DMSO for 4 h in a dark environment. The particles were washed thoroughly to remove residual FITC and DMSO.
000 rpm to remove unabsorbed protamine, and its zeta potential (ζ) measured by Nanozetasizer (ZEN 3690, Malvern Instruments, UK). Pectin solution (0.1% w/v) in 0.15 M NaCl of pH 5 was used to coat the protamine coated MSN. The pectin coated particles 1 mg ml−1 was activated using EDC (2 mM) and NHS (5 mM) in 0.1 M MES buffer at pH 5 for 30 minutes to which 5 mg ml−1 of L-arginine solution in PBS was added. The unreacted reagents were removed by washing steps and zeta potential of the particles measured to confirm tagging of L-arginine to the LbL coated MSN particles to obtain arginine decorated MSN particles (Arg-MSN). Amine-MSN (NH2-MSN) was prepared by refluxing MSN dispersed in dry toluene with aminopropyltrimethoxysilane (APTMS) for 20 h in an inert atmosphere. Fluorescent Arg-MSN and amine MSN were prepared by incubation with 2 mg ml−1 FITC in DMSO for 4 h in a dark environment. The particles were washed thoroughly to remove residual FITC and DMSO.
Characterization of Arg-MSN nanoparticle
The morphology of Arg-MSN such as size and shape was characterized by transmission electron microscopy. The transmission microscopy images of bare and coated MSN were obtained using Technai F30 Transmission Electron Microscope (FEI, The Netherlands) at an operating voltage 300 kV. Particles at a concentration of 0.1 mg ml−1 were briefly sonicated and 5 μl of the dispersion was transferred to a TEM grid. The grids were dried overnight at 40 °C prior to imaging. The surface area of MSN was characterized by Brunauer–Emmett–Teller (BET) method at 76.3 K. The N2 adsorption–desorption measurements was carried on ASAP-200 surface area and porosity analyzer (Micromeritics Instrument Corporation, USA). Fourier transform infrared (FTIR) spectra of the samples (bare MSN, L-arg, Arg-MSN) was recorded in the range 400 cm−1 to 4000 cm−1 by FTIR spectrometer (Bruker, Germany).In vitro anti-bacterial studies and intracellular proliferation assay using Cip Arg-MSN
Salmonella enterica serovar typhimurium (STM) 102 CFU was incubated with free ciprofloxacin and equivalent amount of Cip Arg-MSN for 24 h in LB medium in a 96 well plate. The concentration of antibiotics incubated with STM was 10 μg ml−1, 1 μg ml−1, 0.5 μg ml−1, 0.4 μg ml−1, 0.3 μg ml−1, 0.2 μg ml−1 and 0.1 μg ml−1. After 24 h, aliquots from STM culture were diluted and plated onto Salmonella-Shigella agar plates (SS agar) for determining colony forming units (CFU). The intracellular proliferation assay was carried out according to the reported procedure. 2 × 105 cells of HeLa and RAW 264.7 cells were infected with STM in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (MOI). The cells were washed with PBS after 30 min and incubated with media containing gentamicin (100 μg ml−1) for 1 h to remove extracellular bacteria. The gentamicin containing media (100 μg ml−1) was then replaced with media containing Cip Arg-MSN or free Cip at a concentration of 0.1 μg ml−1, 1 μg ml−1, and 10 μg ml−1 along with lower concentration of gentamicin (10 μg ml−1) and further incubated for 2 h and 16 h. Media containing only gentamicin (10 μg ml−1) was considered as the untreated control (UT). After the incubation period, the cells were lysed with 0.1% Triton-X 100 to release the intracellular bacteria and lysate plated onto SS-agar plates for determining colony forming units (CFU). The fold change in intracellular replication of STM was determined by calculating the ratio of CFU counts at 16 h to 2 h. The intracellular replication assay study was repeated after treatment with 10 μg ml−1 Cip Arg-MSN along with 1 μM iNOS inhibitor 1400w.
