Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Accumulation of camptothecin and 10-hydroxycamptothecin and the transcriptional expression of camptothecin biosynthetic genes in Camptotheca acuminata cambial meristematic and dedifferentiated cells
Yuhua Zhangabc,
Keming Jianga,
Degang Qingd,
Bing Huanga,
Jiayi Jianga,
Shumei Wang*bc and
Chunyan Yan *a
*a
aSchool of Pharmacy, Guangdong Pharmaceutical University, Guangzhou 510006, China. E-mail: ycybridge@163.com; Fax: +86-20-39352052; Tel: +86-20-39352052
bKey Laboratory of Digital Quality Evaluation of Chinese Materia Medica of State Administration of TCM, P. R. China
cEngineering & Technology Research Center for Chinese Materia Medica Quality of the Universities of Guangdong Province, China
dXinJiang Institute of Chinese Materia Medica and Ethnodrug, Urumqi 830002, China
First published on 21st February 2017
Abstract
Camptotheca acuminata Decne (Nyssaceae) is a major natural source of the anticancer drug camptothecin (CPT) and its derivatives. The leaves and stems are two of the main regions that accumulate CPT in C. acuminata. In this study, we successfully isolated cambial meristematic cells (CMCs) and dedifferentiated cells (DDCs) from C. acuminata stems and leaves, identified their characteristic features using micrograph analysis, and assessed their hypersensitivity to γ-irradiation and zeocin. The growth and kinetics curves of CMCs and DDCs were also studied. Furthermore, the accumulation of CPT and 10-hydroxycamptothecin (HCPT)—a more potent and less toxic CPT derivative—was assessed by liquid chromatography, and the transcriptional levels of nine genes encoding key enzymes involved in CPT and HCPT biosynthesis was assessed using quantitative real-time polymerase chain reaction (PCR) in CMCs and DDCs. The results showed that CMCs induced were particularly hypersensitive to γ-irradiation and zeocin and presented with abundant and small vacuoles, whereas DDCs were not. The above morphological and physiological characteristics of CMCs were consistent with previous reports. B5 medium was determined as the best growth medium for CMCs and DDCs. The accumulation of CPT and HCPT was higher in CMCs than DDCs, and the expression of IPI, G10H, ASA1, TSB, TDC1, TDC2, and STR was significantly upregulated in CMCs compared with DDCs; HMGR2 and HMGR3 were downregulated in CMCs. We speculate that the high accumulation of CPT and HCPT in CMCs was due to the upregulation of these seven genes.
Introduction
Camptotheca acuminata Decne belongs to the family of Nyssaceae and genus Camptotheca, and can produce camptothecin (CPT).1 CPT is a terpenoid indole alkaloid (TIA) that exhibits anti-cancer activity by inhibiting DNA topoisomerase I to kill cancer cells.2 However, CPT was not directly used in clinical treatment because of negligible water solubility and severe side effects according to a previous report.3 Conversely, 10-hydroxycamptothecin (HCPT) has been used for clinical research and was approved by the State Food and Drug Administration (SFDA) of China in 2002. In addition, CPT and HCPT are chemical precursors of anticancer drugs such as topotecan and irinotecan, which were approved for the treatment of cervical cancer, ovarian cancer, small-cell lung cancer, and metastatic colorectal cancer by the United States Food and Drug Administration (FDA) in 1994.4 Practically, soil-grown C. acuminata are not only restricted by limited natural resources and the environment, but also produce only 0.2–5.0 mg g−1 dry-weight (DW) CPT in the whole plant and 2–90 μg g−1 DW HCPT in the bark. This illustrates a serious resource crisis that should be alleviated using various approaches to identify alternative sources of CPT and HCPT.5 Sakato et al. first performed studies of C. acuminata cell culture, which produced 2.5 μg g−1 DW CPT.6 Callus cultures of C. acuminata produced 1–2 mg g−1 DW CPT and 80–100 μg g−1 DW HCPT.7,8 Later, some studies were performed to improve CPT production in C. acuminata. C. acuminata seedlings treated with elicitors such as abscisic acid (ABA), methyl jasmonate (MeJA), and salicylic acid (SA) produced 1.33–1.81 mg g−1 DW CPT and 2.25–2.60 mg g−1 DW HCPT.9 Hairy roots of C. acuminata induced by Agrobacterium rhizogenes produced 1.0–1.2 mg g−1 DW CPT and 0.15–0.31 mg g−1 DW HCPT.3,10,11Several attempts have been made to improve the production of CPT, as mentioned above. However, no relevant studies of cultured cambial meristematic cells (CMCs) from C. acuminata have been reported. CMCs are innately undifferentiated cells located in the meristems of plants; they circumvent the dedifferentiation process and serve as the origin of plant vitality.12,13 CMCs are not negatively restricted by the source of the plant, the location, the season of harvest, and the prevailing environmental conditions; therefore, they alleviate the pressure on limited wild plant resources.12 Compared with dedifferentiated cells (DDCs) suspension cultures, CMCs suspension cultures can tolerate shear stress and thereby avoid cell aggregation in air-lift bioreactors for small or large-scale cultures because of the presence of small, abundant vacuoles within each CMC; they can also maintain a stable constant growth rate on an industrial scale.13–15 CMCs can also secrete more natural products into the medium than DDCs. For example, the CMCs of Taxus cuspidata secrete 102 mg kg−1 fresh cell weight paclitaxel after culture for 10 days, whereas DDCs induced from T. cuspidata secrete just 23 mg kg−1 and 39 mg kg−1, respectively.13 Accordingly, CMCs are not only completely suitable for use in industrial production but also yield natural plant products.
