Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Combined biomaterial signals stimulate communications between bone marrow stromal cell and endothelial cell
Yachen Xu,
Zhi Wu,
Xin Dong and
Haiyan Li *
*
Med-X Research Institute, School of Biomedical Engineering, Shanghai Jiao Tong University, 1954 Huashan Road, Shanghai 200030, China. E-mail: haiyan.li@sjtu.edu.cn
First published on 17th January 2017
Abstract
It has been widely reported that chemical, structural or mechanical signals of biomaterials can impact cell behaviors and tissue regeneration, but few studies have investigated the effects of biomaterial signals on cell–cell interactions although communications between cells are critical for tissue regeneration. Our recent studies have shown that chemical signals of bioglass (BG) can stimulate communications in bone marrow stromal cells and endothelial cells, which results in enhanced angiogenesis and osteogenesis. Considering the facts that, in vivo, different biomaterial signals may simultaneously affect cell–cell interactions, in this study, we proposed that combining chemical and structural signals of biomaterials may further improve cell–cell interactions. Results proved that combined structural signals of aligned electrospun nanofibers and chemical signals of BG ionic products could significantly stimulate interactions between co-cultured bone marrow stromal cells and endothelial cells through both of paracrine effects and junctional communications as compared to single type of biomaterial signals. Further study indicated that both chemical signals of BG and structural signals of electrospun nanofibers played important role in stimulating paracrine effects while for improving junctional communication, structural signals of electrospun nanofibers played a more important role than chemical signals of BG, which resulted in enhanced vascularization and osteogenic differentiation in co-cultures. Therefore, applying combined biomaterial signals to activate cell–cell interactions is a promising strategy for enhancing tissue regeneration.
1. Introduction
Numerous studies have reported that biomaterials could impact cell behaviors and tissue regeneration through either their chemical signals, such as ionic products, trace elements and bioactive molecules,1–3 or their structural signals, such as various designed topography of biomaterials,4,5 or their mechanical signals, such as stiffness and mechanical forces of biomaterials.6,7 For example, chemical signals of bioglass (BG), mainly ionic products, have been widely reported to be able to promote osteogenic differentiation of bone marrow stromal cells (BMSCs) and angiogenic differentiation of endothelial cells (ECs).8,9 In addition, a number of studies have demonstrated that the morphology and alignment of electrospun nanofibers have profound effects on morphology and behaviors of BMSCs and ECs, which can stimulate osteogenesis and angiogenesis.10–12 However, most studies only investigated the effects of single type of biomaterial signals on behaviors of single type of cells.Recently, more and more researchers start to realize the fact that tissue regeneration normally involves different types of cells and there are interactions between these different types of cells, which can enhance tissue regeneration.13–15 For example, it has been reported that interactions between BMSCs and ECs can promote vascularization and osteogenesis.16,17 In addition to the effects of biomaterial signals on behaviors of single type of cells, our previous studies demonstrated that chemical signal of biomaterials could influence the communications between different cells, which eventually resulted in promoted tissue regeneration.1,13,14 For an instance, we found that silicate bioactive materials could enhance the cell–cell communications between BMSCs and ECs through stimulating paracrine effects between these two types of cells.14 Chemical signals from ionic products of silicate biomaterials upregulated the vascular endothelial growth factor (VEGF) expression from co-cultured BMSCs. Then, the secreted VEGF activated its receptor KDR in co-cultured ECs and downstream angiogenic factors, which subsequently promoted vascularization. Meanwhile, the activated KDR stimulated the expression of bone morphogenetic protein 2 (BMP-2) in co-cultured ECs, and eventually stimulated the process of osteogenesis. This study suggested that chemical signals of biomaterials could stimulate interactions between different types of cells, which finally enhance tissue regeneration.
However, so far, there are only studies on the effects of chemical signals of biomaterial on cell–cell communication. The effects of structural signals and mechanical signals of biomaterials on cell–cell communication have not been carried out although the effects of these two types of biomaterial signals on behaviors of single type of cells have been widely reported.18–20 In addition, inspired by nature, researchers realized that, during tissue repair or regeneration process, different signals of biomaterials actually affect cell behaviors and tissue regeneration simultaneously. Thus, studying the effects of combined different biomaterial signals on cell–cell behaviors and tissue regeneration is also critical for designing biomaterials with optimized properties in addition to the effects of single type of biomaterial signals on cell–cell communications. The fundamental question to be answered is if the combined biomaterial signals have better stimulatory effects on cell–cell interactions and what specific role of each type of biomaterial signal plays on affecting cell–cell interactions when they are applied together.
Therefore, in the present study, we aimed to investigate the effects of combined biomaterial signals on the communications between BMSCs and ECs as well as the effects of structural signals of biomaterial on cell–cell communications. Specifically, a direct contact co-culture of human bone marrow stromal cells (HBMSCs) and human umbilical vein endothelial cells (HUVECs) was used as the co-culture cell model. Aligned electrospun nanofibers and BG ionic products were used as structural and chemical stimulatory signals of biomaterials, respectively. We focused on investigating the effects of single and combined biomaterials signals on communications between HBMSCs and HUVECs through paracrine effects, gap junction and adherens junction. Our results demonstrated that combined chemical signals of BG ionic products and structural signals of electrospun nanofibers can stimulate paracrine effects and junctional communications as compared to single type of biomaterial signals. After the communications between HBMSCs and HUVECs were stimulated, the vascularization of co-cultured HUVECs and osteogenic differentiation of co-cultured HBMSCs were both enhanced.