10 (MOI). The cells were washed with PBS after 30 min and incubated with media containing gentamicin (100 μg ml−1) for 1 h to remove extracellular bacteria. The gentamicin containing media (100 μg ml−1) was then replaced with media containing Cip Arg-MSN or free Cip at a concentration of 0.1 μg ml−1, 1 μg ml−1, and 10 μg ml−1 along with lower concentration of gentamicin (10 μg ml−1) and further incubated for 2 h and 16 h. Media containing only gentamicin (10 μg ml−1) was considered as the untreated control (UT). After the incubation period, the cells were lysed with 0.1% Triton-X 100 to release the intracellular bacteria and lysate plated onto SS-agar plates for determining colony forming units (CFU). The fold change in intracellular replication of STM was determined by calculating the ratio of CFU counts at 16 h to 2 h. The intracellular replication assay study was repeated after treatment with 10 μg ml−1 Cip Arg-MSN along with 1 μM iNOS inhibitor 1400w.
Cellular uptake studies of Arg-MSN into STM infected cells by confocal microscopy
RAW 264.7 murine macrophage cells were infected with red fluorescence protein expressing STM (STM-RFP) at MOI 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50. Post infection, the cells were treated with FITC tagged Arg-MSN and FITC tagged amine-MSN for 2 h and 4 h to observe the cellular uptake and localization of the nanoparticles by confocal microscopy. The STM infected cells treated with Arg-MSN for 2 h were fixed with paraformaldehyde and stained overnight with 50 μl of 1
50. Post infection, the cells were treated with FITC tagged Arg-MSN and FITC tagged amine-MSN for 2 h and 4 h to observe the cellular uptake and localization of the nanoparticles by confocal microscopy. The STM infected cells treated with Arg-MSN for 2 h were fixed with paraformaldehyde and stained overnight with 50 μl of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 diluted rabbit anti-mCAT 1 antibody (Santacruz) or rat anti-mouse LAMP1 IgG in 2% BSA (bovine serum albumin) and 0.2% saponin. The cells were washed twice with PBS and incubated with the secondary antibody goat anti-rat IgG conjugated to Cy3 or Alexa 647 (Jacksons Lab, 1
100 diluted rabbit anti-mCAT 1 antibody (Santacruz) or rat anti-mouse LAMP1 IgG in 2% BSA (bovine serum albumin) and 0.2% saponin. The cells were washed twice with PBS and incubated with the secondary antibody goat anti-rat IgG conjugated to Cy3 or Alexa 647 (Jacksons Lab, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200). The confocal microscopy images were acquired using a Zeiss LSM 710 (Carl Zeiss Microimaging Inc., Thornwood, USA) using a 63×, 1.4-NA oil immersion objective. Zen 2009 Light Edition software was used for processing and overlaying channels of the image.
200). The confocal microscopy images were acquired using a Zeiss LSM 710 (Carl Zeiss Microimaging Inc., Thornwood, USA) using a 63×, 1.4-NA oil immersion objective. Zen 2009 Light Edition software was used for processing and overlaying channels of the image.
