Open Access Article
Open Access ArticleHierarchically templated beads with tailored pore structure for phosphopeptide capture and phosphoproteomics†
Celina Wierzbicka a,
Silje B. Torsetnesb,
Ole N. Jensen
a,
Silje B. Torsetnesb,
Ole N. Jensen b,
Sudhirkumar Shinde
b,
Sudhirkumar Shinde a and
Börje Sellergren
a and
Börje Sellergren *a
*a
aDepartment of Biomedical Sciences, Faculty of Health and Society, Malmö University, SE 205 06 Malmö, Sweden. E-mail: borje.sellergren@mah.se
bDepartment of Biochemistry and Molecular Biology and VILLUM Center for Bioanalytical Sciences, University of Southern Denmark, DK-5230 Odense M, Denmark
First published on 17th March 2017
Abstract
Two templating approaches to produce imprinted phosphotyrosine capture beads with a controllable pore structure are reported and compared with respect to their ability to enrich phosphopeptides from a tryptic peptide mixture. The beads were prepared by the polymerization of urea-based host monomers and crosslinkers inside the pores of macroporous silica beads with both free and immobilized template. In the final step the silica was removed by fluoride etching resulting in mesoporous polymer replicas with narrow pore size distributions, pore diameters ≈ 10 nm and surface area > 260 m2 g−1. The beads displayed pronounced phosphotyrosine affinity and selectivity in binding tests using model peptides in acetonitrile rich solutions with a performance surpassing solution polymerized bulk imprinted materials. Tests of the beads for the enrichment of phosphopeptides from tryptic digests of twelve proteins revealed both pY/pS and pY/Y selectivity. This was reflected in a nearly 6-fold increase in the enrichment factor of a 23-mer pY-peptide and pY/pS normalized intensity ratios up to 1.5, when comparing the template mesoporous beads with the bulk materials.
Introduction
Protein phosphorylation is a reversible posttranslational modification (PTM) playing a central role in numerous biological events, several of which are directly linked to disease pathogenesis.1 Phosphoproteomics, i.e. the dynamic mapping of the occurrence of this PTM, has become an essential activity in the development of new drugs and diagnostic methods.2–7 Difficulties in analysis of phosphorylated proteins arise from their low abundance (e.g. phosphotyrosine) and transient nature. These challenges were addressed in functional proteomics using phosphopeptide specific affinity enrichment techniques and mass spectrometry, recently in parallel high throughput formats.4,7 Several chemoaffinity-based techniques to concentrate phosphopeptides from complex biological samples have been developed to date, each with their benefits and limitations, the latter generally referring to a lack of side chain selectivity i.e. recognition of pY, pS and pT respectively, a bias towards certain amino acids or sequence motifs and incompatibility with high throughput formats.3 Another limitation is inherent in the bottom up proteomics approach per se.8 Since this relies on the digestion of proteins to small peptides (<3 kDa) by trypsin followed by enrichment and LC-MS based readout of the peptide sequences, it is difficult to pinpoint multiple modifications (e.g. histone acetylation, methylation, phosphorylation) that are far apart from each other. Top down or middle down proteomics analyzing intact proteins or partially digested proteins (3–20 kDa) respectively address these issues.8 Enrichment techniques adapted to larger phosphopeptide targets are therefore needed.Molecular epitope imprinting has in the above contexts emerged as a possible alternative affinity technique.9 Several approaches to develop phosphopeptide selective molecularly imprinted polymers (MIPs) have been reported to date.9–14 In our first report, we introduced an epitope imprinting approach relying on the templating of minimal fragments of the targeted peptides.9 Hence, imprinting of N,C-protected phosphoaminoacids such as pY and pS produced binding sites complementary to pY and pS peptides respectively. The success of this approach depended on the combined use of lipophilic phosphoamino acid salts and urea based host monomer in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry. Incorporation of the resulting ternary complexes in the polymer scaffold followed by template removal left behind binding sites capable of binding the phosphoamino acid guest by quadrupole hydrogen bonding.
2 stoichiometry. Incorporation of the resulting ternary complexes in the polymer scaffold followed by template removal left behind binding sites capable of binding the phosphoamino acid guest by quadrupole hydrogen bonding.
Having addressed the molecular recognition properties of these materials we have recently turned our attention to their morphological properties. The first generation materials were prepared as porous monoliths following the classical approach to produce macroporous polymers.15 This results in materials displaying a wide distribution of pore sizes with a significant number of pores in the micro- and low meso-porous range. As a result, the inner pores of such materials show a size dependent binding preference and are likely to be poorly accessible to larger sized peptides. This will compromise their use for unbiased phosphopeptide enrichment and especially for middle down proteomics. In order to address this weakness, approaches to control the pore sizes and their distribution are required.
