Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ultrasensitive and in situ DNA detection in various pH environments based on a microfiber with a graphene oxide linking layer
Yunyun Huang ,
Bo Yu,
Tuan Guo and
Bai-Ou Guan*
,
Bo Yu,
Tuan Guo and
Bai-Ou Guan*
Guangdong Provincial Key Laboratory of Optical Fiber Sensing and Communications, Institute of Photonics Technology, Jinan University, Guangzhou 210632, China. E-mail: tguanbo@jnu.edu.cn
First published on 24th February 2017
Abstract
Ultrasensitive and in situ DNA detection at different pH values, ranging from 4.3 to 8.5, based on a microfiber with a graphene oxide linking layer was proposed and experimentally demonstrated. The graphene oxide coating over the silica microfiber provides a strong π–π interaction with the detected single-stranded DNA molecules, and induces an amplified surface refractive index modulation over the fiber, and finally leads to a wavelength shift of the optical interference fringe for interrogation. Using this strategy, in situ DNA measurement with a detection limit of up to 10−12 M and a linear response in the pH range from 4.3–8.5 has been achieved. Benefiting from its compact size, high sensitivity, and ease of use, together with remote operational ability, the proposed sensor opens up a multitude of opportunities for quantifying DNA in various hard-to-reach environments. It may supplement the existing DNA detection tools.
1 Introduction
The DNA-based diagnostic test is currently an area of tremendous interest since more and more research has proved that the mutation of genes is responsible for numerous inherited disorders.1–3 Therefore, rapid and simple determination of specific DNA in human, viral and bacterial nucleic acids with low concentration bio-samples using detection methods with low cost, high sensitivity and good selectivity is highly demanded.4 As the gold-standard for DNA detection, the classical polymerase chain reaction (PCR) encouraged the study of DNA detection techniques.5 However, it suffers from involving complex reactions and is hard to apply in vivo or even in situ. Nowadays, many techniques including fluorescence,6 electrochemiluminescence,7 electrochemistry,8 and quartz crystal microbalance9 have been developed for DNA detection. In these methods, labels are usually necessary. However, label-free techniques are highly desired in analysis because they remove the possible effects of labels on the target molecules.10Meanwhile, pH is the main characteristic of biological fluids.11 It plays a critical role in the physiological and pathological processes.12 Therefore, as an essential parameter for cell, enzyme and tissue activities,13 pH is of great concern to all life forms. For the reason that pH gradient differences widely exist in the human body,14 it is well-known that the applicability of in situ DNA biosensors depends strongly upon their stability and sensing activity in solutions at physiological pH values.15 The sensors that are able to show excellent sensing activity at pH values over the important physiological range of 5.0–7.4 will be great candidates for use in biological systems.15 Unfortunately, most current DNA sensors only work in a neutral pH environment.10,16,17 Hence, developing a DNA sensor with a high stability and sensing ability over a broad pH range is of great value in medicine and biochemical research.
Graphene and graphene oxide (GO) are very promising materials for biosensors due to their large specific surface area, low-cost fabrication, and direct interaction with a wide range of biomolecules.18–22 Moreover, they possess high stability in stringent conditions.15 GO possesses both sp2- and sp3-hybridized carbon atoms as well as different functional groups such as epoxy, carboxyl, hydroxyl, and so on.13–16 Biomolecules can be immobilized on its surface through the π-stacking interaction,23–25 or through covalent conjugation between the carboxyl groups of GO and the amino groups of these molecules.26–29 Selective biosensing of single-stranded DNA (ssDNA) based on GO has been demonstrated.30,31 It is found that ssDNA adsorbs strongly on GO, while duplex DNA (dsDNA) cannot bind to it stably.32 It is widely employed in the selective detection of ssDNA in a mixed solution (including dsDNA). Silica fiber is a cost-effective, flexible, and widely available material.33,34 Taking advantage of the miniature size and mechanical flexibility, the optical fiber allows integration with current medical tools for sensing in inaccessible locations. And it could realize the measurement of biological molecules through fiber surface functionalization. The authors have previously demonstrated a silica microfiber biosensor detecting a ssDNA target at pH = 7 based on the surface modification of a conjugated polymer.10 However, the polymer could not bear a stringent pH environment and cannot be applied in acidic or basic solutions.