In this study, C. acuminate CMCs were induced for the first time, and their morphological and physiological characteristics were consistent with CMCs previously reported.13 We then evaluated the accumulation of CPT and HCPT by liquid chromatography (HPLC) and investigated the expression of nine genes in CMCs and DDCs from C. acuminate using quantitative real-time PCR (qRT-PCR). Our results are expected to provide a new promising culture method to improve the content of CPT and its derivatives and thereby develop a sustainable method of obtaining CPT and HCPT in the future.
Results and discussion
Induction of CMCs and DDCs
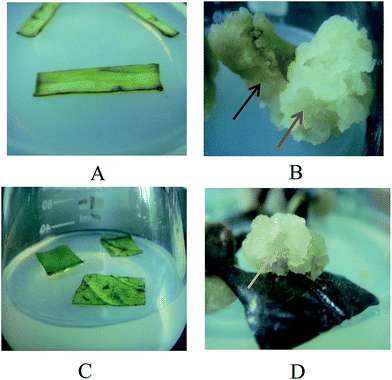
For CMCs, cambium, phloem, cortex, and epidermal stem tissues were peeled off from the xylem and laid on MS medium (Fig. 1A). After 10 days, the cambium started to present evident cell division. However, the phloem, cortex, and epidermis started to exhibit cell division by dedifferentiation by 21 days. After 60 days, compared with dim and irregular cells in phloem, cortex, and epidermal (black arrow), there were some white and loose proliferating cells in the cambium, which were CMCs (red arrow), and a visible split was observed between them (Fig. 1B). A pair of tweezers was used to gently separate the CMCs from the dim and irregular cells in the phloem, cortex, and epidermis. And the CMCs and dim and irregular cells were transferred to Murashige and Skoog (MS) medium supplemented with 0.1 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-D), 0.1 mg L−1 kinetin (KT), and 0.1 mg L−1 naphthalene-acetic acid (NAA). The CMCs of a variety of plant species, including T. cuspidate, Panax ginseng, Ginkao biloba, and Solanum lycopersicon, have been obtained using the above technology.13 After subculture seven times, the phloem, cortex, and epidermal cultures were brown and dead. In contrast, the CMCs were in good condition.For DDCs, wounded leaves were laid on MS medium (Fig. 1C), and cell division appeared in the wounds by dedifferentiation to produce many proliferating cell masses. After 65 days, the actively growing cells were gently separated from the explants and cultured in new MS medium supplemented with 0.1 mg L−1 2,4-D, 0.1 mg L−1 KT, and 0.1 mg L−1 NAA (Fig. 1D, yellow arrow).
Identification of CMCs and DDCs
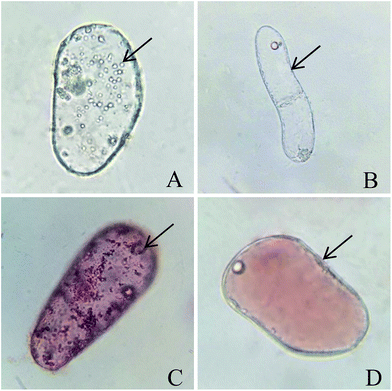
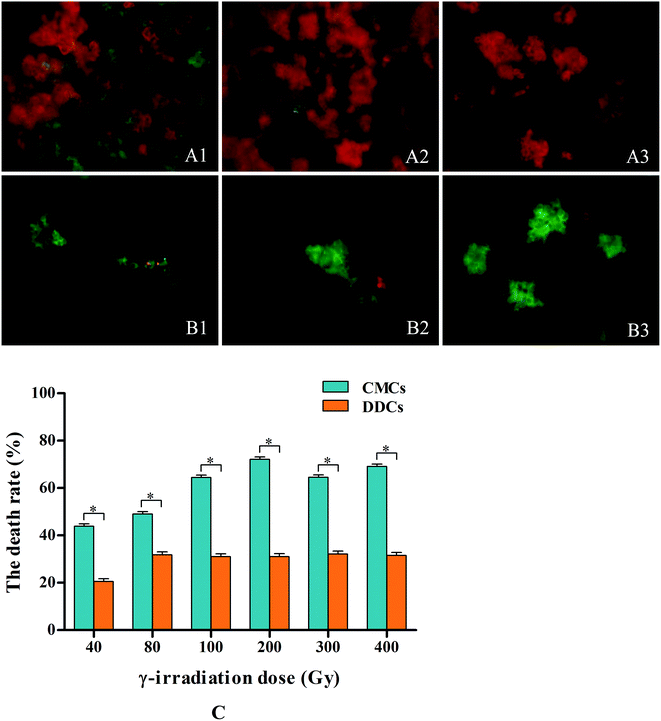