Grafting of thin polymer films or templated synthesis using inorganic supports of known morphology are very promising in this regard.16–18 A prominent example is porous silica which is commonly used as a sacrificial template for the synthesis of mesoporous organic polymer networks with defined particle size, shape and porosity. Hierarchical imprinting takes advantage of this morphology control to obtain imprinted polymer beads exhibiting highly accessible binding sites.19 The molecular template can either be immobilized on the walls19 of the mold or the template can be simply dissolved in the monomer mixture.20 This gives rise to polymer beads featuring enhanced binding site accessibility and faster mass transfer in chromatography. We have here assessed the latter two approaches as means to reduce the size bias in pY selective phosphopeptide enrichments. Micron sized mesoporous polymer beads were thus prepared and compared with a bulk-imprinted benchmark in a mass spectrometry assay of the enrichment of phosphopeptides from tryptic peptide mixtures. We hypothesized that hierarchical imprinting can generate efficient affinity capture for phosphopeptides, particularly of larger phosphopeptides surpassing 3 kDa that are of interest in studies of PTM crosstalk. Our results demonstrate the feasibility of this approach to boost both overall phospho-peptide and pY selectivity.
Experimental
Materials
Pentaerythritol triacrylate (PETA) was from Polysciences (Warrington, PA, USA). N,N′-azo-bis(2,4-dimethyl)valeronitrile (ABDV) was from Wako Chemicals GmbH (Neuss, Germany); 1-hydroxybenzotriazole (HOBt), 1,2,2,6,6-pentamethylpiperidine (PMP), triethylammonium bicarbonate (TEAB) buffer (1 M), acetic anhydride (Ac2O) and formic acid (FA) were from Fluka (Deisenhofen, Germany). Trifluoroacetic acid (TFA), acetonitrile (ACN), methanol (MeOH) were from VWR chemicals. Dry dichloromethane (dry DCM), triethylamine (TEA), ammonium hydrogen difluoride (NH4HF2), 2,5-dihydroxybenzoic acid (DHB), piperidine, ninhydrin, acrylamide, iodoacetamide (IAA) and 1,4-dithiothreitol (DTT) were from Sigma-Aldrich (Milwaukee, USA). Dimethylformamide (DMF), dry acetonitrile (dry ACN) were from Acros Organics. Fmoc-pTyr-OH, Fmoc-pSer-OH, Fmoc-Tyr-OH and Fmoc-Gly-OH were from Bachem GmbH (Weil am Rhein, Germany). Fmoc-Tyr(PO(NMe2)2)-OH, N-ethyldiisopropylamine (DIPEA), (benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (PyBOP) and dichloromethane (DCM) were from Merck KGaA (Dramstadt, Germany). Empore C8 extraction disk was from 3M Bioanalytical Technologies (St. Paul, MN, USA). Trypsin was from Promega (Madison, WI, USA).Amino-functionalized macroporous silica beads (NH2@Si) with an average particle size of 30 μm, a surface area (S) of 45 m2 g−1, average pore diameter (Dp) of 47.5 nm, and a pore volume (Vp) of 0.81 mL g−1 were purchased from Fuji Silysia Chemical Ltd. (Kozoji-cho, Kasugai Aichi, Japan). N-(9-Fluorenylmethoxycarbonyl)-O′-phosphonotyrosine ethyl ester (Fmoc-pTyr-OEt, 2)9 and N-3,5-bis(trifluoromethyl)-phenyl-N′-4-vinylphenylurea (1)21 were synthesized as reported elsewhere.
Apparatus and methods
Silica surface modification
NH2@Si (20 g) was suspended in DMF (100 mL) in a 250 mL round bottomed flask equipped with an overhead stirrer. Next, acetic anhydride was added (20 mL) and the suspension was stirred at room temperature overnight. Thereafter the silica was filtered off, washed with DMF (3 × 50 mL) and MeOH (3 × 50 mL) and dried in vacuum overnight to yield N-acetylated silica (AcNH@Si). Negative ninhydrin test confirmed complete protection of the surface amino groups. The degree of acetylation was confirmed by TGA analysis.Immobilization of pY-template on silica
The pY template was immobilized on the silica surface in a three step process (Fig. S1†).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v; 5 mL) and shaken in a sealed 20 mL scintillation glass vial overnight. The solid was thereafter filtered off, washed with water (3 × 10 mL) and MeOH (3 × 10 mL) and dried in vacuo to yield silica with immobilized template Fmoc-pTyr@Si (Fmoc-pTyr@Si-A–Fmoc-pTyr@Si-E, Table S1†).
1 v/v; 5 mL) and shaken in a sealed 20 mL scintillation glass vial overnight. The solid was thereafter filtered off, washed with water (3 × 10 mL) and MeOH (3 × 10 mL) and dried in vacuo to yield silica with immobilized template Fmoc-pTyr@Si (Fmoc-pTyr@Si-A–Fmoc-pTyr@Si-E, Table S1†).Preparation of pY-imprinted polymers
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 v/v) (3 × 30 mL) followed by solvent extraction with MeOH in a Soxhlet apparatus for 24 h. The course particles were then crushed and sieved. The fraction of particles with the size range 25–50 μm was used in all experiments. Non-imprinted polymer (NIP-B) used as control was prepared in the same way but with the omission of template.
20 v/v) (3 × 30 mL) followed by solvent extraction with MeOH in a Soxhlet apparatus for 24 h. The course particles were then crushed and sieved. The fraction of particles with the size range 25–50 μm was used in all experiments. Non-imprinted polymer (NIP-B) used as control was prepared in the same way but with the omission of template.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 v/v) (3 × 40 mL) and solvent extracted in a Soxhlet apparatus with MeOH for 24 h. The resulting polymer beads (MIP-M, NIP-M) were dried in vacuo overnight.