Herein, a highly sensitive biosensor for in situ detection of label-free ssDNA targets at various pH values of 4.3, 5.5, 7.0, 7.5 and 8.5 based on a silica microfiber is presented. GO thin film was utilized as the linking layer for ssDNA adsorption. Using GO as a catcher, the selectivity is realized by the selective interaction between GO and ssDNA, while the sensitivity is obtained through the surface aggregation of ssDNA on the microfiber. Moreover, the adaptation of the GO layer to a broad pH range adapts the biosensor to the physiological environment. This sensing method has potential applications for the in situ ultrasensitive detection of ssDNA in the human body, supplementing the existing DNA detection tools.
2 Materials and methods
2.1 Evanescent wave microfiber tapered interferometer
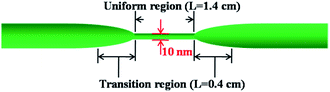
The silica fiber used in our work to fabricate the microfiber was supplied by CorActive High-Tech Inc. (UVS-INT-PREMIUM, 100536). The microfiber structure fabrication followed the procedure described previously.35 Briefly, the optical fiber was tapered down to a micron-scale diameter using a flame-heated drawing technique. The flame with a width of 5 mm scanned across the fiber once, accompanied by slowly stretching it with two linear stages. A microfiber with a uniform region where the diameter and length were 10 μm and 1.4 cm, respectively, was fabricated (Fig. 1). The transition region was 0.4 cm in length. Coupling and recombination of modes in the microfiber are allowed by this geometry. Entering into the tapered down region, the fundamental core mode excites a fundamental one and a higher order one. Mode beating is mainly between the HE11 and the HE12 modes because they have similar azimuthal symmetry and the smallest phase mismatch, although more than two modes may be excited. Normally, the external RI sensitivity of the interferometer can be expressed by formula (1).34
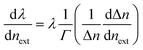 | (1) |
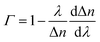 is the dispersion factor, which characterizes the effect of the variation of the index difference with wavelength. When the external RI increases, the effective indexes of both the fundamental and higher-order modes rise accordingly. The higher-order mode has a higher change rate due to its larger energy fraction evanescent field. As the index difference between the two modes decreases with the external RI, the term in the bracket is also negative. As a result, the transmission dips red-shift with increasing RI. On the other side, both the dispersion parameter Γ and dΔn/dnext are strongly dependent on the silica fiber diameter. Using thinner microfibers, the sensitivity can be greatly enhanced due to the stronger evanescent-field interaction and reduced dispersion factor.
is the dispersion factor, which characterizes the effect of the variation of the index difference with wavelength. When the external RI increases, the effective indexes of both the fundamental and higher-order modes rise accordingly. The higher-order mode has a higher change rate due to its larger energy fraction evanescent field. As the index difference between the two modes decreases with the external RI, the term in the bracket is also negative. As a result, the transmission dips red-shift with increasing RI. On the other side, both the dispersion parameter Γ and dΔn/dnext are strongly dependent on the silica fiber diameter. Using thinner microfibers, the sensitivity can be greatly enhanced due to the stronger evanescent-field interaction and reduced dispersion factor.