2. Materials and methods
2.1. Electrospun nanofibrous scaffolds
The mixture of poly(D,L-lactide) (PDLLA, Mw = 45 kDa) (Jinan Daigang Biomaterial, China) and polycaprolactone (PCL, Mw = 80 kDa) (Sigma, USA) with a mass ratio of 50/50 was dissolved in hexafluoroisopropanol (HFIP) (Aladdin, China) at the concentration of 4.8% (w/v), and then stirred for 6 h to obtain a homogeneous solution. The feed rate of the solution was 0.02 ml m−1 by using a syringe pump (Baoding Longer Precision Pump LSP01-1A, China), and the applied voltage was 8 kV. The distance between the tip of needle and the collector was 15 cm, and the collecting time was 1 h. To prepare electrospun scaffolds with aligned nanofibers, high-speed roller (rotating speed = 2000 rpm) was used as collectors. All the experiments were conducted at room temperature and the relative humidity was about 40–60%. All the electrospun scaffolds were vacuum dried for 24 h to completely remove any residual solvent.Microstructure of the electrospun scaffolds with aligned nanofibers were observed by a scanning electron microscope (SEM, S-4800, Hitachi, Japan). Surface wettability of the scaffolds were evaluated by measuring the static water contacting angles using a Kruss GmbH DSA 100 Mk 2 goniometer (Hamburg, Germany) equipped with a DataPhysics OCA20 CA system. Electrospun scaffolds were cut into squares with dimensions of 10 mm × 10 mm and 25 × 25 mm for 24-well plates and 6-well plates, respectively. The obtained scaffold squares were sterilized after being soaked in 75% alcohol for 20 min for further cell culture.
2.2. BG ion extracts
BG powders were provided by Shanghai Institute of Ceramics, Chinese Academy of Science. BG ion extracts were prepared according to the methods reported in literatures adapted from ISO10993-1 procedures.21,22 Briefly, 1 g of BG powders was soaked in 5 ml of serum-free endothelial culture medium (ECM) (Sciencell, USA) and mesenchymal stem cell culture medium (MSCM) (Cyagen, China), respectively. After being incubated for 24 h in a humidified 37 °C/5% CO2 incubator, the supernatant was then collected and sterilized through a filter (Millipore, 0.22 μm). For further use, BG ion extracts were diluted with total ECM (endothelial cell basal medium + 5% fetal bovine serum (FBS) + 1% endothelial cell growth supplement + 1% penicillin–streptomycin (P/S)) and total MSCM (MSCM + 10% FBS + 1% P/S + 1% L-glutamine) at ratios of 1/128, respectively. The concentrations of Ca, Si and P in the diluted ion extracts were detected by inductively coupled plasma atomic emission spectroscopy (ICP-AES; Optima 3000DV, PerkinElmer, USA).2.3. Cell isolation and culture
Human umbilical vein endothelial cells (HUVECs) were isolated from human umbilical cord veins according to the method reported previously,23 and human bone marrow stromal cells (HBMSCs) were purchased from Cyagen Co. Ltd (Guangzhou, China). The use of human umbilical cord veins was approved by the donor, and an Institutional Review Committee of Shanghai Jiao Tong University, School of Biomedical Engineering approved all these protocols. Total ECM was used as HUVECs' culture medium, while total MSCM was used as HBMSCs' culture medium. The culture medium was replaced every 3 days, and only early passages (passages 2–7) of the HUVECs and HBMSCs were used in this study.2.4. Effects of combined biomaterial signals on HBMSC–HUVEC co-cultures
The BG ion extracts diluted at 1/128 (recorded as 1/128BG) and electrospun scaffolds with aligned nanofibers were used to determine the effects of BG's chemical signals and nanofibers' structural signals on HUVECs and HBMSCs. Total ECM and total MSCM without BG ion extracts were used as control medium for HUVECs and HBMSCs, respectively. Cells seeded on coverslips were regarded as non-structure groups.The HBMSC–HUVEC direct contact co-culture model was established according to previous study.14 HBMSCs were first seeded on coverslips and electrospun scaffolds with aligned nanofibers at a density of 8 × 104 cells per well in 24-well plates and 2 × 105 cells per well in 6-well plates, respectively. After cultured with control medium for 12 h, HUVECs were seeded on HBMSCs at a density of 1.2 × 105 cells per well in 24-well plates and 3 × 105 cells per well in 6-well plates, respectively. Meanwhile, the culture medium was replaced by BG ion extracts diluted with mixed total MSCM and total ECM (v/v = 1/1) at 1/128. The mixed medium of total MSCM and total ECM (v/v = 1/1) was considered as control medium.