20 v/v) (3 × 40 mL) and solvent extracted in a Soxhlet apparatus with MeOH for 24 h. The resulting polymer beads (MIP-M, NIP-M) were dried in vacuo overnight.Solid phase extraction
Polymer particles (20 mg) were packed in single fritted SPE cartridges (ISOLUTE, Biotage) and protected with a frit on top. The polymers were first conditioned with 95% ACN + 0.1% FA (2 × 1 mL) followed by loading of an equimolar mixture of Fmoc-pTyr-OH, Fmoc-pSer-OH and Fmoc-Tyr-OH (1 mL, each at 100 μM concentration) in 95% ACN + 0.1% FA. The washing step was performed with 80% ACN + 0.1% FA (1 mL) and it was followed by elution with 80% MeOH + 1% TFA (1 mL). Flow-through (FT), washing (W) and elution (E) fractions were analyzed by reversed phase HPLC to determine the unbound amount of each compound. The column was Synergi 4 μm POLAR-RP 80 Å (Phenomenex, 75 × 2 mm). Mobile phases were (A) H2O + 0.1% TFA and (B) MeOH + 0.1% TFA. A linear gradient method of 40% B to 80% B in 15 min at a flow rate of 0.6 mL min−1 was used. The injection volume was 20 μL and the detection was performed by UV absorbance measurement at 265 nm. The resulting peak areas were used to calculate the amount of bound analytes (B) on the polymer according to eqn (1),| B = (C0 − c)v/m | (1) |
Binding isotherms
The polymers (10 mg each) were separately mixed with 0.5 mL of Fmoc-pTyr-OH at 0.05, 0.1, 0.25, 0.50, 1.0 and 1.5 mM concentration in 95% ACN and shaken vigorously for 2 h. Next, the samples were centrifuged and the supernatant analyzed by reversed phase HPLC using method described above to determine concentration of unbound Fmoc-pTyr-OH. The amount of bound amino acid per unit mass of polymer (B) was calculated according to eqn (1). Each experiment was performed three times. Binding curves were constructed by plotting B against free concentration c and were subsequently fitted by non-linear regression in the GraphPad Prism 7 software (GraphPad Software, La Jolla, CA, USA) to a Langmuir mono-site model:| B = Bmaxc/(Kd + c) | (2) |
Equilibrium binding tests
The polymers (10 mg each) were suspended in 0.5 mL of a mixture of VpYI, VpSI and VYI (each 20 μM) in 95% ACN + 0.1% FA and shaken vigorously for 2 h. Next the samples were centrifuged and the supernatant (400 μL) was dried (Genevac EZ-2 evaporator), dissolved in 15% ACN (200 μL) and analyzed by reversed phase HPLC. The column was Prodigy 5 μm ODS-3 100 Å (Phenomenex, 150 × 4.6 mm). Mobile phases were (A) H2O + 0.1% TFA and (B) ACN + 0.1% TFA. A linear gradient method of 15% B to 20% B in 10 min at a flow rate of 1.5 mL min−1 was used. The injection volume was 100 μL and the detection was performed by UV absorbance measurement at 210 nm. The amount of bound peptide per unit mass of polymer (B) was calculated according to eqn (1). Each experiment was performed three times. The test was repeated for octapeptides, using instead a mixture of DRVpYIHPF, DRVpSIHPF and DRVYIHPF (each 20 μM) in 95% ACN + 0.1% TFA. The method for HPLC analysis was a linear gradient of 20% B to 30% B in 10 min with other parameters remaining unchanged.Extraction of phosphopeptides from protein digest
The protein digest (20 μL, 1 pmol μL−1) and peptide mixture (5 μL, 1 pmol μL−1 of each peptide, see Table 2) were diluted to 1 mL with loading solvent (95% ACN + 1% FA) (diluted digest). Polymers (2 mg each) were mixed with 100 μL of diluted digest and agitated in a Thermomixer at 25 °C (1300 rpm) for 30 min. The polymer suspension was transferred to a pipette tip microcolumn (200 μL) protected with a C8 plug and passed through the column with the aid of a syringe and the flow-through fraction was collected. Thereafter washing was performed with 95% ACN + 1% FA (100 μL). The flow-through and washing fractions were pooled. Finally elution was performed with 80% MeOH + 0.1% TFA (100 μL) and the elution fraction saved. The combined flow-through and washing (FTW) and elution (E) fractions and a fraction of the sample before MIP treatment were dried in a speedvac and redissolved in 0.1% TFA (5 μL). Each sample (0.5 μL) was spotted together with the DHB matrix (0.5 μL) on a MALDI target plate, dried and analyzed by MALDI-TOF/TOF mass spectrometry.