2.2 Reagents and apparatus
All chemicals and solvents supplied by Aladdin were of analytical grade and were used without further purification. The GO was prepared by oxidizing graphite using a modified Hummer’s method.20 The carboxylated GO (G-COOH) was synthesized via the method presented in the work of Huang et al.36 The DNA oligonucleotides (ssDNA 5′ > CA TCA ATG TAT CTT ATC ATG TCT GGA < 3′) were synthesized and purified by Sangon Inc. (Shanghai, China). NaOH and HCl were used to adjust the pH value of the DNA solution. The DNA solution was diluted to concentrations of 10−6 M down to 10−18 M, respectively.2.3 Characterization
The surface morphology of the GO coated microfiber was observed using a scanning electron microscope (SEM, ULTRA 55, ZEISS, BRUKER). The morphology of the GO was examined using transmission wavelength spectra (TEM, JEM-2010HR TEM). The laser confocal microscope photos were taken using a laser confocal microscope (LSM 510 META).2.4 Immobilization of GO membrane onto optical fiber sensor surface
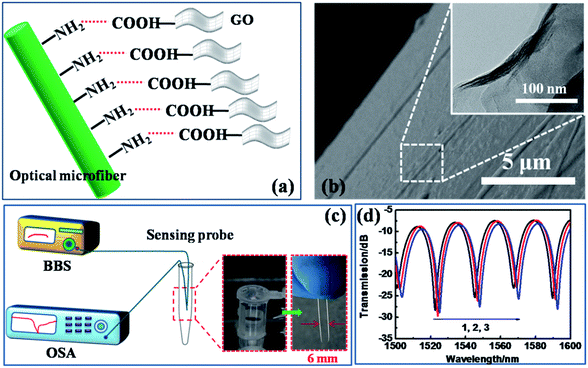
The functionalization of the optical microfiber by GO was performed as shown in Fig. 2(a). The optical fiber was cleaned for 30 min in a bath with a piranha solution consisting of 30% H2O2 and concentrated H2SO4 at a volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 to generate reactive hydroxyl groups, then in a 5% solution of (3-aminopropyl)triethoxysilane (APTES) in acetone for 30 min to generate amino groups. The surface amino modified fiber was then immersed in the G-COOH dispersion containing N-3-dimethyl-aminopropyl-N′-ethylcarbodimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) at a mass ratio of 2
3 to generate reactive hydroxyl groups, then in a 5% solution of (3-aminopropyl)triethoxysilane (APTES) in acetone for 30 min to generate amino groups. The surface amino modified fiber was then immersed in the G-COOH dispersion containing N-3-dimethyl-aminopropyl-N′-ethylcarbodimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) at a mass ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The pH value of the solution was adjusted using hydrochloric acid to 4.5 and the temperature was kept at 25 °C.36 Then, the GO functionalized microfiber was pulled out of the solution, washed, and dried under vacuum.
1. The pH value of the solution was adjusted using hydrochloric acid to 4.5 and the temperature was kept at 25 °C.36 Then, the GO functionalized microfiber was pulled out of the solution, washed, and dried under vacuum.
2.5 Experimental setup and optical configuration