2.5. Quantitative real-time polymerase chain reaction (Q-RT-PCR)
After being cultured for 3 days, cells in 6-well plates were washed using cold phosphate buffered saline (PBS) and HUVECs were separated from HBMSCs in co-cultures. To separate HUVECs from HBMSCs, co-cultured cells in 6-well plates were washed with PBS and collected by trypsinization, and then magnetic beads coupled with an antibody against CD31 (Invitrogen, USA) and Magnetic Separation Rack (Invitrogen, USA) were used according to the method established by Guillotin et al.24 The separated HUVECs and HBMSCs were named co-HUVEC and co-HBMSC, respectively.For RNA extraction, an E. Z. N. A total RNA kit I (OMEGA, Bio-tek, USA) was used according to the instructions. The concentration of RNA was measured by a Nanodrop 1000 reader (Thermo Scientific, USA) and cDNA was synthesized using a ReverTra Ace-α kit (Toyobo, Japan) according to the instructions. cDNA was diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 with sterilized deionized water. And then 4.2 μl of diluted cDNA was mixed with 5.8 μl of SYBR-Green (ToYOBO, Japan) and primers (Sangon Biotech, China). Primers of connexin 43 (Cx43), neural-cadherin (N-cad), vascular endothelial cadherin (VE-cad), VEGF, VEGF receptor 2 (KDR), endothelial nitric oxide synthase (eNOS), alkaline phosphatase (ALP) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used at a final concentration of 400 nM. GAPDH was used as a housekeeping gene. The sequences for primers are listed in Table 1. The mixture was finally loaded in a 384-well plate and analyzed by 7900 Real-time PCR system (Applied Biosystems, USA), which performed 40 cycles (95 °C for 15 s, 60 °C for 15 s, 72 °C for 45 s) followed by a 1 min denaturation at 95 °C. Each reaction was performed in triplicate, and data were analyzed by ΔΔCt method. The data were then normalized to GAPDH gene expression of each condition and compared to the corresponding gene expression in control samples.
20 with sterilized deionized water. And then 4.2 μl of diluted cDNA was mixed with 5.8 μl of SYBR-Green (ToYOBO, Japan) and primers (Sangon Biotech, China). Primers of connexin 43 (Cx43), neural-cadherin (N-cad), vascular endothelial cadherin (VE-cad), VEGF, VEGF receptor 2 (KDR), endothelial nitric oxide synthase (eNOS), alkaline phosphatase (ALP) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used at a final concentration of 400 nM. GAPDH was used as a housekeeping gene. The sequences for primers are listed in Table 1. The mixture was finally loaded in a 384-well plate and analyzed by 7900 Real-time PCR system (Applied Biosystems, USA), which performed 40 cycles (95 °C for 15 s, 60 °C for 15 s, 72 °C for 45 s) followed by a 1 min denaturation at 95 °C. Each reaction was performed in triplicate, and data were analyzed by ΔΔCt method. The data were then normalized to GAPDH gene expression of each condition and compared to the corresponding gene expression in control samples.
| Gene | Gene bank | Primer sequences | Tm (°C) |
|---|---|---|---|
| GAPDH | NM_002046 | F: 5′-GATTTGGTCGTATTGGGCG-3′ | 60 |
| R: 5′-CTGGAAGATGGTGATGG-3′ | |||
| Connexin 43 | NC_000017.11 | F: 5′-GGA GGG AAG GTG TGG CTG TC-3′ | 62 |
| R: 5′-GGC AGG GCT CAG CGC ACC AC-3′ | |||
| N-cadherin | NC_000007.14 | F: 5′-GGT GGA GGA GAA GAA GAC CAG-3′ | 58 |
| R: 5′-GGC ATC AGG CTC CAC AGT-3′ | |||
| VE-cadherin | NC_000002.12 | F: 5′-ATGTTCGTGCAGCTGTTTACCA-3′ | 58 |
| R: 5′-TCTGTGACACAGTGGCCATAGG-3′ | |||
| VEGF165 | AB_021221 | F: 5′-TGCGGATCAAACCTCACCA-3′ | 58 |
| R: 5′-CAGGGATTTTTCTTGTCTTGCT-3′ | |||
| KDR | NM_002253 | F: 5′-GTGATCGGAAATGACACTGGAG-3′ | 60 |
| R: 5′-CATGTTGGTCACTAACAGAAGCA-3′ | |||
| eNOS | NM_001160111.1 | F: 5′-TGTCCAACATGCTGCTGGAAATTG-3′ | 55 |
| R: 5′-AGGAGGTCTTCTTCCTGGTGATGCC-3′ | |||
| ALP | NC_000018.10 | F: 5′-AGCCCTTCACTGCCATCCTGT-3′ | 58 |
| R: 5′-ATTCTCTCGTTCACCGCCCAC-3′ |
2.6. Live/dead staining
To evaluate cell viability to BG and electrospun nanofibrous scaffolds, a live/dead viability cytotoxicity kit (Invitrogen, USA) was applied to co-cultures after being cultured for 3 days according to the supplier's procedure. The living co-cultured cells arranged on the surfaces of coverslips and electrospun scaffolds with aligned nanofibers were observed and photographed using a CCD camera (Leica DFC 420C) equipped with a fluorescence microscope (Leica DM2500 M).2.7. Immunofluorescence staining
Immunofluorescence staining of von Willebrand factor (vWF) was applied on co-cultures to observe the distribution of HUVECs and tubule formation. Immunofluorescence staining of Cx43, N-cad, VE-cad and KDR were applied on co-cultures to detect the location and expression of Cx43, N-cad, VE-cad and KDR.After being co-cultured for 3 days, the cells co-cultured on electrospun scaffolds with aligned nanofibers and coverslips in 24-well plates were washed twice with PBS and fixed with 4% (w/v) paraformaldehyde (PFA) (Dingguo Chemical Reagent Co. Ltd, China) at room temperature for 15 min. And then, the co-cultured cells were permeabilized with methanol for 5 min and blocked with PBS containing 1% (w/v) bovine serum albumin (BSA) for 1 h at 37 °C. Next, primary antibody solution containing rabbit anti-vWF, rabbit anti-Cx43, mouse anti-N-cad, mouse anti VE-cad or mouse anti-KDR antibody was added to the co-cultures and incubated at 37 °C for 2 h, all these antibodies were purchased from Abcam Co, Ltd (USA) and diluted in PBS–0.5% BSA at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. After being incubated overnight at 4 °C, the co-cultures were washed twice with PBS and Alexa 488 goat anti-mouse IgG (Invitrogen, USA) or Alexa 488 goat anti-rabbit IgG (Invitrogen, USA) secondary antibody (diluted in PBS–0.5% BSA at 1/1000) was used to incubate the cells at 37 °C for 1 h. Finally, 1 μg ml−1 4-6-diamidino-2-phenylindole (DAPI) (Invitrogen, USA) was added to co-cultures for 10 min at room temperature to reveal nuclei. After staining, the electrospun scaffolds or coverslips with cells were removed and mounted on slides. The stained co-cultures were observed with a confocal microscope (Leica TCS SP5, Germany) and images were taken by a CCD camera (Leica DFC 420C, Germany).