Results and discussion
Preparation of materials
The purpose of the study was to investigate the influence of porosity parameters in phosphotyrosine (pY) imprinted materials on their static and dynamic peptide binding properties. We therefore decided to compare the performance of our previously reported pY-MIP (used as benchmark) featuring a broad pore size distribution, with controlled pore size formats based on silica templating (Fig. 1).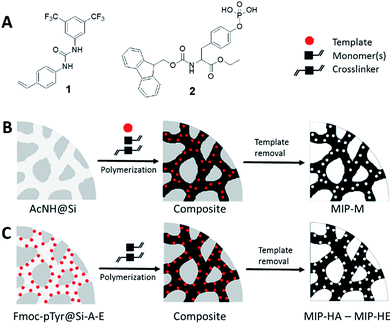 | ||
| Fig. 1 (A) Structures of urea-based functional monomer (1) and Fmoc-pTyr-OEt template (2). Schematic representation of preparation of (B) MIP-M and (C) hierarchically imprinted MIPs (MIP-HA–MIP-HE). | ||
The benchmark polymer was prepared using the functional monomer 1 and the bis-PMP salt of Fmoc-pTyr-OEt dissolved in ACN in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric ratio. Acrylamide was added as a supplementary monomer to provide additional hydrogen bond stabilization and pentaerythritoltriacrylate (PETA) was used as crosslinking monomer. Conventional azo-initiated thermal polymerization at 50 °C subsequently afforded the imprinted and non-imprinted polymers which were subsequently crushed and sieved to a 25–36 μm particle size fraction. The particles were subjected to template removal by washing with acidified MeOH, followed by extraction with MeOH and drying.
1 stoichiometric ratio. Acrylamide was added as a supplementary monomer to provide additional hydrogen bond stabilization and pentaerythritoltriacrylate (PETA) was used as crosslinking monomer. Conventional azo-initiated thermal polymerization at 50 °C subsequently afforded the imprinted and non-imprinted polymers which were subsequently crushed and sieved to a 25–36 μm particle size fraction. The particles were subjected to template removal by washing with acidified MeOH, followed by extraction with MeOH and drying.
The silica-templated polymers were subsequently prepared following two alternative approaches (Fig. 1). In both approaches we used mesoporous (Dp ≈ 50 nm) spherical silica microparticles as vessels for polymerization in two variants i.e. with free or immobilized template. In the former approach (Fig. 1B) endcapped silica was allowed to soak in the prepolymerization mixture (identical to the one used to prepare the benchmark polymer i.e. containing pY template, monomers and initiator) whereafter the particles were thermally cured at 50 °C. This contrasted with the latter approach (Fig. 1C) where the template had been pre-immobilized on the surface of the silica pore walls (Fig. S1†). After 24 h curing the silica mold was dissolved by treatment with an aqueous solution of NH4HF2 leaving behind organic polymer beads with a size and morphology reflecting those of the original silica (Fig. S3 and S4†).
A crucial parameter in the latter approach is the surface coverage with immobilized template. A high coverage may lead to template clustering which precludes the formation of separate binding sites whereas a low coverage leads to materials featuring insufficient binding capacity. We therefore studied this parameter in more detail and compared polymers prepared from five different silica templates with increasing template coverages (Table S1†).
The polymers were characterized by elemental analysis (Table S3†), optical and scanning electron microscopy (SEM) (Fig. S3 and S4†), IR spectroscopy (Fig. S6†), swelling measurements and nitrogen sorption analysis (Table 1, Fig. S5†). This confirmed their identity and near identical chemical compositions. The SEM images confirmed that the silica-templated materials retained the spherical shape and size of the silica scaffold after etching. This indicates that the residual beads originated from polymer formed in the silica pore system.
| Polymer | S (m2 g−1) | Vp (mL g−1) | Dp (nm) | Swelling (mL mL−1) |
|---|---|---|---|---|
| a The BET specific surface area (S), specific pore volume (Vp) and average pore diameter (Dp) were calculated from the nitrogen adsorption isotherms whereas the swelling was determined by soaking 100 mg of packed bed of polymer particles in ACN as described in the experimental section. | ||||
| MIP-B | 72 | 0.06 | 3.4 | 2.3 |
| NIP-B | 248 | 0.48 | 8.5 | 1.4 |
| MIP-M | 264 | 0.84 | 12.4 | 2.1 |
| NIP-M | 325 | 1.22 | 15.1 | 1.7 |
| MIP-HE | 326 | 0.94 | 10.4 | 1.9 |
| NIP-HE | 201 | 0.89 | 17.0 | 1.9 |
Otherwise only the nitrogen sorption technique, which provides information concerning the porous properties of the materials, and associated swelling tests gave evidence for differences between the polymers (Table 1, Fig. S5†).
Thus, all polymers except MIP-B exhibited a mesoporous morphology with surface areas larger than 200 m2 g−1 and relatively narrow pore size distributions (Fig. S5†) with average pore diameters exceeding 10 nm. This contrasted with MIP-B, featuring nearly no dry state porosity but significant swelling in acetonitrile. In view of the contrasting MIP/NIP properties in this case, the origin of this effect is template related. It has previously been attributed to a template induced delay in the phase separation.9
We gratefully noted that the silica-templated materials featured significantly increased surface areas and average pore sizes. It remained to be investigated how this morphology difference translated into the peptide binding properties of the materials.
Binding isotherms and imprinting efficiency
In order to compare the affinity and capacity of the materials we recorded the binding isotherms of all materials for the template analogue Fmoc-pTyr-OH in 95% ACN (Fig. 2A and S2†). This solvent was also used in the polymerization step and will hence promote polymer chain conformers present during imprinting, this in turn enhancing imprinting efficiency. Generally, binding to the imprinted polymers exceeded by far that to the corresponding non-imprinted polymers, the latter displaying binding capacities not exceeding 8 μmol g−1. Comparing different isotherm models23 it was clear from the correlation coefficients that the data for all imprinted polymers fitted well to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model (Table S4†). Hence, the imprinted sites appear uniform in this concentration interval.24 This is in agreement with imprinting stemming from a near stoichiometric monomer–template complex formation. Indeed, 1H NMR titrations has revealed strong interactions between the two species with Ka exceeding 10
1 binding model (Table S4†). Hence, the imprinted sites appear uniform in this concentration interval.24 This is in agreement with imprinting stemming from a near stoichiometric monomer–template complex formation. Indeed, 1H NMR titrations has revealed strong interactions between the two species with Ka exceeding 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1.