The experimental setup permitted the sensor to operate in the transmission mode. The sensing microfiber was excited using a broadband source (BBS) emitting light over the wavelength range of 1250–1650 nm and its interference spectra were monitored using an optical spectrum analyzer (OSA) with a minimum wavelength resolution of 0.02 nm.3 Results and discussion
3.1 Surface functionalization of the silica microfiber
The surface functionalization scheme of the microfiber is illustrated in Fig. 2(a). The carboxylated GO with carboxyl groups bound on to the amino group-modified silica fiber surface. The SEM image in Fig. 2(b) identifies that the GO has been successfully deposited on the fiber surface, and the inset presents the further zoomed-in surface drape, imaged by TEM.36 The transmission of the microfiber interferometer was recorded using the experimental setup illustrated in Fig. 2(c). The sensing region was connected to the OSA and a BBS emitter with a normal silica optical fiber. Due to the miniaturized size of the fiber probe, the detected sample solution volume could be less than 2 ml, paving the way to in situ and even in vivo detection. The deposition process of GO on the fiber surface is shown in Fig. 2(d). The gradual deposition induces a slight red-shift of the wavelength due to the increasing RI of the fiber surface (before coating: surface RI = 1.33 (bare fiber in water); after coating: surface RI = 1.36 (fiber coated with GO)). This result testifies the successful GO functionalization over the fiber surface.3.2 Environmental stability of the sensor
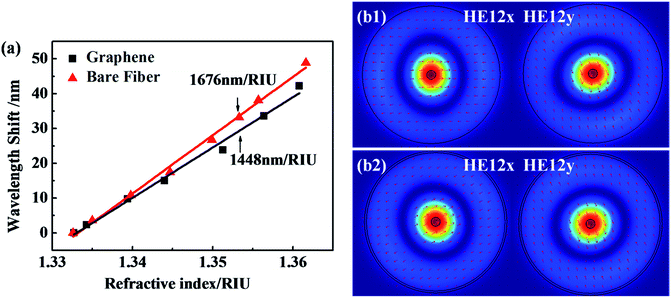
Fig. 3 shows the sensitivity of the GO functionalized fiber with the RI ranging from 1.33 to 1.36, which covers the RI modulation range of the DNA solutions. A bare optical fiber-based interferometer has an estimated sensitivity of 1676 nm per RIU. After GO coating, the fiber presents a RI sensitivity of 1448 nm per RIU. Fig. 3(b) presents the composition of the constituent modes analyzed by numerical mode simulation (COMSOL) software. The HE12 mode shows a similar evanescent field over the fiber surface after coating (see Fig. 3(b)). Therefore, after GO coating on the fiber surface, the sensor shows a similar evanescent-field interaction with the surrounding medium,37,38 presenting good environmental stability.3.3 Mechanism of the DNA sensor’s selectivity
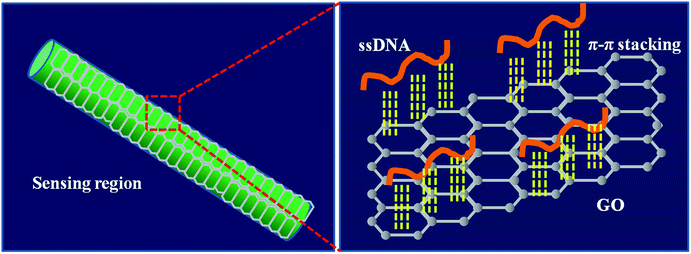
The schematic of GO catching the ssDNA over the fiber surface is shown in Fig. 4. In the DNA solution, ssDNA molecules were in the dissociated state, resulting in a uniform concentration throughout the solution. However, after GO coating on the fiber surface, the ssDNA selectively attached onto the GO surface through π–π interactions, while dsDNA freely stayed in solution.32,39,40 This catching process resulted in the gathering of ssDNA on the fiber surface, amplifying the RI change of the fiber surface.3.4 Sensitivity of the DNA sensor