Cellular endocytosis and trafficking studies of Arg-MSN
RAW 264.7 cells (2 × 105 per well) were incubated with 50 μg ml−1 of FITC tagged Arg-MSN for 2 h and cellular uptake inhibitors; amiloride (2 mM), chlorpromazine (25 μM), genistein (100 μM) and cytochalasin D (4 μM) were added to the cell culture media 1 h prior to addition of the FITC tagged particles. The percentage cell uptake was studied by flow cytometry by comparing relative percentage of cells showing FITC signal with Arg-MSN treatment with samples treated with both Arg-MSN and pharmacological inhibitors (BD FACSVerse). In vitro toxicity of the inhibitors on RAW 264.7 and HeLa cells was analyzed by flow cytometry determination of percentage of cells positive for propidium iodide (PI) stain.In vivo resolution of infection in Salmonella infected mice after Cip Arg-MSN treatment
In vivo experiments were approved by institutional animal ethics committee (registration no. 48/1999/CPCSEA) of Indian Institute of Science. The handling and experimentation on mice were carried out strictly as per the institutional guidelines. Salmonella burden was determined in liver, spleen and mesenteric lymph nodes (MLN) of BALB/c mice (n = 3) after oral infection (106 CFU per mice) and oral administration with Cip Arg-MSN and Cip at a concentration of 10 mg kg−1. The organs were aseptically isolated 5 days post infection and treatment, weighed and homogenized in PBS. The homogenate was plated on SS agar plates to determine the bacterial burden. The weight of the isolated spleens was measured to determine STM induced splenomegaly. For histopathological examination, liver and spleen isolated from infected mice after 3 days of antibiotic treatment were fixed in 10% formalin, embedded in paraffin, sectioned and stained by hematoxylin and eosin (H&E). The pathologist was blind to the identity of the pathology slides. Qualitative examination of inflammation in spleen and liver was assessed and reported.In vivo survival studies using Salmonella infected mouse model system
BALB/c mice (6–8 weeks) were orally infected with 108 STM CFU per mice and two treatment regimens for 3 day oral administration of ciprofloxacin at 10 mg per kg per day and Cip Arg-MSN with 10 and 5 mg per kg per day was initiated 12 h post infection. Infected mice treated with PBS and Arg-MSN alone was used as controls. Another set of survival studies was carried out to assess impact of 10 mg per kg per day Cip Arg-MSN and free Cip treatment on infected mice with advanced stage of Salmonella infection. The infected mice were administered antibiotics 5 days post infection when they show clinical signs of infection like hair loss and lethargy. The morbidity and mortality of both sets of mice were observed for 15 days post infection. The survival studies were carried out on mice aged above 52 weeks to assess the effect of Salmonella infection in geriatric populations upon treatment with Cip Arg-MSN. Treatment regimens with ciprofloxacin at 20 mg kg−1, 10 mg kg−1 and 5 mg kg−1 and Cip Arg-MSN with 10 and 5 mg kg−1 were tested on the aged mice and their survival checked for 15 days (n = 5). All the animal experiments were approved and carried out with due care in accordance with Institutional animal ethics committee regulations.Statistical analysis
Data is presented as means ± SEM. Data were analyzed using t-test analysis (Graphpad, Prism, USA). The difference between values for the treatments was considered to be statistically significant if p < 0.05 (* < 0.05, ** < 0.001, *** < 0.0001).Conclusions
Arginine decorated nanoparticles (Arg-MSN) were investigated as a possible drug carrier and targeting agent against intravacuolar infections using Salmonella as a model pathogen. The arginine nanocarriers were synthesized and characterized for size, surface area and drug loading. The particles exhibited pH dependent surface potential that may result in better efficacy as the particle encounters different pH environments in the cell. The particles were evaluated for cytotoxicity and hemolytic potential. The Arg-MSN was observed to be preferentially internalized by infected cells and co-localized with the intravacuolar pathogen showing its targeting capabilities. The slow release of the antibiotic coupled with intravacuolar