Observed under an optical microscope, single CMC presented with abundant and small vacuoles, which is the notable characteristic feature of CMCs (Fig. 2A).12 However, single DDC just had a large vacuole (Fig. 2B). Treating with 0.1% neutral red stained the vacuoles red, there were abundant and small red vacuoles in single CMC (Fig. 2C) compared with a large red vacuole in single DDC (Fig. 2D). The above properties of the CMCs obtained from C. acuminata are characteristic of cambium stem cells.13,15CMCs and DDCs were observed and photographed under a fluorescent inverted microscope after treatment with different doses of γ-irradiation and incubation for 24 h in the dark. Thereafter, images were processed by Adobe Photoshop software (Fig. 3A1–A3 and B1–B3). Fig. 4C shows that the cell death rate was significantly higher in CMCs than DDCs after treatment with different doses of γ-irradiation (P < 0.05). After incubation for 24 h in the dark, the cell death rate of CMCs gradually increased with increasing doses of γ-irradiation (40–200 Gy), and reached 72.1% with 200 Gy. However, DDCs cell death reached only 31.1% after exposure to 200 Gy of γ-irradiation. These results suggest that CMCs were significantly more sensitive to γ-irradiation than DDCs.
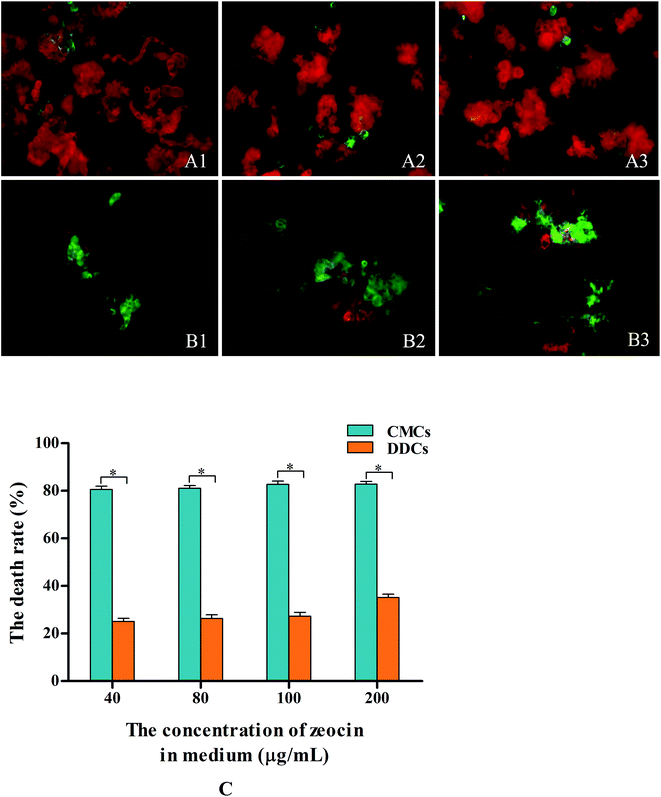
CMCs and DDCs were incubated with different concentration zeocin for 24 h in the dark, and then photographed (Fig. 4A1–A3 and B1–B3). The cell death rate of CMCs was increased significantly (P < 0.05) compared with that of DDCs treated with different concentrations of zeocin, as shown in Fig. 5C. After incubation in medium containing 40 μg mL−1 zeocin for 24 h, CMCs had a high death rate of 80.5%. The cell death rate of CMCs had little increase at doses of 80–200 μg mL−1 zeocin, and reached 82.8% with 200 μg mL−1. However, the maximum DDCs death rate was 35.1% after incubation with different concentrations of zeocin for 24 h. These results suggest that CMCs were particularly hypersensitive to zeocin, whereas DDCs were not.
The morphologic and physiologic characteristics of CMCs, including microscopic analysis revealing small, abundant vacuoles, hypersensitivity to γ-irradiation and the radiomimetic drug zeocin, and increased cell death rates, have been analyzed in a variety of plant species.12–16 The above traits of CMCs from C. acuminate are completely consistent with the CMCs reported previously.