000 M−1.
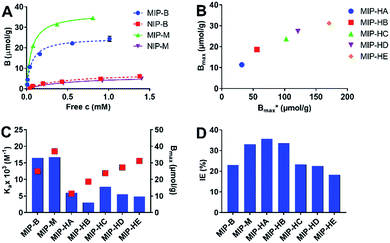 | ||
| Fig. 2 (A) Binding isotherms of Fmoc-pTyr-OH (concentration range 0.05–1.5 mM) for MIP-B (blue circles), NIP-B (red squares), MIP-M (green triangles), NIP-M (purple triangles). The results are averages of three replicates with error bars representing standard deviation. (B) Nominal (Bmax*) versus measured (Bmax) capacity of MIPs. (C) Association constants (Ka, blue bars) and binding capacities (Bmax, red squares) for Fmoc-pTyr-OH interacting with imprinted polymers in 95% ACN. The binding parameters were obtained by fitting of the binding data in (A) and Fig. S2† to a Langmuir mono-site binding model. (D) Imprinting efficiency (IE) of MIPs. | ||
Fig. 2C shows the binding constants and capacity estimated by non-linear curve fitting of the data in Table S4† assuming a Langmuir mono-site binding model. The silica-templated polymer MIP-M prepared using free template displayed the most promising performance after ranking of the polymers in terms of both binding constant and saturation capacity (Fig. 2C). This is likely to be the result of a more open pore system (Table 1) offering improved accessibility for the guest to the imprinted sites. However, a direct comparison of saturation capacities may be misleading given the different template loads used when synthesizing the materials. Instead it is interesting to compare the imprinting efficiency which relates the experimentally determined capacity to the nominal value based on the actual load of template. The silica-templated materials (e.g. MIP-M, MIP-HA) show the highest values approaching 35% imprinting efficiency, to be compared with an efficiency of 23% for the benchmark material MIP-B (Fig. 2D, Table S4†). Turning to the hierarchically imprinted materials two striking features should be noted. First of all, the template immobilization seems to compromise the binding affinity as seen in the ca. three times lower binding constants for these materials compared to the benchmark MIP-B and MIP-M. Immobilization prevents imprinting memory with respect to the C-terminal substituent of the template. Nevertheless, the absence of a clear trend when comparing MIP-H(A–E) indicates that the quality of the binding sites are similar for these materials, also in agreement with the strong monomer template interactions (vide supra). The second striking feature is the apparent correlation between the measured capacity and the nominal value (Fig. 2B). Also this observation is in agreement with the presence of stable monomer template complexes. It is interesting to note that the efficiency decreases with template coverage. This we attribute to template clustering and site coalescence as a result thereof.
Investigation of binding selectivity
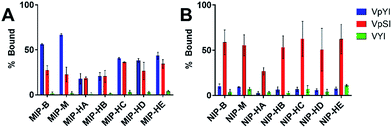
In order to probe the binding selectivity of the materials a competitive solid-phase extraction (SPE) experiment was performed. An equimolar mixture of three amino acids (Fmoc-pTyr-OH, Fmoc-pSer-OH and Fmoc-Tyr-OH) was loaded onto each material packed in SPE cartridges (see experimental part) followed by a washing and an elution step. The amount of each analyte in each fraction was quantified by reversed phase HPLC analysis (Fig. 3 and S7†). Non-imprinted materials showed an overall weak retention of the analytes, all of which were quantitatively recovered in the flow-through and wash fractions (Fig. S7†). Nevertheless, as can be seen in the wash fraction recoveries, a weak preference for the phosphorylated analytes, notably pY, was present. This effect was clearest for the materials prepared in presence of free template (MIP-B, MIP-M). When considering the imprinted materials all of them showed a pronounced selectivity for phosphorylated amino acids with a clear preference for phosphotyrosine. MIP-B, MIP-M and MIP-H(C–E) retained phosphotyrosine very strongly while phosphoserine was mainly found in the wash fractions. The retention of phosphotyrosine on the hierarchically imprinted materials increased with the order of increasing template load, a result corroborated by the measured saturation capacities.To gain insight into the potential phosphopeptide preference of the materials we designed a model system based on the peptide hormone angiotensin II (DRVYIHPF) including different phosphorylated analogues and truncated sequences. The ability of the receptors to discriminate pY and pS in the same sequence context and in peptides of different sizes was thus investigated by comparing the retention of the phosphorylated peptides DRVpYIHPF, DRVpSIHPF, VpYI, VpSI, with the nonphosphorylated peptides DRVYIHPF and VYI.
We first investigated binding of the shorter sequences VpYI, VpSI and VYI. A ternary mixture of these peptides was thus equilibrated with each of the materials followed by measurement of bound and unbound fractions (Fig. 4). The uptakes measured for the imprinted materials showed a pronounced phosphopeptide preference, in the case of MIP-B and MIP-M accompanied by a notable pY selectivity. An interesting reversal of the binding preference was observed for all the non-imprinted materials which exhibited a strong pS selectivity. We believe this to be related to the different basicities of the alkyl versus aryl phosphate groups (pY: pK2 = 5.80; pS: pK2 = 6.19).