100. After being incubated overnight at 4 °C, the co-cultures were washed twice with PBS and Alexa 488 goat anti-mouse IgG (Invitrogen, USA) or Alexa 488 goat anti-rabbit IgG (Invitrogen, USA) secondary antibody (diluted in PBS–0.5% BSA at 1/1000) was used to incubate the cells at 37 °C for 1 h. Finally, 1 μg ml−1 4-6-diamidino-2-phenylindole (DAPI) (Invitrogen, USA) was added to co-cultures for 10 min at room temperature to reveal nuclei. After staining, the electrospun scaffolds or coverslips with cells were removed and mounted on slides. The stained co-cultures were observed with a confocal microscope (Leica TCS SP5, Germany) and images were taken by a CCD camera (Leica DFC 420C, Germany).
2.8. ALP staining assay
To observe the osteogenic differentiation of mono-cultured and co-cultured cells, ALP staining was applied after being cultured for 3 days. Cells were washed twice with PBS and fixed by 4% PFA for 15 min at room temperature. And then the cells were incubated in the Naphthol Fastblue ready-to-use solution (Naphthol/Fastblue, 166 μl/4 ml, Sigma) in dark at 37 °C for 40 min according to the manufacturer's instruction. After incubation, cells were washed with PBS and photographed by digital camera.2.9. NO staining assay
To determine the NO synthesis in direct contact co-cultures, diaminofluorescein-2 (DAF-2) (Sekisui Medical, USA) was using as a fluorescent indicator to reveal the NO staining. Briefly, DAF-2 was dissolved in dimethyl sulfoxide to obtain 1 mM stock solutions. After being co-cultured for 3 days, the cells were washed twice with PBS and incubated at 37 °C for 1 h in PBS containing 10 μM DAF-2. After staining, the co-cultures were observed and photographed by a fluorescence microscope (Leica DMI 3000B, Germany) equipped a CCD camera (Leica DFC 420C, Germany).2.10. Statistical analysis
Data were expressed as means ± standard deviation. Three independent experiments were carried out for validity, and at least three samples per test were taken for statistical analysis. Statistical significance between groups was calculated using two-tailed analysis of variance (ANOVA) and performed with a Student's t test program. The differences were considered significant when p < 0.05 (*, #, % or Δ) or p < 0.01 (**, ##, %% or ΔΔ).3. Results
3.1. Characterization of electrospun nanofibers and BG ion extracts
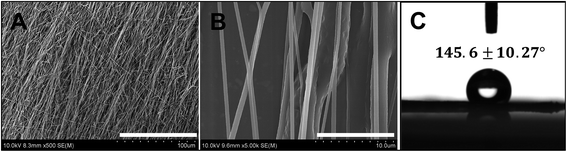
Fig. 1A and B show SEM images of electrospun scaffold with aligned nanofibers. It can be seen that the nanofibers are uniform in diameter, arranged regularly and oriented directionally. Fig. 1C shows that the electrospun scaffold with aligned nanofibers is hydrophobic, and the water contact angle is 145.6 ± 10.27°. The concentrations of ions in BG ionic products were detected and are shown in Table 2. There is little difference between the concentrations of Ca ion in different medium. However, the concentrations of Si and P ions were significantly increased when BG ionic products were added no matter in ECM or MSCM. In particular, the concentrations of Si increased almost 3 times in ECM, as the data detected were 0.41 ± 0.04 μg ml−1 in control ECM but 1.14 ± 0.07 μg ml−1 in BG containing ECM. While the concentrations of Si increased more than 3 times in MSCM, as the data detected were 0.27 ± 0.03 μg ml−1 in control MSCM but 0.94 ± 0.11 μg ml−1 in BG containing MSCM.| Ca (μg ml−1) | P (μg ml−1) | Si (μg ml−1) | |
|---|---|---|---|
| a p < 0.01 compared with data of the same ion in ECM + 5% FBS.b p < 0.01 compared with data of the same ion in MSCM + 10% FBS. | |||
| ECM + 10% FBS | 61.70 ± 0.21 | 21.7 ± 0.28 | 0.41 ± 0.04 |
| BG 1/128 in ECM | 61.39 ± 0.17 | 23.81 ± 0.12a | 1.14 ± 0.07a |
| MSCM + 10% FBS | 73.39 ± 0.57 | 35.88 ± 0.14 | 0.27 ± 0.03 |
| BG 1/128 in MSCM | 73.35 ± 0.41 | 37.68 ± 0.18b | 0.94 ± 0.11b |
3.2. Effects of combined biomaterial signals on cell viability and capillary tube formation in HBMSC–HUVEC co-culture
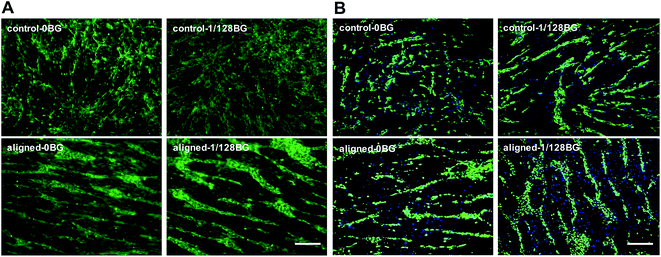