Optimization of the growth medium for CMCs and DDCs
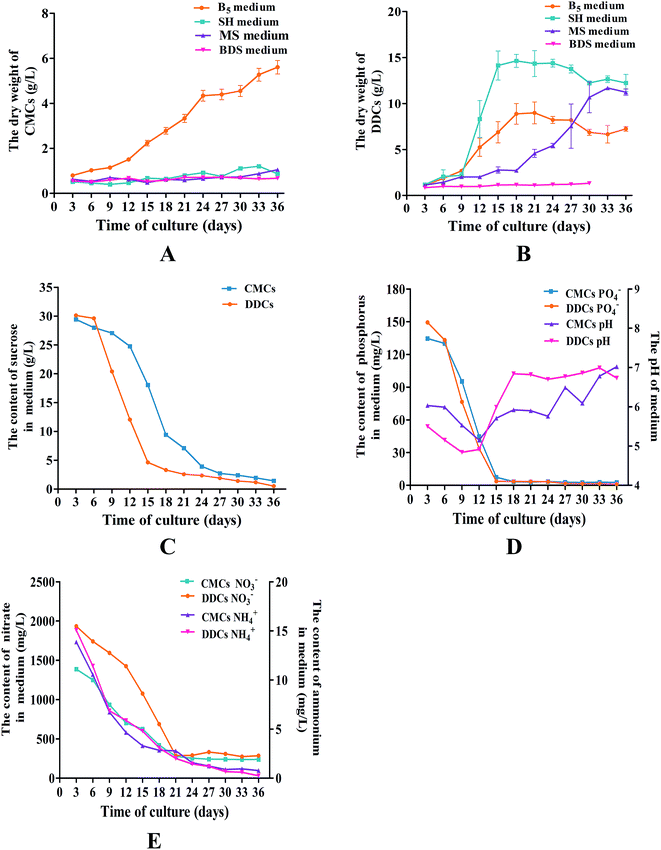
To optimize the growth medium, CMCs and DDCs were subcultured in MS, B5, BDS, or SH medium supplemented with 0.1 mg L−1 2,4-D, 0.1 mg L−1 6-benzyl aminopurine (6-BA), and 30 g L−1 sucrose. After 36 days, the dry weight of the cultures was used to establish growth curves, which were made up of the lag phase, the logarithmic phase, and the plateau phase. For CMCs, B5 was obviously the best choice of growth medium. The logarithmic phase of cultures occurred at days 12–24. Thereafter, the growth of the cultures entered the plateau phase. Ultimately, the cultures had a total DW of 5.0 ± 0.8 g L−1 in the plateau phase, which was an approximate 6.3-fold increase compared with the DW at day 3 (Fig. 5A). For DDCs, cultures grown in SH medium had a better growth state, with a logarithmic phase at days 9–15 (Fig. 5B). However, the growth of DDCs decreased slowly after subculture some times. As a result, B5 medium was selected as the growth medium for DDCs. Ultimately, B5 medium supplemented with 1.0 mg L−1 NAA, 0.5 mg L−1 2,4-D, 0.05 mg L−1 KT, and 30 g L−1 sucrose was used as the subculture medium for both CMCs and DDCs, and the cells were subcultured every 21 days.The growth kinetics of CMCs and DDCs
According to the sucrose, PO43−, NH4+ and NO3− content in the media and pH of media, additional information was obtained about the CMCs and DDCs from C. acuminata. The consumption of sucrose was consistent with the logarithmic phase of the cultures. Sucrose, which is used as the energy source for cell division, was quickly consumed when the cultures were about to enter the logarithmic phase after 6 days in culture in both CMCs and DDCs (Fig. 5C). Sucrose was consumed slowly when the cultures entered the plateau phase. Phosphorus is a constituent of membrane phospholipids and nucleic acids, and was consumed quickly when CMCs and DDCs were subcultured for 6–15 days (Fig. 5D). The pH of the media fluctuated between 5.0–7.0 during the growth period in both CMCs and DDCs (Fig. 5D). Ammonium and nitrate were immediately utilized to synthesis proteins when CMCs and DDCs were cultured (Fig. 5E). The nitrate content in the medium was stable after CMCs and DDCs were cultured for 21 days. However, ammonium levels were ultimately almost exhausted at the end of the plateau phase in both CMCs and DDCs. This demonstrates that the subculture media had a good buffering capacity and satisfied the demands for sucrose, phosphorus, ammonium, and nitrate during the growth of CMCs and DDCs.Quantification of CPT and HCPT
CPT and HCPT have anti-cancer activity, and are used for the treatment of a diverse range of cancers.2,3 CPT and HCPT were detected in both CMCs and DDCs from C. acuminata. The concentration of CPT in CMCs (2.55 μg g−1 DW) was 1.55-fold higher than that in DDCs (1.65 μg g−1 DW). Similarly, the concentration of HCPT in CMCs (25.59 μg g−1 DW) was 2.50-fold higher than that in DDCs (10.22 μg g−1 DW). Significantly, CMCs from C. acuminata were capable of yielding more CPT and HCPT than DDCs.Expression of CPT and HCPT biosynthesis genes
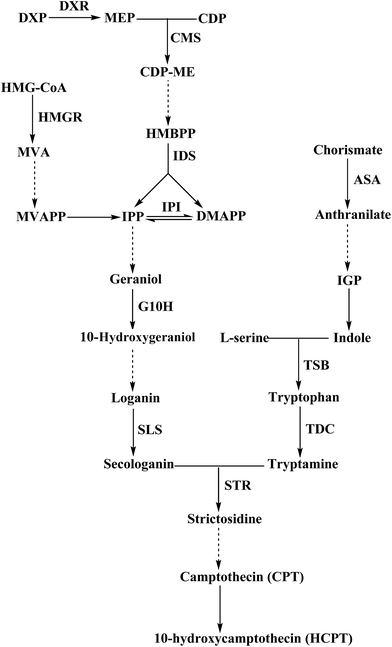
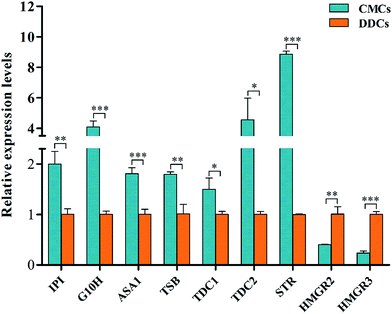