targeting makes the Arg-MSN attractive carrier system. The Arg-MSN particles also exhibited co-localization with the cationic amino acid transporter (mCAT-1) and further evidenced by exhibiting receptor mediated cellular uptake mechanism. In vitro and in vivo studies with ciprofloxacin loaded Arg-MSN particles showed better activity than free drug alone. The nanodelivery system improved the antibacterial efficacy by 3–4 times thus allowing reduction in antibiotic dose requirement. The Arg-MSN system was found to enhance the host cell defense system by activating the iNOS pathway, producing reactive nitrogen species. The concerted action of improved antibiotic delivery, intracellular targeting and production of reactive NO aided in efficiently eliminating the intravacuolar pathogen. The consequence of such a coordinated anti-bacterial activity against the pathogen may lead to decreased probability of drug resistance and better therapeutic outcomes. This approach can be extended to other intravacuolar pathogens exhibiting such specific intracellular nutritional requirements.Funding sources
This work was supported by DAE SRC Outstanding Research Award (DAE0195) and DBT-IISc partnership program. Infrastructure support from ICMR (Center for Advanced Study in Molecular Medicine), DST (FIST), and UGC (special assistance) is acknowledged. This study was partly supported by the “Infosys Foundation”.Acknowledgements
Authors would like to thank the Dr Ramachandra and Ms Rosa Samuel from Central Animal Facility, Ms Pannaga, Ms Saima, and Ms Anusha from MCBL and Divisional confocal imaging facility, Ms Shravani from Advanced Facility for Microscopy and Microanalysis (AFMM), Department of Materials Engineering, Department of Microbiology and Cell Biology (MCBL) and the Centre for Biosystems Science and Engineering (BSSE). We thank the facility and help extended by Ms Leepika from Flow cytometry facility. Ms Rinsha and Sreenath are acknowledged for providing assistance with hemolysis study and confocal imaging respectively. We also acknowledge Dr Madhavi, consultant pathologist for assistance in interpreting histopathological sections. Financial assistance from DBT and LSRB is gratefully acknowledged.References
- R. L. Ochiai, C. J. Acosta, M. C. Danovaro-Holliday, D. Baiqing, S. K. Bhattacharya, M. D. Agtini, Z. A. Bhutta, D. G. Canh, M. Ali, S. Shin, J. Wain, A.-L. Page, M. J. Albert, J. Farrar, R. Abu-Elyazeed, T. Pang, L. Claudia, M. Galindo, V. Seidlein and J. Clemens, Bull. W. H. O., 2008, 86, 260–268 CrossRef PubMed.
- World Heal. Organ. Drug-Resistant Salmonella, http://www.who.int/mediacentre/factsheets/fs139/en/print.html, 2005, WHO website.
- K. Ray, B. Marteyn, P. J. Sansonetti and C. M. Tang, Nat. Rev. Microbiol., 2009, 7, 333–340 CrossRef CAS PubMed.
- S. Helaine, A. M. Cheverton, K. G. Watson, L. M. Faure, S. A. Matthews and D. W. Holden, Science, 2014, 343, 204–208 CrossRef CAS PubMed.
- A. Slattery, A. H. Victorsen, A. Brown, K. Hillman and G. J. Phillips, J. Bacteriol., 2013, 195, 647–657 CrossRef CAS.
- R. O. Darouiche and R. J. Hamill, Antimicrob. Agents Chemother., 1994, 38, 1059–1064 CrossRef CAS PubMed.
- S. M. Lehar, T. Pillow, M. Xu, L. Staben, K. K. Kajihara, R. Vandlen, L. DePalatis, H. Raab, W. L. Hazenbos, J. Hiroshi Morisaki, J. Kim, S. Park, M. Darwish, B.-C. Lee, H. Hernandez, K. M. Loyet, P. Lupardus, R. Fong, D. Yan, C. Chalouni, E. Luis, Y. Khalfin, E. Plise, J. Cheong, J. P. Lyssikatos, M. Strandh, K. Koefoed, P. S. Andersen, J. A. Flygare, M. Wah Tan, E. J. Brown and S. Mariathasan, Nature, 2015, 527, 323–328 CrossRef CAS PubMed.
- J. A. Crump and E. D. Mintz, Clin. Infect. Dis., 2010, 50, 241–246 CrossRef PubMed.
- T. T. Chau, J. I. Campbell, C. M. Galindo, N. Van Minh Hoang, T. S. Diep, T. T. T. Nga, N. Van Vinh Chau, P. Q. Tuan, A. L. Page, R. L. Ochiai, C. Schultsz, J. Wain, Z. A. Bhutta, C. M. Parry, S. K. Bhattacharya, S. Dutta, M. Agtini, B. Dong, Y. Honghui, D. D. Anh, D. G. Canh, A. Naheed, M. J. Albert, R. Phetsouvanh, P. N. Newton, B. Basnyat, A. Arjyal, T. T. P. La, N. N. Rang, L. T. Phuong, P. Van Be Bay, L. von Seidlein, G. Dougan, J. D. Clemens, H. Vinh, T. T. Hien, N. T. Chinh, C. J. Acosta, J. Farrar and C. Dolecek, Antimicrob. Agents Chemother., 2007, 51, 4315–4323 CrossRef PubMed.