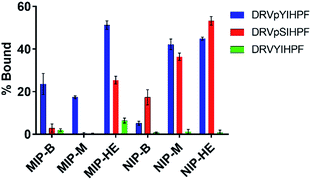
Turning to the native octapeptides (DRVpYIHPF, DRVpSIHPF and DRVYIHPF) we narrowed the investigation to include MIP-B, MIP-M and MIP-HE. Equilibrium binding tests using an equimolar mixture of the three peptides led to the binding results shown in Fig. 5.
Considering first the uptakes measured for the pY peptide we note that these are significantly lower on the imprinted materials prepared using free template (MIP-B and MIP-M: ca. 22 and 18% respectively) compared to the uptakes displayed by the hierarchically surface imprinted material (MIP-HE: 50%). Moreover, the uptakes shown by the former materials are lower than those measured for the tripeptides. All in all, this indicates that the binding sites in these materials are more buried and therefore less accessible. This also explains the high binding specificity which is reflected in the lack of uptake of the pS peptide. On the other hand, MIP-HE, featuring the surface imprinted sites, did bind this peptide, albeit to a lower extent than the pY peptide. The non-imprinted materials NIP-M and NIP-HE showed either no side chain selectivity or a preference for pS peptide (NIP-B).
Phosphopeptide enrichment from complex peptide mixtures
To validate the ability of the controlled pore size materials to enrich phosphopeptides from complex peptide mixtures we turned to protein digests. Hence, a sample originating from the tryptic digestion of twelve proteins (of which three contain a high abundance of serine-phosphorylated amino acids i.e. ovalbumin, α-casein and β-casein)25 was spiked with eight pY peptides one pS peptide and two Y peptides (Table 2). The size of the peptides ranged from 8 to 31 amino acids with charges spanning from −5 to +0.1 and GRAVY index from −1.27 to −0.26 (for the parent nonphosphorylated peptides). Phosphotyrosine peptides represented less than 1% (by mass) of all peptides in the sample. The polymers (2 mg) were equilibrated with the digest sample in the loading buffer for 30 minutes. Thereafter, the particle suspensions were transferred with pipette to tip columns and the packing then protected with a C8 plug. The flow-through (FT) fraction was collected and pooled with the fraction collected in the washing step (W). Thereafter, elution was performed and the resulting fraction (E) collected and saved. All fractions were dried, redissolved and analyzed by MALDI-MS with respect to the presence of tyrosine-, serine- and non-phosphorylated peptides.| Peptide sequence | Sizea | [M + H]+ | Net chargeb | GRAVY indexc |
|---|---|---|---|---|
| a Number of amino acids.b Net charge at pH 7.c GRAVY index for the parent nonphosphorylated peptide. | ||||
| DRVYIHPF | 8 | 1046.54 | 0.1 | −0.32 |
| DRVpSIHPF | 8 | 1050.48 | 0.1 | −0.26 |
| GADDSYYTAR | 10 | 1118.47 | −1 | −1.24 |
| DRVpYIHPF | 8 | 1126.51 | 0.1 | −0.32 |
| GADDSYpYTAR | 10 | 1198.44 | −1 | −1.24 |
| GADDSpYpYTAR | 10 | 1278.41 | −1 | −1.24 |
| GSTAENAEpYLR | 11 | 1290.54 | −1 | −0.98 |
| MHLPSPTDSNFpYR | 13 | 1644.68 | +1 | −1.00 |
| GSHQISLDNPDpYQQDFFPK | 19 | 2316.00 | −1 | −1.27 |
| RPAGSVQNPVpYHNQPLNPAPSRD | 23 | 2594.23 | +2 | −1.25 |
| YSSDPTGALTEDSIDDTFLPVPEpYINQSVPK | 31 | 3478.58 | −5 | −0.55 |
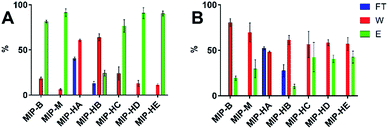
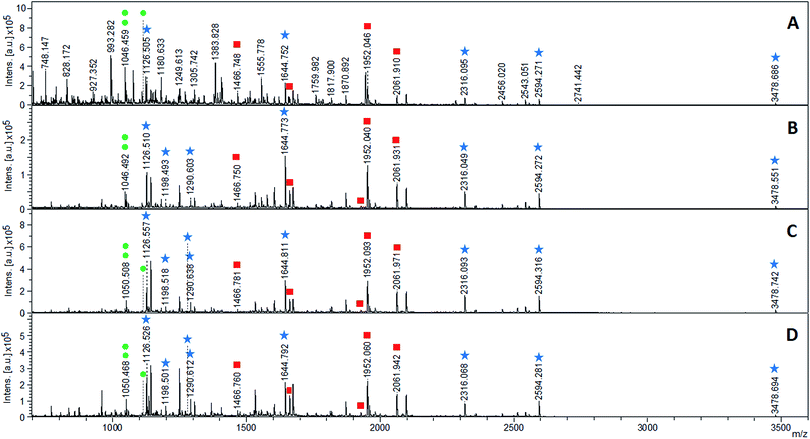
As can be seen in Fig. 6A, five out of the eight spiked pY peptides along with four pS peptides from the protein digest and the spiked pS peptide were identified in the crude sample before enrichment (Fig. 6A, Table S5†). The sample clean-up performed with the MIPs allowed the reduction of ion signals stemming from nonphosphorylated peptides to a great extent (Fig. 6B–D, Table S5†) for improved detection of phosphopeptides.