The CPT and HPCT biosynthesis pathway in C. acuminata is a very complex process that involves many vital steps that are controlled by the key enzymes isopentenyl diphosphate isomerase (IPI), geraniol-10-hydroxylase (G10H), anthranilate synthase (ASA), tryptophan synthase (TSB), tryptophan decarboxylase (TDC), strictosidine synthase (STR), and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) (Fig. 6). The genes encoding the above enzymes have all been isolated from C. acuminata.17–23 To gain insight into the molecular events underlying the significant difference between CPT and HCPT concentrations in CMCs and DDCs, the transcriptional expression of IPI, G10H, ASA1, TSB, TDC1, TDC2, STR, HMGR2 and HMGR3 was monitored using qRT-PCR (Fig. 7). The relative expression levels of HMGR2, HMGR3, TSB, TDC1, TDC2, and STR were normalized to the reference housekeeping gene actin as an internal standard. In contrast, the relative expression levels of IPI, G10H, and ASA1 were normalized to the reference housekeeping gene 18S as the internal standard. The expression levels of IPI, G10H, ASA1, TSB, TDC1, TDC2, STR, HMGR2, and HMGR3 were significantly different between CMCs and DDCs (P < 0.01, P < 0.001, P < 0.001, P < 0.01, P < 0.05, P < 0.05, P < 0.001, P < 0.01, and P < 0.001, respectively). The expression of IPI, ASA1, TSB and TDC1 were significantly higher in CMCs, with a 1.99-, 1.80-, 1.78-, and 1.47-fold increase compared with DDCs, respectively. The expression levels of TDC2 and G10H were 4.56- and 4.07-fold higher in CMCs compared with DDCs, respectively. In particular, the expression of STR was 8.86-fold higher in CMCs than DDCs. In contrast, HMGR2 and HMGR3 levels were 2.51- and 4.28-fold higher in DDCs compared with CMCs, respectively.Obviously, the transcriptional expression of IPI, G10H, ASA1, TSB, TDC1, TDC2 and STR in CMCs was remarkably upregulated, whereas HMGR2 and HMGR3 were not. The effects of the expressions of the above genes on terpenoid synthesis have been reported. For example, Kajiwara et al. reported that expression of IPI could enhance isoprenoid biosynthesis in Escherichia coli.24 The co-overexpression of G10H and 1-deoxy-D-xylulose synthase (DXS) or ASA and DXS resulted in an increase in several TIAs in Catharanthus roseus hairy roots.25 It was also reported that the G10H activity was correlated to the accumulation of TIAs in C. roseus.26 The patterns of STR expression roughly correlated with CPT accumulation in some organs of Ophiorrhiza pumila.27 HMGR catalyzes the conversion of 3-hydroxy-methylglutaryl-CoA (HMG-CoA) to mevalonate (MVA), and this conversion is the first key step in the MVA pathway in plants. In addition, the HMGR gene plays a key role in the biosynthesis of triterpenoid in many medical plants, and its overexpression can improve the production of triterpenoid.28–30 We speculate that the high accumulation of CPT and HCPT in CMCs could likely be attributed to the effects of the upregulation of the above mentioned seven genes, which offset the downregulation of HMGR2 and HMGR3.
Experimental
Induction of CMCs and DDCs
The stems and leaves of C. acuminata were obtained from Yaowang Mountain, Guangzhou University of Chinese Medicine, Guangzhou, China. The stems were washed immediately in running tap water for 30 min, and the surfaces were disinfected with 75% ethanol for 1 min. They were then washed with sterilized distilled water (dH2O), before 1% NaOCl and a drop of Tween-20 were added, and mixed vigorously for 4 min. After an additional wash with dH2O, 0.1% HgCl2 and a drop of Tween-20 were added, and the stems were mixed vigorously for 4 min and washed with dH2O. The leaves were sterilized as described above, but were maintained in HgCl2 for only 2 min due to their tender nature.For CMCs, stems were cut into 1 cm (length) segments. The cambium, phloem, cortex, and epidermal tissue were peeled off from the xylem and laid on 100 mL Erlenmeyer flasks containing 40 mL of MS medium supplemented with 0.1 mg L−1 2,4-D, 0.1 mg L−1 KT, 0.1 mg L−1 NAA, and 30 g L−1 sucrose. The pH of the medium was adjusted to 5.75 before the addition of 7.5 g L−1 agar, which was followed by sterilization at 121 °C for 20 min. The explants were cultured at 25 ± 1 °C in the dark. For DDCs, leaves were cut and laid on MS medium as described above.