- T. Butler, Clin. Microbiol. Infect., 2011, 17, 959–963 CrossRef CAS PubMed.
- H. Sarantis and S. Grinstein, Cell Host Microbe, 2012, 12, 419–431 CAS.
- E. P. Thi, U. Lambertz and N. E. Reiner, PLoS Pathog., 2012, 8, 1–4 Search PubMed.
- R. R. Isberg, T. J. O'Connor and M. Heidtman, Nat. Rev. Microbiol., 2009, 7, 13–24 CrossRef CAS PubMed.
- C. Manske and H. Hilbi, Front. Cell. Infect. Microbiol., 2014, 4, 1–10 Search PubMed.
- S. Ronneau, S. Moussa, T. Barbier, R. Conde-Álvarez, A. Zuniga-Ripa, I. Moriyon and J.-J. Letesson, Crit. Rev. Microbiol., 2015, 1–19 Search PubMed.
- S. Steele, J. Brunton, B. Ziehr, S. Taft-Benz, N. Moorman and T. Kawula, PLoS Pathog., 2013, 9, e1003562 CAS.
- I. Tattoli, M. T. Sorbara, D. Vuckovic, A. Ling, F. Soares, L. A. M. Carneiro, C. Yang, A. Emili, D. J. Philpott and S. E. Girardin, Cell Host Microbe, 2012, 11, 563–575 CAS.
- Y. Abu Kwaik and D. Bumann, Cell. Microbiol., 2013, 15, 882–890 CrossRef CAS PubMed.
- P. Das, A. Lahiri, A. Lahiri and D. Chakravortty, PLoS Pathog., 2010, 6, e1000899 Search PubMed.
- J. MacMicking, Q. W. Xie and C. Nathan, Annu. Rev. Immunol., 1997, 15, 323–350 CrossRef CAS PubMed.
- M. T. Talaue, V. Venketaraman, M. H. Hazbón, M. Peteroy-Kelly, A. Seth, R. Colangeli, D. Alland and N. D. Connell, J. Bacteriol., 2006, 188, 4830–4840 CrossRef CAS PubMed.
- M. A. Peteroy-Kelly, V. Venketaraman, M. Talaue, A. Seth and N. D. Connell, Infect. Immun., 2003, 71, 1011–1015 CrossRef CAS PubMed.
- P. Das, A. Lahiri, A. Lahiri, M. Sen, N. Iyer, N. Kapoor, K. N. Balaji and D. Chakravortty, PLoS One, 2010, 5, e15466 CAS.
- Y. M. Lvov, M. M. DeVilliers and R. F. Fakhrullin, Expert Opin. Drug Delivery, 2016, 13, 977–986 CrossRef CAS PubMed.
- M. R. Dzamukova, E. A. Naumenko, Y. M. Lvov and R. F. Fakhrullin, Sci. Rep., 2015, 5, 10560 CrossRef CAS PubMed.
- F.-X. Xiao, M. Pagliaro, Y.-J. Xu and B. Liu, Chem. Soc. Rev., 2016, 45, 3088–3121 RSC.
- G. Rydzek, Q. Ji, M. Li, P. Schaaf, J. P. Hill, F. Boulmedais and K. Ariga, Nano Today, 2015, 10, 138–167 CrossRef CAS.
- M. Nepal, S. Thangamani, M. N. Seleem and J. Chmielewski, Org. Biomol. Chem., 2015, 13, 5930–5936 CAS.
- J. Kuriakose, V. Hernandez-Gordillo, M. Nepal, A. Brezden, V. Pozzi, M. N. Seleem and J. Chmielewski, Angew. Chem., Int. Ed. Engl., 2013, 52, 9664–9667 CrossRef CAS PubMed.
- E. K. Lei, M. P. Pereira and S. O. Kelley, Angew. Chem., Int. Ed. Engl., 2013, 52, 9660–9663 CrossRef CAS PubMed.