In order to investigate the effects of combined biomaterial signals on cell viability and capillary tube formation, live/dead staining and vWF staining were performed after HBMSCs and HUVECs were co-cultured with combined structural signals of aligned electrospun nanofibers and chemical signals of BG ionic products for 3 days, respectively, and the results are shown in Fig. 2. The green fluorescence in Fig. 2A is derived from living HBMSCs and HUVECs, while the green fluorescence in Fig. 2B is only derived from HUVECs. As shown in the images of live/dead staining, both of HBMSCs and HUVECs cultured under different conditions are attached and grew well. It can be seen that the cells cultured with combined structural signals of aligned nanofibers and chemical signals of 1/128BG showed the longest and densest tube formation of HUVECs in co-cultures as compared to the single structural or chemical signal. The structural signals of electrospun nanofibers could obviously influence cell distribution, as the cells cultured on coverslips with non-structure formed a random and reticular structure, while the cells cultured on electrospun scaffolds with aligned nanofibers revealed an obvious stratification, where the lower cells directionally covered the surface of the scaffolds along the electrospun nanofibers, and the upper cells formed a tube-like network. The vWF staining further confirmed that the tube-like network was contributed by HUVECs, indicating the initial capillary tube formation. In addition to structural signals of aligned electrospun nanofibers, chemical signals of BG also stimulated the tube-like network formation of HUVECs to some degree, as the 1/128BG groups showed longer and denser tubes than 0BG groups.3.3. Effects of combined biomaterial signals on vascular growth factor expression in HBMSC–HUVEC co-culture
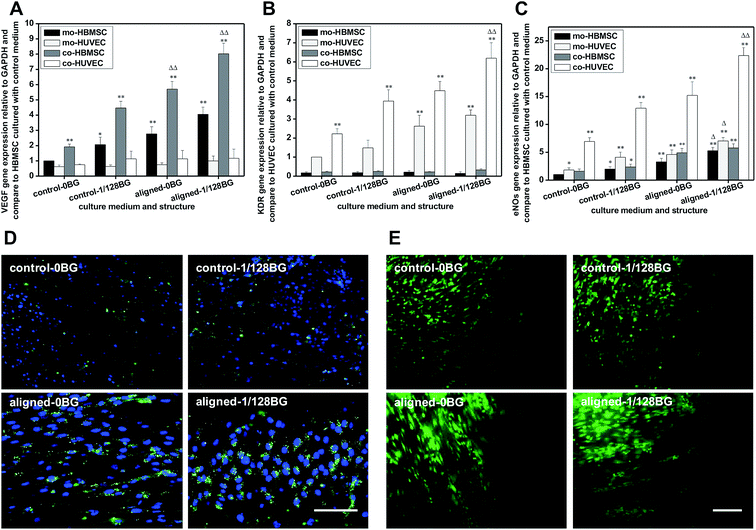
In order to investigate the effects of combined structural and chemical signals of biomaterials on the vascularization in co-cultures through paracrine effects between HBMSCs and HUVECs, the related expression of angiogenetic genes VEGF, KDR and eNOS were measured and the results are shown in Fig. 3. It can be seen from Fig. 3A, as compared to control group, aligned nanofibers with medium containing 1/128 BG resulted in a 4 time-increase of the expression of VEGF in co-HBMSC. In contrast, when cells were cultured with different structural signals but same chemical signals, as compared to control group, electrospun scaffolds with aligned nanofibers resulted in about 2.5 times increase of the VEGF expression in co-HBMSC. Meanwhile, when cells were cultured with same structural signals, the chemical signal of 1/128BG stimulated the expression of VEGF in co-HBMSC for about 2 times. In addition to VEGF, aligned nanofibers with medium containing 1/128 BG also resulted in 3 times for KDR and 4 times for eNOS expression in co-HUVEC as compared to control group, respectively (Fig. 3B and C). In contrast, when cells were cultured with different structural signals but same chemical signals, as compared to control group, electrospun scaffolds with aligned nanofibers significantly increased the expression of KDR and eNOS in co-HUVEC for about 2.5 times. Meanwhile, when cells were cultured with same structural signals, 1/128BG significantly upregulated the expression of KDR and eNOS in co-HUVEC for about 2 times. Based on the above results, it can be concluded the combined structural signals of aligned electrospun nanofibers and chemical signals of BG ionic products have an improved stimulatory effect on paracrine effects between HBMSCs and HUVECs as compared to single structural signals or chemical signals of biomaterials, which further stimulated the vascularization in HBMSC–HUVEC co-cultures. Besides, these results also demonstrated that, not only the chemical signals of biomaterials but also the structural signals of biomaterials have stimulatory effects on paracrine effects between HBMSCs and HUVECs.3.4. Effects of combined biomaterial signals on osteogenic differentiation in HBMSC–HUVEC co-culture
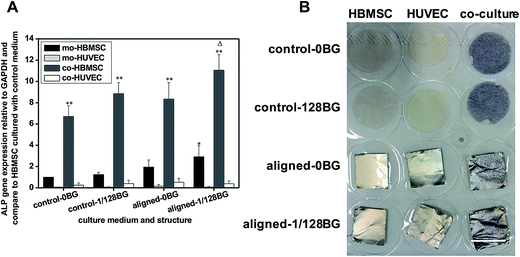
In order to find out the effects of combined biomaterial signals on osteogenic differentiation in HBMSC–HUVEC co-cultures, the expression of osteogenic gene ALP was further analyzed, and the results are shown in Fig. 4. It can be seen that, the combined structural signals of aligned electrospun nanofibers and chemical signals of BG ionic products significantly increased the expression of ALP in co-HBMSC for about 2 times as compared to control, while the single structural signals and chemical signals only improved the expression of ALP in co-HBMSC for about 1.2–1.5 times, which indicated an improved stimulatory effect of combined biomaterial signals on osteogenesis in HBMSC–HUVEC co-cultures as compared to single structural or chemical signals.3.5. Effects of combined biomaterial signals on junctional communications in HBMSC–HUVEC co-culture