Identification of CMCs and DDCs
Optimizing the growth medium for CMCs and DDCs
CMCs and DDCs were inoculated into 250 mL Erlenmeyer flasks containing 80 mL of MS, B5, BDS, or SH medium supplemented with 0.1 mg L−1 2,4-D, 0.1 mg L−1 6-BA, and 30 g L−1 sucrose. The Erlenmeyer flasks were agitated at 110 rpm and 25 °C in the dark. Cultures were assessed every 3 days to day 36 to establish growth curves. All cultures were analyzed in triplicate, and the values are expressed as means ± SDs.The growth kinetics of CMCs and DDCs
According to the CMCs and DDCs growth curves obtained in different media described in Section 2.3, both CMCs and DDCs were inoculated into 250 mL Erlenmeyer flasks containing 80 mL B5 medium supplemented with 0.1 mg L−1 2,4-D, 0.1 mg L−1 6-BA, and 30 g L−1 sucrose. The Erlenmeyer flasks were agitated at 110 rpm and 25 °C in the dark. Liquid cultures were removed every 3 days to day 36 to study the growth kinetics of CMCs and DDCs. After the liquid media were filtered, the sucrose, NH4+, NO3− and PO43− contents in the filtrate were measured using phenol–sulfuric acid, phenol–hypochlorite, salicylic acid–sulfuric acid, and phosphomolybdate blue methods, respectively, as reported previously.31–34 The pH of the filtrates was also measured using a pH meter (FE20, METTLER TOLEDO, Switzerland).Extraction and quantification of CPT and HCPT
After culture for 10 days in the dark, CMCs and DDCs cultures were dried at 55 °C and crushed separately. One gram of the above dried cultures was soaked in 50 mL methanol for 12 h, followed by ultrasonic extraction for 2.5 h. The methanol extracts were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and the supernatants were collected. Then, the residues were extracted with an equal volume methanol twice. The extracts were concentrated and evaporated to dryness. Next, 50 mL of water was added and three extractions were performed using an equal volume of chloroform. The chloroform extracts were evaporated to dryness, dissolved in 3 mL methanol, filtered using a 0.45 μm Acrodisc syringe filter, and analyzed by HPLC. HPLC was performed using a C18 column (250 × 4.6 mm, 5 μm; Phenomenex Synergi, USA) with an Agilent 1260HPLC system (Agilent Technologies, Santa Clara, CA, USA) equipped with an ultraviolet detector. The mobile phase consisted of a mixture of MeOH/H2O (50
000 rpm for 10 min and the supernatants were collected. Then, the residues were extracted with an equal volume methanol twice. The extracts were concentrated and evaporated to dryness. Next, 50 mL of water was added and three extractions were performed using an equal volume of chloroform. The chloroform extracts were evaporated to dryness, dissolved in 3 mL methanol, filtered using a 0.45 μm Acrodisc syringe filter, and analyzed by HPLC. HPLC was performed using a C18 column (250 × 4.6 mm, 5 μm; Phenomenex Synergi, USA) with an Agilent 1260HPLC system (Agilent Technologies, Santa Clara, CA, USA) equipped with an ultraviolet detector. The mobile phase consisted of a mixture of MeOH/H2O (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v). The flow rate was maintained at 1.0 mL min−1, the injection volume was 10 μL, the detection wavelength was 254 nm, and the column was maintained at 25 °C. The concentrations of CPT and HCPT were determined using standard curves. The standard curves were as follows:
50, v/v). The flow rate was maintained at 1.0 mL min−1, the injection volume was 10 μL, the detection wavelength was 254 nm, and the column was maintained at 25 °C. The concentrations of CPT and HCPT were determined using standard curves. The standard curves were as follows:| CPT, Y = 4202.2X − 0.9626 (R2 = 0.9999) |
| HCPT, Y = 708.86X + 5.5547 (R2 = 0.9992), |
Expression of CPT and HCPT biosynthesis genes
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C. The supernatants were transferred to new tubes; an equal volume of isopropyl alcohol was added, mixed very well, settled for 20 min, and centrifuged for 10 min at 12
000 rpm at 4 °C. The supernatants were transferred to new tubes; an equal volume of isopropyl alcohol was added, mixed very well, settled for 20 min, and centrifuged for 10 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C. After removing the supernatant, the precipitates were dissolved in 1 mL of a 3
000 rpm and 4 °C. After removing the supernatant, the precipitates were dissolved in 1 mL of a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of alcohol
1 ratio of alcohol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DEPC, washed, and centrifuged for 5 min at 12
DEPC, washed, and centrifuged for 5 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C. After removing the supernatant the RNA pellet was air-dried at 25 °C, verified by running on an agarose gel, dissolved in 30 μL of DEPC-treated ddH2O, and stored at −80 °C. For cDNA synthesis, 1 μg of total RNA was used for reverse transcription using Bestar qPCR RT Kit (DBI, Germany) according to the manufacturer's instructions. Twenty microliters of the cDNA was used as a template for qRT-PCR.