- S. Boonyarattanakalin, J. Hu, S. A. Dykstra-Rummel, A. August and B. R. Peterson, J. Am. Chem. Soc., 2007, 129, 268–269 CrossRef CAS PubMed.
- B. J. Edagwa, D. Guo, P. Puligujja, H. Chen, J. McMillan, X. Liu, H. E. Gendelman and P. Narayanasamy, FASEB J., 2014, 28, 5071–5082 CrossRef CAS PubMed.
- D. L. Clemens, B.-Y. Lee, M. Xue, C. R. Thomas, H. Meng, D. Ferris, A. E. Nel, J. I. Zink and M. A. Horwitz, Antimicrob. Agents Chemother., 2012, 56, 2535–2545 CrossRef CAS PubMed.
- P. Narayanasamy, B. L. Switzer and B. E. Britigan, Sci. Rep., 2015, 5, 8824 CrossRef CAS PubMed.
- V. Kiruthika, S. Maya, M. K. Suresh, V. Anil Kumar, R. Jayakumar and R. Biswas, Colloids Surf., B, 2015, 127, 33–40 CrossRef CAS PubMed.
- N. M. Zaki and M. M. Hafez, AAPS PharmSciTech, 2012, 13, 411–421 CrossRef CAS PubMed.
- P. Jena, S. Mohanty, R. Mallick, B. Jacob and A. Sonawane, Int. J. Nanomed., 2012, 7, 1805–1818 CAS.
- D. P. Gnanadhas, M. Ben Thomas, M. Elango, A. M. Raichur and D. Chakravortty, J. Antimicrob. Chemother., 2013, 68, 2576–2586 CrossRef CAS PubMed.
- M. N. Seleem, P. Munusamy, A. Ranjan, H. Alqublan, G. Pickrell and N. Sriranganathan, Antimicrob. Agents Chemother., 2009, 53, 4270–4274 CrossRef CAS PubMed.
- K. Radhakrishnan, S. Gupta, D. P. Gnanadhas, P. C. Ramamurthy, D. Chakravortty and A. M. Raichur, Part. Part. Syst. Charact., 2013, 1–10 Search PubMed.
- E. Yamamoto and K. Kuroda, Bull. Chem. Soc. Jpn., 2016, 89, 501–539 CrossRef CAS.
- V. Malgras, Q. Ji, Y. Kamachi, T. Mori, F. K. Shieh, K. C. W. Wu, K. Ariga and Y. Yamauchi, Bull. Chem. Soc. Jpn., 2015, 88, 1171–1200 CrossRef CAS.
- M. Karimi, A. Ghasemi, P. Sahandi Zangabad, R. Rahighi, S. M. Moosavi Basri, H. Mirshekari, M. Amiri, Z. Shafaei Pishabad, A. Aslani, M. Bozorgomid, D. Ghosh, A. Beyzavi, A. Vaseghi, A. R. Aref, L. Haghani, S. Bahrami and M. R. Hamblin, Chem. Soc. Rev., 2016, 45, 1457–1501 RSC.
- Y. Lvov, G. Decher and H. Moehwald, Langmuir, 1993, 9, 481–486 CrossRef CAS.
- I. L. Radtchenko, G. B. Sukhorukov, S. Leporatti, G. B. Khomutov, E. Donath and H. Möhwald, J. Colloid Interface Sci., 2000, 230, 272–280 CrossRef CAS PubMed.
- H. Wanyika, Afr. J. Pharm. Pharmacol., 2011, 5, 2402–2410 CAS.
- M. Thommes, Chem.-Ing.-Tech., 2010, 82, 1059–1073 CrossRef CAS.
- D. Chakravortty, J. Exp. Med., 2002, 195, 1155–1166 CrossRef CAS PubMed.
- T. dos Santos, J. Varela, I. Lynch, A. Salvati and K. A. Dawson, PLoS One, 2011, 6, e24438 CAS.