MIP-B was able to extract seven out of the eight spiked pY peptides whereas spherical MIPs (MIP-M and MIP-HE) extracted all spiked pY peptides. The combined flow-through and washing fractions from the MIPs, on the other hand, contained four (MIP-B and MIP-M) or two (MIP-HE) pS peptides and only two (MIP-B, MIP-M) or three (MIP-HE) of the eight pY peptides (Table S5, Fig. S8†).
Peptide ion signal intensities in MALDI-MS are dependent on the ionization efficiency of each of the analytes and the solvent/matrix composition.26 This precludes its use for direct quantitative analysis unless special precautions are taken and carefully optimized internal standards are used. Changes in the relative abundance of a given compound can however be determined by comparing normalized signal intensities.26 Hence, we calculated the relative signal intensities of the pY, pS and Y peptides respectively to the total intensity of all selected peptides in a given fraction (Table S6,† Fig. 7).
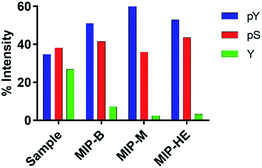 | ||
| Fig. 7 Normalized signal intensities for the pY, pS and Y peptides found in the elution fractions given as percentage of accumulated intensity of selected ions in a given fraction. | ||
The increase in the relative signal intensities of the pY peptides together with a decrease of the nonphosphorylated peptide signals show that the MIPs in principle work as expected and confirm our previous observations.10 A more refined analysis of the data allows further conclusions concerning the relative performance of the materials. Hence, we compared the normalized signal intensities collectively for all assigned pY and pS peptides (Fig. 8A) and of all assigned pY and Y peptides (Fig. 8B). This showed that the overall pY/pS ratio increased significantly after enrichment. On the other hand, the highest phosphopeptide specific enrichment factor (pY/Y) was obtained using the templated materials MIP-M and MIP-HE. Hence, the nonphosphorylated peptides were depleted from the sample treated with these MIPs. MIP-M here proved to be the most successful in removing the Y peptide (angiotensin).
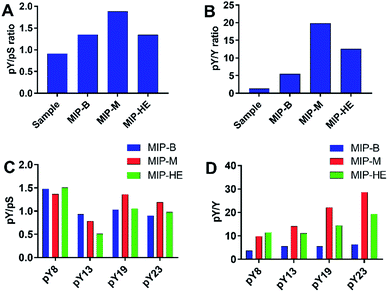 | ||
| Fig. 8 Ratios of normalized signal intensities of the pY, pS and Y peptides for nontreated and MIP-treated samples (A and B) and with respect to peptide size for the treated samples (C and D). | ||
Given the aim of enriching larger phosphopeptide fragments we were interested in how the selectivity and enrichment responded to the size of the peptide. For this purpose we compared groups of peptides (one pY, one pS and one nonphosphorylated peptide) of similar size with the pY being of size 8, 14, 19 and 24 amino acids. The octapeptides corresponding to the angiotensin II model system (vide supra) offers the most stringent test for the pY/pS selectivity since it features peptides of identical sequences at the C and N terminus. In spite of the sequence similarity the MIPs showed a marked pY selectivity (pY/pS ≈ 1.5) also in the more complex digest sample. Turning to the size dependent phosphospecific enrichment (Fig. 8C and D) we observed clear differences between the materials.
Whereas the benchmark material MIP-B showed the lowest pY/Y ratio and only a minor increase of the ratio with peptide size, both the mesoporous materials MIP-M and MIP-HE showed a clear trend of increasing enrichment efficiency with increasing size of the peptide. This we attribute to the larger average pore diameter of these materials and a relative increase in the number of sites associated with such pores. In addition to size exclusion effects, it is important to consider the surface chemistry of the materials. The silica template defines not only the average size and distribution of pores in the polymer replica but also the surface chemistry of these pores. Hence, as we have previously shown by the Engelhardt test,18 surface hydrophilicity is affected by the type of silica surface modification, e.g. use of endcapped silica as template leads to a more hydrophobic material than a material synthesized from a rehydroxylated silica template. The acetamide endcapped silicas used in the current study may have resulted in a more hydrophilic surface compared to that present in the benchmark material. Such material will display a lower degree of nonspecific peptide binding.
Conclusions
In conclusion, we have shown that silica-templated MIP synthesis and surface imprinting are useful approaches to bias the phosphopeptide preferences to larger peptide sizes (>2 kDa) as well as to suppress nonspecific binding of nonphosphorylated peptides. Further optimization of the synthesis parameters is in progress to adapt the materials to middle down and top down phosphoproteomics workflows.Acknowledgements
This work was supported by the EU-funded Marie Curie ITN project PEPMIP (PITN-GA-2010-264699). Work in the ONJ laboratory is supported by a generous grant from the VILLUM Foundation to the VILLUM Center for Bioanalytical Sciences at SDU, Odense.Notes and references
- S. A. Johnson and T. Hunter, Nat. Methods, 2005, 2, 17–24 CrossRef CAS PubMed.