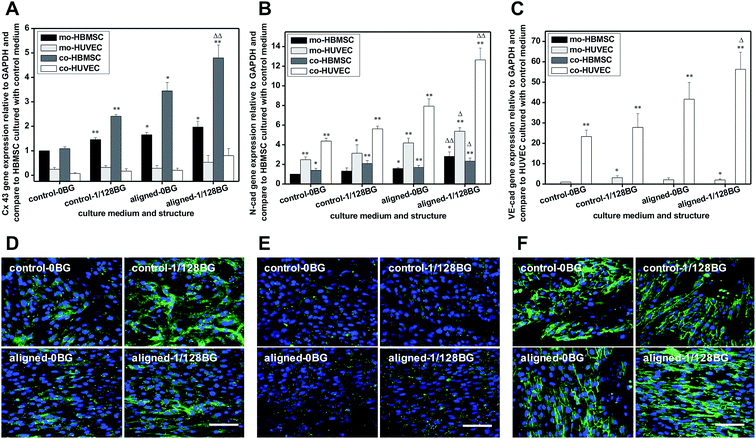
To confirm the effects of combined chemical and structural signals of biomaterials on cell–cell interactions through junctional communication, we further analyzed the expression of a gap junction protein of Cx43 as well as the adherens junction proteins of N-cad and VE-cad in HBMSC–HUVEC co-cultures. Fig. 5 shows the expression of Cx43, N-cad and VE-cad in HBMSC–HUVEC co-cultures. When the cells were stimulated with combined structural and chemical signals, as compared to the control group, the expression of Cx43 was upregulated for about 4.5 times in co-HBMSC, while the expression of N-cad was increased for about 2.5 times in co-HBMSC and about 3 times in co-HUVEC. The expression of VE-cad was upregulated for about 2.5 times in co-HUVEC. In contrast, when the cells were cultured with different structural signals but same chemical signals, as compared to the control group, electrospun scaffolds with aligned nanofibers significantly upregulated the gene expression of Cx43 for about 3.5 times in co-HBMSC, the gene expression of N-cad was increased for about 2 times in co-HBMSC and 2.5 times in co-HUVEC, and the gene expression of VE-cad was upregulated for about 2 times in co-HUVEC. In addition, when cells were cultured with same structural signals, 1/128BG medium increased the expression of Cx43 for about 2.5 times in co-HBMSC as compared to 0BG, the expression of N-cad was increased for about 1.5 times in co-HBMSC and about 1.5 times in co-HUVEC. Meanwhile, the expression of VE-cad was increased for 1.5 times. The results indicated that the combined structural and chemical signals possessed a stronger stimulatory effects on junctional cell–cell communication as compared to the single stimulation of structural or chemical signal. In addition, as compared to the stimulatory effects of structural signals and those of chemical signals, the structural signals of aligned electrospun nanofibers showed higher stimulatory effects on the junctional cell–cell communication through Cx43, N-cad and VE-cad.4. Discussion
In the present study, we used aligned electrospun nanofibers and BG ionic products as structural and chemical stimulatory signals of biomaterials, respectively, and studied the effects of the combination of the two signals on cell–cell communications in a direct contact co-cultures of HBMSCs and HUVECs model. The results demonstrated that combined structural signals of aligned electrospun nanofibers and chemical signals of BG ionic products can significantly stimulate communications between HBMSCs and HUVECs, which results in improved vascularization of co-HUVECs and osteogenic differentiation of co-HBMSCs.It has been widely reported that paracrine effects, gap junctions and adherens junction play important roles in cell–cell interactions between BMSCs and ECs.24–26 During communications between BMSCs and ECs through paracrine effects, the co-cultured BMSCs normally upregulated the expression of vascular growth factors to activate the receptors of growth factors in co-cultured ECs and initiated the vascularization. Reciprocally, the activated co-cultured ECs released osteogenic growth factors to stimulate the osteogenic differentiation of co-cultured BMSCs.26–28 In this study, the same paracrine effects between co-HBMSCs and co-HUVECs were found when the cells were co-cultured with control medium, single type of biomaterial signals as well as combined biomaterial signals. As compared to control group, both of single structural signals of electrospun nanofibers and chemical signals of BG ionic products could stimulate the paracrine effects between HBMSCs and HUVECs, and the stimulatory effects of chemical signals of BG were similar to those of structural signals of electrospun nanofiber. Interestingly, combined structural signals of electrospun nanofibers and chemical signals of BG ionic products show stronger stimulatory effects on the paracrine effects than either chemical signals of BG or structural signals of electrospun nanofibers. We reason that the structural signals of aligned electrospun nanofibers and chemical signals of BG ionic products may influence cell–cell communications in different ways as there is not only paracrine effects but also junctional communication between the two types of cells.