- G. Romero, D. J. Sanz, Y. Qiu, D. Yu, Z. Mao, C. Gao and S. E. Moya, J. Mater. Chem. B, 2013, 1, 2252–2259 RSC.
- A. J. Griffin and S. J. McSorley, Mucosal Immunol., 2011, 4, 371–382 CrossRef CAS PubMed.
- K. G. Watson and D. W. Holden, Cell. Microbiol., 2010, 12, 1389–1397 CrossRef CAS PubMed.
- P. D. Barnes, M. A. Bergman, J. Mecsas and R. R. Isberg, J. Exp. Med., 2006, 203, 1591–1601 CrossRef CAS PubMed.
- J. D. Söderholm, Nat. Nanotechnol., 2015, 10, 298–299 CrossRef PubMed.
- J. J. Powell, E. Thomas-McKay, V. Thoree, J. Robertson, R. E. Hewitt, J. N. Skepper, A. Brown, J. C. Hernandez-Garrido, P. A. Midgley, I. Gomez-Morilla, G. W. Grime, K. J. Kirkby, N. A. Mabbott, D. S. Donaldson, I. R. Williams, D. Rios, S. E. Girardin, C. T. Haas, S. F. A. Bruggraber, J. D. Laman, Y. Tanriver, G. Lombardi, R. Lechler, R. P. H. Thompson and L. C. Pele, Nat. Nanotechnol., 2015, 10, 361–369 CrossRef CAS PubMed.
- O. Schulz and O. Pabst, Trends Immunol., 2013, 34, 155–161 CrossRef CAS PubMed.
- O. Balland, H. Pinto-Alphandary, S. Pecquet, A. Andremont and P. Couvreur, J. Antimicrob. Chemother., 1994, 33, 509–522 CrossRef CAS PubMed.
- O. Balland, H. Pinto-Alphandary, A. Viron, E. Puvion, A. Andremont and P. Couvreur, J. Antimicrob. Chemother., 1996, 37, 105–115 CrossRef CAS PubMed.
- E. Fattal, M. Youssef, P. Couvreur and A. Andremont, Antimicrob. Agents Chemother., 1989, 33, 1540 CrossRef CAS PubMed.
- J. Z. S. Chiu, I. G. Tucker, B. J. McLeod and A. McDowell, Eur. J. Pharm. Biopharm., 2015, 89, 48–55 CrossRef CAS PubMed.
- J.-H. Yeom, B. Lee, D. Kim, J. Lee, S. Kim, J. Bae, Y. Park and K. Lee, Biomaterials, 2016, 104, 43–51 CrossRef CAS PubMed.
- M. E. Page-Clisson, H. Pinto-Alphandary, E. Chachaty, P. Couvreur and A. Andremont, Pharm. Res., 1998, 15, 544–549 CrossRef CAS.
- Y. Zhang and M. Hensel, Nanoscale, 2013, 5, 9296 RSC.
- I. Chico-Calero, M. Suárez, B. González-Zorn, M. Scortti, J. Slaghuis, W. Goebel and J. A. Vázquez-Boland, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 431–436 CrossRef CAS PubMed.
- H. Marquis, H. G. Bouwer, D. J. Hinrichs and D. A. Portnoy, Infect. Immun., 1993, 61, 3756–3760 CAS.
- J. Stritzker, J. Janda, C. Schoen, M. Taupp, S. Pilgrim, I. Gentschev, P. Schreier, G. Geginat and W. Goebel, Infect. Immun., 2004, 72, 5622–5629 CrossRef CAS PubMed.
- A. Cersini, A. M. Salvia and M. L. Bernardini, Infect. Immun., 1998, 66, 549–557 CAS.
- A. Cersini, M. C. Martino, I. Martini, G. Rossi and M. L. Bernardini, Infect. Immun., 2003, 71, 7002–7013 CrossRef CAS PubMed.
- F. R. Noriega, G. Losonsky, C. Lauderbaugh, F. M. Liao, J. Y. Wang and M. M. Levine, Infect. Immun., 1996, 64, 3055–3061 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1–S5. See DOI: 10.1039/c6ra27868j |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2017 |