000 rpm at 4 °C. After removing the supernatant the RNA pellet was air-dried at 25 °C, verified by running on an agarose gel, dissolved in 30 μL of DEPC-treated ddH2O, and stored at −80 °C. For cDNA synthesis, 1 μg of total RNA was used for reverse transcription using Bestar qPCR RT Kit (DBI, Germany) according to the manufacturer's instructions. Twenty microliters of the cDNA was used as a template for qRT-PCR.| Gene names | Primer sequence (5′ to 3′) | Product sizes (bp) |
|---|---|---|
| IPI | Forward: AAGGTAACATTCCCTCTGGC | 114 |
| Reverse: CTTCCTTTGTGCAGCATTTC | ||
| G10H | Forward: ATGGATGTATTTATTGCTGCC | 141 |
| Reverse: TTCTACTAACTTGCCTTTGCC | ||
| ASA1 | Forward: GGAATGAAACGCTTGGAAAT | 111 |
| Reverse: CTTCAAAGGAGGACCGTAATG | ||
| TSB | Forward: CTGCACTATCGCCAGAGAGAT | 175 |
| Reverse: TTGGTCTCCAGATAGAGATCG | ||
| TDC1 | Forward: ACTGAATCTCCGGCATCCGTT | 208 |
| Reverse: TCAGAATGCTCTCGAATGGCT | ||
| TDC2 | Forward: AGCGGA ACTTGAGCTGGAGAT | 353 |
| Reverse: CTGCCACGTCAGCTTCTATCT | ||
| STR | Forward: TACCTTTGACTTGCCCCTTG | 168 |
| Reverse: ATTGGTGCTAAGAATGTGTTGTT | ||
| HMGR2 | Forward: AGACAAGAAGCCAGCAGCAGT | 182 |
| Reverse: TAAACCCACCAAGAGCACCAG | ||
| HMGR3 | Forward: AGGAGGTTGTGAGGAAGGTGT | 470 |
| Reverse: TGATCTGTTGTATTTCATGTGGC |
Conclusions
In the present study, CMCs of C. acuminata were isolated for the first time. The characteristic features of C. acuminate CMCs, including abundant and small vacuoles within each cell and hypersensitivity to γ-irradiation and zeocin, were consistent with CMCs reported previously. The growth curves and kinetics of CMCs and DDCs were detected simultaneously. Furthermore, HPLC analysis showed that the accumulation of CPT and HCPT was higher in CMCs than DDCs. Additionally, transcriptional analyses determined that the expression of IPI, G10H, ASA1, TSB, TDC1, TDC2, and STR was upregulated in CMCs compared with DDCs. The accumulation of CPT and HCPT in CMCs could likely be attributed to the upregulation of these seven genes which offset the effects of HMGR2 and HMGR3 downregulation. In conclusion, CMCs may be a good choice for obtaining CPT and HCPT, and this may offer an attractive and cost-effective option for the industrial production of CPT and HCPT.Acknowledgements
This study was funded by the National Natural Science Foundation of China (No. 81673557, 81102779 and 81274060), the Guangdong Natural Science Foundation (No. 9451022401003453), the Pearl River S&T Nova Program of Guangzhou (No. 2013J2200035), the Innovation Program of the University of Guangdong Province (No. 2014KTSCX118), the Science and Technology Program of Guangdong Province (No. 2014A050503067 and 2015A020211032) and the High-level Talents Program of the University of Guangdong Province.Notes and references
- M. E. Wall, M. C. Wani, C. E. Cook, K. H. Palmer, A. T. McPhail and G. A. Sim, J. Am. Chem. Soc., 1966, 88, 3888–3890 CrossRef CAS.
- Y.-H. Hsiang, R. Hertzberg, S. Hecht and L. F. Liu, J. Biol. Chem., 1985, 260, 14873–14878 CAS.
- A. Lorence, F. Medina-Bolivar and C. L. Nessler, Plant Cell Rep., 2004, 22, 437–441 CrossRef CAS PubMed.
- V. J. Venditto and E. E. Simanek, Mol. Pharmaceutics, 2010, 7, 307–349 CrossRef CAS PubMed.
- M. López-Meyer, C. L. Nessler and T. D. McKnight, Planta Med., 1994, 60, 558–560 CrossRef.
- K. Sakato, H. Tanaka, N. Mukai and M. Misawa, Agric. Biol. Chem., 1974, 38, 217–218 CrossRef CAS.
- A. J. V. Hengel, M. P. Harkes, H. J. Wichers, P. G. M. Hesselink and R. M. Buitelaar, Plant Cell, Tissue Organ Cult., 1992, 28, 11–18 CrossRef.