Gap junction communications have been known to allow metabolic cooperation between adjacent cells and control cell behaviors.29,30 Cx43 has been reported as an important gap junction molecular and there are intracellular channels in Cx43 that provide direct cytoplasmic connections between apposed cells,29 which allows the passage of ions and small molecules. Adherens junction also plays an important role in the communication between BMSCs and ECs,31–33 among which N-cad and VE-cad are major ones that can adhere cells together and stimulate functional differentiation of cells.34–36 Cx43 is expressed in both BMSCs and ECs and these two types of cells can talk to each other through the gap junctional channel constituted by Cx43, which results in stimulation of osteoblastic differentiation.24,37,38 In addition, co-cultures of BMSCs and ECs could increase the expression of N-cad and VE-cad, which can subsequently upregulate the early osteoblastic differentiation of BMSCs and stimulate vascularization of ECs.31,33 In this study, both structural signals of aligned electrospun nanofibers and chemical signals of BG ionic products showed stimulatory effects on gap junctional and adherens junctional communications. Interestingly, in contrast to the similar effects of two types of biomaterial signals on paracrine effects between HBMSCs and HUVECs, structural signals of aligned electrospun nanofibers played much higher stimulatory effects on gap junction and adherens junctions than chemical signals of BG ionic products, which indicated that structural signals stimulated cell–cell communications mainly through junctional pathways. When these two signals were combined, the stimulatory effects were significantly improved. We reason that it is aligned electrospun nanofibers which makes cells attach and grow directionally, and the oriented regularity in distribution of cells subsequently brings out more contacts between different cells, which finally results in enhanced junctional communications. Different from the contact dependence of junctional communications, paracrine effect is achieved through diffusible factors, which does not require direct contact between cells. Therefore, chemical signals of BG ionic products and structural signals of electrospun nanofibers has its specific role in cell–cell communications. The chemical signals of BG ionic products mainly stimulate cell–cell communications through diffusible factors, as the ions released from BG can participate the cell metabolism directly,39 while the structural signals of aligned electrospun nanofibers mainly stimulate cell–cell interactions through junctional communications, as the aligned nanofibers can remodel the skeletons and nuclei of cells and guide cells along the direction of nanofibers,40,41 which results in enhanced cell–cell contacts and adhesion and subsequently improved the junctional communications.
5. Conclusion
In the current study, we proved that the combined signals of structural signals of aligned electrospun nanofibers and chemical signals of BG ionic products could significantly enhance interactions between HBMSCs and HUVECs as compared to single structural signals of aligned electrospun nanofibers or chemical signals of BG ionic products. Both paracrine effects and junctional communications of gap junction of Cx43 as well as adherens junctions of N-cad and VE-cad were stimulated by the combined signals, which results in enhanced vascularization and osteogenic differentiation. In addition, for different communication ways, different biomaterial signals show different stimulatory effects. The chemical signals of BG ionic products have similar stimulatory effects as structural signals of electrospun nanofibers for paracrine effects while structural signals of electrospun nanofibers shows higher stimulatory effects for junctional communications as compared to chemical signals of BG ionic products. These results suggest that it is feasible to stimulate cell–cell interactions with different stimulatory biomaterial signals. Furthermore, combining of co-cultures of different cells and combined signals of different biomaterials may be a potential complex for enhancing tissue regeneration.Conflict of interest
The authors declare that all of the experiments comply with the current laws of China, and an Institutional Review Committee of Shanghai Jiao Tong University, School of Biomedical Engineering approved all experimental protocols.Acknowledgements
This work was supported by Natural Science Foundation of China (Grant No. 81190132 and 31470918), the Innovation Program of Shanghai Municipal Education Commission (Grant No. 14ZZ032), the National Key Research and Development Program of China (Grant No. 2016YFC1100201) and the 2016 “SMC-Chenxing” Talent Program of Shanghai Jiao Tong University.References
- H. Li and J. Chang, Acta Biomater., 2013, 9, 5379 CrossRef CAS PubMed
.
- N. Kong, K. Lin, H. Li and J. Chang, J. Mater. Chem. B, 2014, 2, 1100 RSC
.
- W. Ji, Y. Sun, F. Yang, J. J. van den Beucken, M. Fan, Z. Chen and J. A. Jansen, Pharm. Res., 2011, 28, 1259 CrossRef CAS PubMed
.
- M. R. Williamson, R. Black and C. Kielty, Biomaterials, 2006, 27, 3608 CrossRef CAS PubMed
.
- J. S. Choi, S. J. Lee, G. J. Christ, A. Atala and J. J. Yoo, Biomaterials, 2008, 29, 2899 CrossRef CAS PubMed
.
- S. Chien, S. Li and Y. J. Shyy, Macromol. Chem. Phys., 1998, 31, 162 CAS
.
- X. Wang, J. Mater. Chem. B, 2015, 4, 37 RSC
.
- L.-C. Gerhardt and A. R. Boccaccini, Materials, 2010, 3, 3867 CrossRef CAS
.
- A. A. Gorustovich, J. A. Roether and A. R. Boccaccini, Tissue Eng., Part B, 2010, 16, 199 CrossRef CAS PubMed
.
- A. Doustgani, E. Vasheghani-Farahani and M. Soleimani, Nanomed. J., 2013, 1, 20 CAS
.