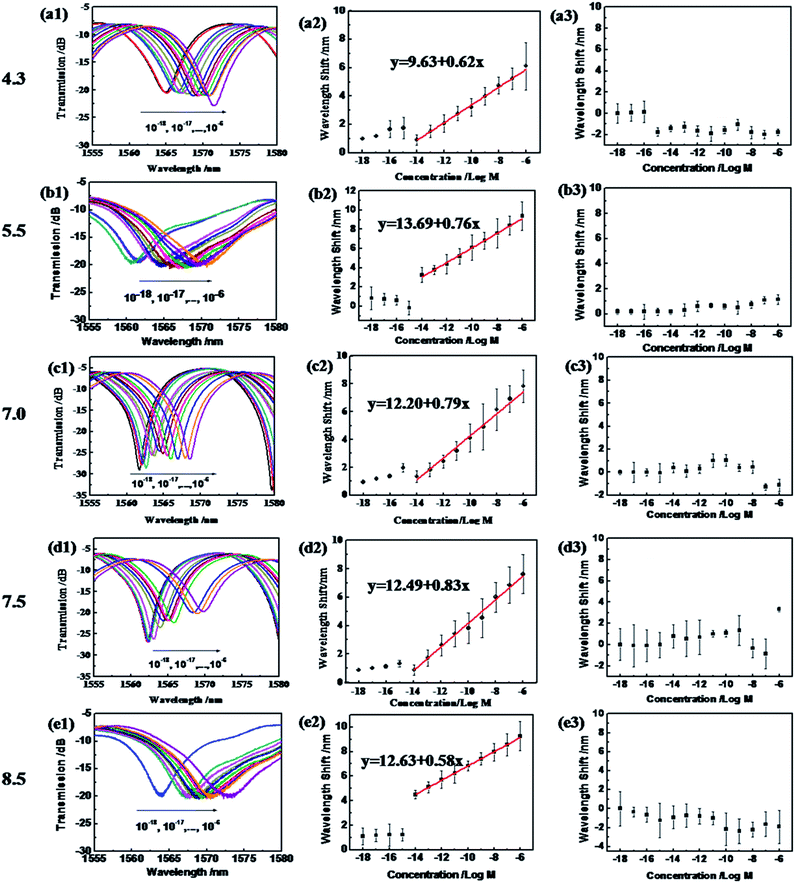
The as-prepared sensor shows a ssDNA detection ability from pH 4.3–8.5, as shown in Fig. 5. In the blank solution at pH 4.3, the GO coated microfiber showed a transmission peak centered at 1564 nm. When transported into the ssDNA solution, with a concentration from 10−18 to 10−15 M, it presented a very slight shift accordingly (hard to distinguish using the OSA with a resolution of 0.02 nm). However, when the ssDNA concentration increased to 10−14 M, an obvious red-shift of 0.93 nm was recorded, and is shown in Fig. 5(a1). With the ssDNA concentration increasing from 10−12 to 10−6 M, the sensor showed a regular red-shift in transmission wavelength (Fig. 5(b1)). The relationship between the wavelength and the ssDNA concentration is almost linear, and a linear fitting of the data gives| pH 4.3: Δλ = 9.63 + 0.62x | (2) |
It is revealed that the sensitivity of the ssDNA sensor is 0.94 nm/log M, and the limit of detection (LOD) is 4.84 × 10−12 M at pH 4.3 (according to evaluation work of the LOD by White et al.41). Meanwhile, at pH 5.5, 7.0, 7.5 and 8.5, similar linear dependencies appeared between the wavelength and the ssDNA concentration, which linear fitting of the data gives as:
| pH 5.5: Δλ = 13.69 + 0.76x | (3) |
| pH 7.0: Δλ = 12.20 + 0.79x | (4) |
| pH 7.5: Δλ = 12.49 + 0.83x | (5) |
| pH 8.5: Δλ = 12.63 + 0.58x | (6) |
The sensitivity of the ssDNA sensor at pH 5.5, 7.0, 7.5 and 8.5 is 0.76 nm/log M, 0.79 nm/log M, 0.83 nm/log M and 0.58 nm/log M, respectively. And the LOD in these four pH environments reaches 3.95 × 10−12 M, 3.80 × 10−12 M, 3.61 × 10−12 M, and 5.17 × 10−12 M, which is much lower than those reported in our previous work: 10−10 M using a conjugated polymer as the DNA catcher10 and 5 × 10−7 M based on the reflective microfiber Bragg grating,16 and Pollet’s work achieving 2 × 10−6 M based on fiber optic surface plasmon resonance biosensing.42 Moreover, the detection in this work can be performed across a wide pH range, from 4.3 to 8.5, which covers the pH range of bodily fluids (except gastric juice) for in situ DNA detection.
However, when employing the naked silica optical fiber without the GO coating for ssDNA concentration detection in the same pH range, the wavelength shifts appeared irregular, as shown in Fig. 5(a3–e3). Therefore, this proves the special role of the GO coating in the sensitivity and selectivity of the sensor versus ssDNA.
When the detection is finished, the GO coating could be removed by a piranha solution in 30 min and can be re-coated by immersing for 30 min. After that, the biosensor can be applied in a new DNA detection process, showing the reusability of the fiber.