- T. E. Thingholm, T. J. Jorgensen, O. N. Jensen and M. R. Larsen, Nat. Protoc., 2006, 1929–1935 CrossRef CAS PubMed.
- N. M. Riley and J. J. Coon, Anal. Chem., 2016, 88, 74–94 CrossRef CAS PubMed.
- S. J. Humphrey, S. B. Azimifar and M. Mann, Nat. Biotechnol., 2015, 33, 990–995 CrossRef CAS PubMed.
- K. Engholm-Keller and M. R. Larsen, Proteomics, 2013, 13, 910–931 CrossRef CAS PubMed.
- O. Pagel, S. Loroch, A. Sickmann and R. P. Zahedi, Expert Rev. Proteomics, 2015, 12, 235–253 CrossRef CAS PubMed.
- C. J. Tape, J. D. Worboys, J. Sinclair, R. Gourlay, J. Vogt, K. M. McMahon, M. Trost, D. A. Lauffenburger, D. J. Lamont and C. Jørgensen, Anal. Chem., 2014, 86, 10296–10302 CrossRef CAS PubMed.
- B. A. Garcia, J. Am. Soc. Mass Spectrom., 2010, 21, 193–202 CrossRef CAS PubMed.
- M. Emgenbroich, C. Borrelli, S. Shinde, I. Lazraq, F. Vilela, A. J. Hall, J. Oxelbark, E. De Lorenzi, J. Courtois, A. Simanova, J. Verhage, K. Irgum, K. Karim and B. Sellergren, Chem.–Eur. J., 2008, 14, 9516–9529 CrossRef CAS PubMed.
- J. Chen, S. Shinde, M.-H. Koch, M. Eisenacher, S. Galozzi, T. Lerari, K. Barkovits, P. Subedi, R. Krüger, K. Kuhlmann, B. Sellergren, S. Helling and K. Marcus, Sci. Rep., 2015, 5, 11438 CrossRef PubMed.
- Y. Chen, D. Li, Z. Bie, X. He and Z. Liu, Anal. Chem., 2016, 88, 1447–1454 CrossRef CAS PubMed.
- L. Xu, Y. Hu, F. Shen, Q. Li and X. Ren, J. Chromatogr. A, 2013, 1293, 85–91 CrossRef CAS PubMed.
- J. Haginaka, H. Tabo and H. Matsunaga, Anal. Chim. Acta, 2012, 748, 1–8 CrossRef CAS PubMed.
- Q. Li, F. Shen, X. Zhang, Y. Hu, Q. Zhang, L. Xu and X. Ren, Anal. Chim. Acta, 2013, 795, 82–87 CrossRef CAS PubMed.
- B. Sellergren and A. J. Hall, in Molecularly imprinted polymers. Man-made mimics of antibodies and their applications in analytical chemistry, ed. B. Sellergren, Elesevier Science B.V., Amsterdam, 2001, vol. 23, pp. 21–57 Search PubMed.
- C. Sulitzky, B. Rückert, A. J. Hall, F. Lanza, K. Unger and B. Sellergren, Macromolecules, 2002, 35, 79–91 CrossRef CAS.
- M. R. Halhalli, C. S. A. Aureliano, E. Schillinger, C. Sulitzky, M. M. Titirici and B. Sellergren, Polym. Chem., 2012, 3, 1033–1042 RSC.
- M. R. Halhalli, E. Schillinger, C. S. A. Aureliano and B. Sellergren, Chem. Mater., 2012, 24, 2909–2919 CrossRef CAS.
- M. M. Titirici, A. J. Hall and B. Sellergren, Chem. Mater., 2003, 15, 822–824 CrossRef CAS.
- E. Yilmaz, O. Ramström, P. Möller, D. Sanchez and K. Mosbach, J. Mater. Chem., 2002, 12, 1577–1581 RSC.
- J. Hall Andrew, P. Manesiotis, M. Emgenbroich, M. Quaglia, E. De Lorenzi and B. Sellergren, J. Org. Chem., 2005, 70, 1732–1736 CrossRef CAS PubMed.
- S. Kjellström and O. N. Jensen, Anal. Chem., 2004, 76, 5109–5117 CrossRef PubMed.
- K. A. Connors, Binding constants. The measurement of molecular complex stability, John Wiley & Sons, New York, 1987 Search PubMed.
- K. D. Shimizu, in Molecularly Imprinted Materials: Science and Technology, 2005, pp. 419–434 Search PubMed.
- M. R. Larsen, T. E. Thingholm, O. N. Jensen, P. Roepstorff and T. J. D. Jørgensen, Mol. Cell. Proteomics, 2005, 4, 873–886 CAS.
- O. Haglund, Qualitative Comparison of Normalization Approaches in MALDI-MS, MSc thesis, Royal Institute of Technology, Stockholm, Sweden, 2008.
Footnote |
| † Electronic supplementary information (ESI) available: Binding isotherms, SEM images and optical micrographs, nitrogen sorption, TGA and elemental analysis, ATR-FTIR and MALDI spectra, MALDI peak intensity analysis. See DOI: 10.1039/c7ra00385d |
| This journal is © The Royal Society of Chemistry 2017 |