- C. Xu, Biomaterials, 2004, 25, 877 CrossRef CAS PubMed
.
- H. Xu, H. Li, Q. Ke and J. Chang, ACS Appl. Mater. Interfaces, 2015, 7, 8706 CAS
.
- H. Li and J. Chang, Acta Biomater., 2013, 9, 6981 CrossRef CAS PubMed
.
- H. Li, K. Xue, N. Kong, K. Liu and J. Chang, Biomaterials, 2014, 35, 3803 CrossRef CAS PubMed
.
- S. J. Bidarra, C. C. Barrias, M. A. Barbosa, R. Soares, J. Amedee and P. L. Granja, Stem Cell Res., 2011, 7, 186 CrossRef PubMed
.
- S. Fuchs, A. Hofmann and C. Kirkpatrick, Tissue Eng., 2007, 13, 2577 CrossRef CAS PubMed
.
- H. Yu, P. J. Vandevord, L. Mao, H. W. Matthew, P. H. Wooley and S. Y. Yang, Biomaterials, 2008, 30, 508 CrossRef PubMed
.
- B. Kim, X. Zhang, H. Borteh, Z. Li, J. Guan and Y. Zhao, J. Micromech. Microeng., 2012, 22, 085001 CrossRef
.
- D. E. Anderson and M. T. Hinds, Biochem. Biophys. Res. Commun., 2012, 427, 159 CrossRef CAS PubMed
.
- C. L. Ives, S. G. Eskin and L. V. Mcintire, In Vitro Cell. Dev. Biol., 1986, 22, 500 CAS
.
- H. Li and J. Chang, Biomaterials, 2004, 25, 5473 CrossRef CAS PubMed
.
- C. Wu, C. Jiang, W. Zhai, S. Ni and J. Wang, J. Biomed. Mater. Res., Part B, 2006, 78, 47 CrossRef PubMed
.
- L. Bordenave, C. Baquey, R. Bareille, F. Lefebvre, C. Lauroua, V. Guerin, F. Rouais, N. More, C. Vergnes and J. M. Anderson, J. Biomed. Mater. Res., 1993, 27, 1367 CrossRef CAS PubMed
.
- B. Guillotin, R. Bareille, C. Bourget, L. Bordenave and J. Amédée, Bone, 2008, 42, 1080 CrossRef CAS PubMed
.
- M. Grellier, L. Bordenave and J. Amedee, Trends Biotechnol., 2009, 27, 562 CrossRef CAS PubMed
.
- F. Villars, B. Guillotin, T. Amédée, S. Dutoya, L. Bordenave, R. Bareille and J. Amédée, Am. J. Physiol.: Cell Physiol., 2002, 282, C775 CrossRef CAS PubMed
.
- H. Li, R. Daculsi, M. Grellier, R. Bareille, C. Bourget, M. Remy and J. Amedee, PLoS One, 2012, 6, 575 Search PubMed
.
- H. Li, R. Daculsi, M. Grellier, R. Bareille, C. Bourget and J. Amedee, Am. J. Physiol.: Cell Physiol., 2010, 299, 422 CrossRef PubMed
.
- H. A. Dbouk, R. M. Mroue, M. E. El-Sabban and R. S. Talhouk, Cell Commun. Signaling, 2009, 7, 1 CrossRef PubMed
.
- J. P. Stains and R. Civitelli, Biochim. Biophys. Acta, 2006, 1719, 69 CrossRef PubMed
.
- S. L. Ferrari, K. Traianedes, M. Thorne, M. H. Lafage-Proust, P. Genever, M. G. Cecchini, V. Behar, A. Bisello, M. Chorev and M. Rosenblatt, J. Bone Miner. Res., 2000, 15, 198 CrossRef CAS PubMed
.
- E. Haÿ, J. Lemonnier, D. Modrowski, A. Lomri, F. Lasmoles and P. J. Marie, J. Cell. Physiol., 2000, 183, 117 CrossRef
.
- P. J. Marie, J. Cell. Physiol., 2002, 190, 297 CrossRef CAS PubMed
.
- P. Carmeliet and D. Collen, Ann. N. Y. Acad. Sci., 2000, 902, 249–264 CrossRef CAS PubMed
.
- J. S. Alexer, O. W. Blaschuk and F. R. Haselton, J. Cell. Physiol., 1993, 156, 610 CrossRef PubMed
.
- A. Aplin, A. Howe, S. k. Alahari and R. Juliano, Pharmacol. Rev., 1998, 50, 197 CAS
.
- P. J. Bouletreau, S. M. Warren, J. A. Spector, Z. M. Peled, R. P. Gerrets, J. A. Greenwald and M. T. Longaker, Plast. Reconstr. Surg., 2002, 109, 2384 CrossRef PubMed
.
- D. S. Wang, M. Miura, H. Demura and K. Sato, Endocrinology, 1997, 138, 2953 CAS
.
- I. D. Xynos, A. J. Edgar, L. D. Buttery, L. L. Hench and J. M. Polak, Biochem. Biophys. Res. Commun., 2000, 276, 461 CrossRef CAS PubMed
.
- R. L. Juliano and S. Haskill, J. Cell Biol., 1993, 120, 577 CrossRef CAS PubMed
.
- M. J. Dalby, M. O. Riehle, S. J. Yarwood, C. D. Wilkinson and A. S. Curtis, Exp. Cell Res., 2003, 284, 272 CrossRef
.
| This journal is © The Royal Society of Chemistry 2017 |