3.5 Wide pH adaptation of the DNA sensor
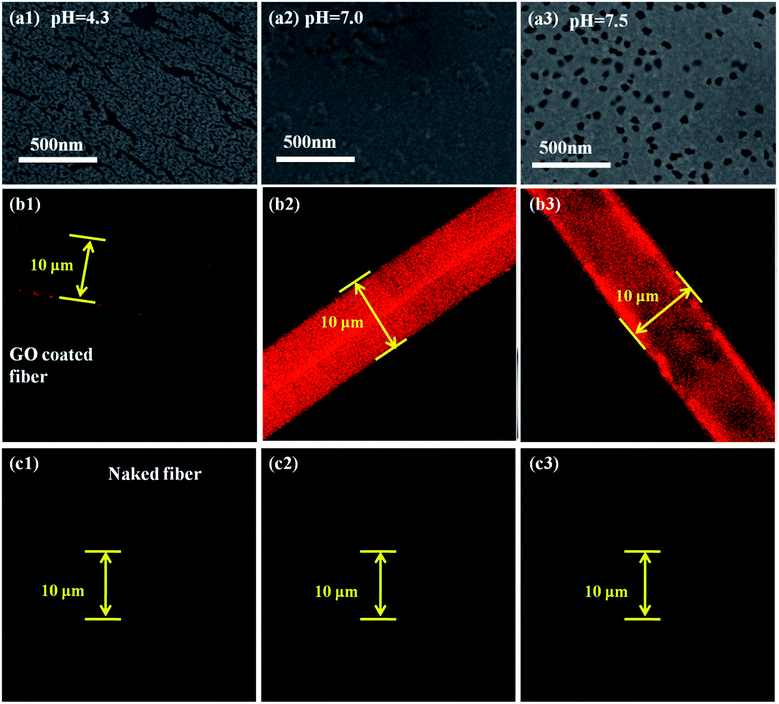
To reveal why the as-prepared DNA sensor has a wide pH adaptation, SEM images and laser confocal images of the as-prepared DNA sensor with fluorescent ssDNA at pH = 4.3, 7.0 and 7.5 are shown in Fig. 6. It is reported that the pH-dependent adsorption may be due to the different degrees of hydrogen-bonding interaction and electrostatic attraction between the two species under different pH conditions.43 At pH 4.3, about 50–75% of the –COOH groups are deprotonated to produce –COO−.44 And the not-deprotonated –COOH might induce the GO to slightly aggregate as photographed in Fig. 6(a1). The slight aggregation of the GO resulted in the uneven attachment of DNA, as shown in Fig. 6(b1). However, the slight aggregation did not greatly affect the ability to catch DNA. Therefore, the DNA sensor operated well in this acidic environment. And in neutral conditions, 85% of the –COOH groups were deprotonated to produce –COO−, and this resulted in a basically even GO surface on the fiber, as shown in Fig. 6(a2). Under the effects of the hydrogen bonds and the electrostatic attraction between the GO and ssDNA,45 the maximum amount of ssDNA gathered on the fiber surface, as shown in Fig. 6(b2). And in the weak basic condition (pH 7.5), most of the –COOH groups were deprotonated to produce –COO−. And the electrostatic repulsion between the negatively charged GO sheets46 kept it uniform on the fiber surface (Fig. 6(a3)), resulting in a large quantity of ssDNA adsorption (Fig. 6(b3)). In the case of the naked optical fiber without the GO coating, no ssDNA attachment occurs in all pH environments. This also explains the irregularity in ssDNA molecule detection using the naked silica fiber. It explains the broad pH adaptation of our GO coated DNA sensor, while a lot of other sensitive membranes such as conjugated polymers10 could not bear stringent environments.4 Conclusions
Based on the strong π–π interaction between ssDNA and GO, the RI change signal of ssDNA on the tapered fiber surface is amplified. Depending on the pH adaptation of GO, the LOD of the DNA sensor reaches 10−12 M and it presents a good linear response over a wide pH range, from 4.3–8.5. The wide pH range adaptation and high sensitivity of the proposed GO functionalized microfiber biosensor provides the potential ability to quantify ssDNA in situ and even possibly in vivo.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 51403077), the National Science Fund for Distinguished Young Scholars of China (No. 61225023), the Research Fund for the Doctoral Program of Higher Education (No. 20114401110006), the Guangdong Natural Science Foundation (No. S2013030013302, 2014A030313387), the Youth Science and Technology Innovation Talents of Guangdong (No. 2014TQ01X539), and Fundamental Research Funds for the Central Universities (No. 21615446). And we acknowledge Otsuka Electronics Co., Ltd for the measurement of the surface RI of the GO conjugated fiber.References
- M. O. Malley and R. G. Hutcheon, J. Am. Med. Dir. Assoc., 2007, 8, 332–334 CrossRef PubMed.
- J. V. Borsel and J. A. Tetnowski, Journal of Fluency Disorders, 2007, 32, 279–296 CrossRef PubMed.
- C. Chen, W. T. D. Zhai, D. Lu, H. B. Zhang and W. G. Zheng, Mater. Res. Bull., 2011, 46, 583–587 CrossRef CAS.