- H. Wiedenfeld, M. Furmanowa, E. Roeder, J. Guzewska and W. Gustowski, Plant Cell, Tissue Organ Cult., 1997, 49, 213–218 CrossRef CAS.
- G. Kai, X. Teng, L. Cui, S. Li, X. Hao, M. Shi and B. Yan, International Journal of Science, 2014, 3, 86–95 Search PubMed.
- X. Ni, S. Wen, W. Wang, X. Wang, H. Xu and G. Kai, J. Appl. Pharm. Sci., 2011, 01, 85–88 Search PubMed.
- W. Wang, Y. Lu, L. Li, J. Wang and G.-Y. Kai, Acta Bot. Boreali-Occident. Sin., 2008, 28, 2416–2422 CAS.
- C. Frankenstein, D. Eckstein and U. Schmitt, Dendrochronologia, 2005, 23, 57–62 CrossRef.
- E.-K. Lee, Y.-W. Jin, J. H. Park, Y. M. Yoo, S. M. Hong, R. Amir, Z. Yan, E. Kwon, A. Elfick, S. Tomlinson, F. Halbritter, T. Waibel, B.-W. Yun and G. J. Loake, Nat. Biotechnol., 2010, 28, 1213–1217 CrossRef CAS PubMed.
- N. Fulcher and R. Sablowski, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 20984–20988 CrossRef CAS PubMed.
- B.-W. Yun, Z. Yan, R. Amir, S. Hong, Y.-W. Jin, E.-K. Lee and G. J. Loake, Biotechnol. Genet. Eng. Rev., 2012, 28, 47–60 CrossRef CAS PubMed.
- T. Furukawa, M. J. Curtis, C. M. Tominey, Y. H. Duong, B. W. L. Wilcox, D. Aggoune, J. B. Hays and A. B. Britt, DNA Repair, 2010, 9, 940–948 CrossRef CAS PubMed.
- R. J. Burnett, I. E. Maldonado-Mendoza, T. D. McKnight and C. L. Nessler, Plant Physiol., 1993, 103, 41–48 CrossRef CAS PubMed.
- M. López-Meyer and C. L. Nessler, Plant J., 1997, 11, 1167–1175 Search PubMed.
- H. Lu and T. D. McKnight, Plant Physiol., 1999, 120, 43–51 CrossRef CAS PubMed.
- H. Lu, E. Gorman and T. D. McKnight, Planta, 2005, 221, 352–360 CrossRef CAS PubMed.
- I. E. Maldonado-Mendoza, R. M. Vincent and C. L. Nessler, Plant Mol. Biol., 1997, 34, 781–790 CrossRef CAS PubMed.
- X. Pan, M. Chen, Y. Liu, Q. Wang, L. Zeng, L. Li and Z. Liao, DNA Sequence, 2008, 19, 98–105 CrossRef CAS PubMed.
- Y. Sun, H. Luo, Y. Li, C. Sun, J. Song, Y. Niu, Y. Zhu, L. Dong, A. Lv, E. Tramontano and S. Chen, BMC Genom., 2011, 12, 533–544 CrossRef CAS PubMed.
- S. Kajiwara, P. D. Fraser, K. Kondo and N. Misawa, Biochem. J., 1997, 324, 421–426 CrossRef CAS PubMed.
- C. A. M. Peebles, G. W. Sander, E. H. Hughes, R. Peacock, J. V. Shanks and K.-Y. San, Metab. Eng., 2011, 13, 234–240 CrossRef CAS PubMed.
- G. Collu, A. A. Garcia, R. V. D. Heijden and R. Verpoorte, Plant Sci., 2002, 162, 165–172 CrossRef CAS.
- Y. Yamazaki, A. Urano, H. Sudo, M. Kitajima, H. Takayama, M. Yamazaki, N. Aimi and K. Saito, Phytochemistry, 2003, 62, 461–470 CrossRef CAS PubMed.
- Q.-Y. Gai, J. Jiao, M. Luo, W. Wang, C.-J. Zhao, Y.-J. Fu and W. Ma, Ind. Crops Prod., 2016, 84, 350–357 CrossRef CAS.
- S. J. Hey, S. J. Powers, M. H. Beale, N. D. Hawkins, J. L. Ward and N. G. Halford, Plant Biotechnol. J., 2006, 4, 219–229 CrossRef CAS PubMed.
- Y.-K. Kim, J. K. Kim, Y. B. Kim, S. Lee, S.-U. Kim and S. U. Park, J. Agric. Food Chem., 2013, 61, 1928–1934 CrossRef CAS PubMed.
- P. S. Chen, T. Y. Toribara and H. Warner, Anal. Chem., 1956, 28, 1756–1758 CrossRef CAS.
- M. Dubois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- U. Hecht and H. Mohr, Physiol. Plant., 1990, 78, 379–387 CrossRef CAS.
- M. W. Weatherburn, Anal. Chem., 1967, 39, 971–974 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2017 |