- F. Lucarelli, G. Marrazza, A. P. F. Turner and M. Mascini, Biosens. Bioelectron., 2004, 19, 515–530 CrossRef CAS PubMed.
- R. K. Saiki, S. Scharf, F. Faloona, K. B. Mullis, G. T. Horn, H. A. Erlich and N. Arnheim, Science, 1985, 230, 1350–1354 CAS.
- R. Yang, J. Jin, Y. Chen, N. Shao, H. Kang, Z. Xiao, Z. Tang, Y. Wu, Z. Zhu and W. Tan, J. Am. Chem. Soc., 2008, 130, 8351–8358 CrossRef CAS PubMed.
- X. Y. Wang, P. G. He and Y. Z. Fang, J. Lumin., 2010, 130, 1481–1484 CrossRef CAS.
- H. X. Chang, Y. Shi and N. L. Guan, Anal. Chem., 2007, 79, 5000–5115 CrossRef PubMed.
- Y. H. Fei, X. Y. Jin, Z. S. Wu, S. B. Zhang, G. L. Shen and R. Q. Yu, Anal. Chim. Acta, 2011, 691, 95–102 CrossRef CAS PubMed.
- Y. Huang, Z. Tian, L. Sun, D. Sun, J. Li, Y. Ran and B. Guan, Opt. Express, 2015, 23, 26962–26968 CrossRef PubMed.
- G. Kaur and S. K. Tripathi, Mater. Chem. Phys., 2014, 143, 514–523 CrossRef CAS.
- C. Ding and Y. Tian, Biosens. Bioelectron., 2015, 65, 183–190 CrossRef CAS PubMed.
- Z. Wu, M. Gao, T. Wang, X. Wan, L. Zheng and C. A. Huang, Nanoscale, 2014, 6, 3868–3874 RSC.
- U. Shamim, S. Hanif, A. Albanyan, F. W. J. Beck, B. Bao, Z. Wang, S. Banerjee, F. H. Sarkar, R. M. Mohammad, S. M. Hadi and A. S. Azmi, J. Cell. Physiol., 2012, 227, 1493–1500 CrossRef CAS PubMed.
- Y. Tao, Y. Lin, Z. Huang, J. Ren and X. Qu, Adv. Mater., 2013, 25, 2594–2599 CrossRef CAS PubMed.
- D. Sun, T. Guo, Y. Ran, Y. Huang and B.-O. Guan, Biosens. Bioelectron., 2014, 61, 541–546 CrossRef CAS PubMed.
- B.-C. Yin, Y.-M. Guan and B.-C. Ye, Chem. Commun., 2012, 48, 4208–4210 RSC.
- E. Morales-Narváez and A. Merkoçi, Adv. Mater., 2012, 24, 3298–3308 CrossRef PubMed.
- D. R. Dreyer, S. Park, C. W. Bielawski and R. S. Ruoff, Chem. Soc. Rev., 2010, 39, 228–240 RSC.
- D. Chen, H. Feng and J. Li, Chem. Rev., 2012, 112, 6027–6053 CrossRef CAS PubMed.
- V. Georgakilas, M. Otyepka, A. B. Bourlinos, V. Chandra, N. Kim, K. C. Kemp, P. Hobza, R. Zboril and K. S. Kim, Chem. Rev., 2012, 112, 6156–6214 CrossRef CAS PubMed.
- Y. Wang, Z. Li, J. Wang, J. Li and Y. Lin, Trends Biotechnol., 2011, 29, 205–212 CrossRef CAS PubMed.
- E. Wihaya, N. Maalouli, R. Boukherroub, S. Szunerits and J.-P. Vilcot, Proc. SPIE, 2012, 8424, 84240R CrossRef.
- P. Subramanian, A. Lesniewski, I. Kaminska, A. Vladas, A. Vasilescu, J. Niedziolka-Jonsson, E. Pichonat, H. Happy, R. Boukherroub and S. Szunerits, Biosens. Bioelectron., 2013, 50, 239–243 CrossRef CAS PubMed.
- P. Subramanian, F. Barka-Bouaifei, J. Bouckaert, N. Yamakawa, R. Boukherroub and S. Szunerits, ACS Appl. Mater. Interfaces, 2014, 6, 5422–5431 CAS.
- J. Zhang, Y. Sun, B. Xu, H. Zhang, Y. Gao, H. Zhang and D. Song, Biosens. Bioelectron., 2013, 45, 230–236 CrossRef CAS PubMed.
- J. Zhang, Y. Sun, Q. Wu, Y. Gao, H. Zhang, Y. Bai and D. Song, Colloids Surf., B, 2014, 116, 211–218 CrossRef CAS PubMed.
- N.-F. Chiu, T.-Y. Huang, H.-C. Lai and K.-C. Liu, Nanoscale Res. Lett., 2014, 9, 445 CrossRef PubMed.
- N.-F. Chiu and T.-Y. Huang, Sens. Actuators, B, 2014, 197, 35–42 CrossRef CAS.
- X. Liu, R. Aizen, R. Freeman, O. Yehezkeli and I. Willner, ACS Nano, 2012, 6, 3553–3563 CrossRef CAS PubMed.
- Y. V. Stebunov, O. A. Aftenieva, A. V. Arsenin and V. S. Volkov, ACS Appl. Mater. Interfaces, 2015, 7, 21727–21734 CAS.
- Y. Liu, X. Dong and P. Chen, Chem. Soc. Rev., 2012, 41, 2283–2307 RSC.
- H. Gu, D. Li, L. Xue, Y. Zhang and Y. Long, Analyst, 2015, 140, 7934–7938 RSC.
- L. Sun, J. Li, Y. Tan, S. Gao, L. Jin and B. Guan, Opt. Express, 2013, 21, 26714–26720 CrossRef PubMed.
- T. Ramanathan, A. A. Abdala, S. Stankovich, D. A. Dikin, M. Herrere-Alonso, R. D. Pinger, D. H. Adamson, H. C. Schniepp, X. Chen, R. S. Ruoff, S. T. Nguyen, I. A. Aksay, R. K. Prud’homme and L. C. Crinso, Nat. Nanotechnol., 2008, 3, 327–331 CrossRef CAS PubMed.
- Y. Huang, W. Lin, K. Chen, W. Zhang, X. Chen and M. Zhang, Phys. Chem. Chem. Phys., 2014, 16, 11584–11589 RSC.
- Y. Tan, L. Sun, L. Jin, J. Li and B. Guan, Opt. Express, 2013, 21, 154–164 CrossRef CAS PubMed.
- Y. Ran, L. Jin, J. Li and B. Guan, IEEE Photonics J., 2013, 5, 7100208 CrossRef.
- Z. Tang, H. Wu, J. R. Cort, G. W. Buchko, Y. Zhang, Y. Shao, I. A. Akay, J. Liu and Y. Liu, Small, 2010, 6, 1205–1209 CrossRef CAS PubMed.
- L. Gao, C. Lian, Y. Zhou, L. Yan, Q. Li, C. Zhang, L. Chen and K. Chen, Biosens. Bioelectron., 2014, 60, 22–29 CrossRef CAS PubMed.
- I. M. White and X. Fan, Opt. Express, 2008, 16, 1020–1028 CrossRef PubMed.
- J. Pollet, F. Delport, K. P. F. Janssen, K. Jans, G. Maes, H. Pfeiffer, M. Wevers and J. Lammertyn, Biosens. Bioelectron., 2009, 25, 864–869 CrossRef CAS PubMed.
- X. Y. Yang, X. Y. Zhang, Z. F. Liu, Y. F. Ma, Y. Huang and Y. S. Chen, J. Phys. Chem. C, 2008, 112, 17554–17558 CAS.
- D. Nuvoli, L. Valentini, V. Alzari, S. Scognamillo, S. B. Bon, M. Piccinini, J. Illescas and A. Mariani, J. Mater. Chem., 2011, 21, 3428–3431 RSC.
- C. Tao, J. Wang, S. Qin, Y. Lv, Y. Long, H. Zhu and Z. Jiang, J. Mater. Chem., 2012, 22, 24856–224861 RSC.
- X. Fan, G. Jiao, W. Zhao, P. Jin and X. Li, Nanoscale, 2013, 5, 1143–1152 RSC.
| This journal is © The Royal Society of Chemistry 2017 |