Open Access Article
Open Access ArticlePoly-γ-glutamic acid coupled Pseudomonas putida cells surface-displaying metallothioneins: composited copper(II) biosorption and inducible flocculation in aqueous solution†
Penggao Hua,
Zhongna Zhanga,
Fei Shena,
Xun Yua,
Mingshun Lia,
Hong Nib and
Lin Li *a
*a
aState Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan 430070, China. E-mail: lilin@mail.hzau.edu.cn; Fax: +86-27-8728-0670; Tel: +86-27-8728-6952
bCollege of Life Sciences, Hubei University, Wuhan 430062, China
First published on 28th March 2017
Abstract
Poly-γ-glutamic acid (γ-PGA) is a biodegradable macromolecular polymer that is capable of adsorbing heavy metal ions through its side chains. In this study, a new biocomposite biosorbent that incorporates Cu2+ adsorption and recovery from aqueous solutions was developed through immobilization of a glutamine-binding protein GlnBP and cyanobacterial metallothionein SmtA onto the surface of Pseudomonas putida cells using an InaQ-N protein anchor, followed by coupling the cells with side chain-activated γ-PGA. The surface localization of the fusion protein (InaQ)2–GlnBP–(SmtA)2 was confirmed by SDS-PAGE/Western blot, immunofluorescence microscopy and fluorescence-activated cell sorting assays. The engineered biocomposite MB546-PGA was capable of Cu2+ adsorption across a pH range of 3.5–5.5 and a relatively broad temperature range of 20–60 °C, with a maximum adsorption capacity for Cu2+ of 145.99 mg g−1. The pseudo-second-order equation was applicable to the sorption data. Moreover, MB546-PGA exhibited an inducible flocculation capability following Cu2+ adsorption. The infrared spectroscopy data showed that the hydroxyl and amino groups of MB546-PGA were involved in Cu2+ adsorption, and γ-PGA likely bound to the cells through its carbonyl and amino groups. Given the features of the high capacity of Cu2+ adsorption and the easily inducible flocculation, the developed biosorbent shows potential for use in the remediation and purification of polluted or waste waters.
Introduction
Metal pollution caused by various heavy metals from industrial processes has been a serious worldwide environmental threat. Copper is a commonly used heavy metal that is normally found in wastewater and effluents from electroplating, mining and various electronic industries. Because of the wide use, severe toxicity and non-degradability, copper ions ultimately accumulate in humans through the food chain, which poses a severe risk of health related problems to humans.1,2 Remediation of Cu-like heavy metal pollution often involves chemical, physical, and biological approaches.3,4 In the past few decades, biological remediation based on cell membrane-grafted metal-reacting chemical groups, which leads to the biosorption of metal ions onto the surface of intact cells or biocomposites, has attracted increasing interest because of its cost-effectiveness, high removal efficiency, and requirements for only moderate reaction conditions.5 Unfortunately, the conventional biosorption processes normally encounter difficulty in the separation of the biomasses from the effluents and in the desorption of the metals from the biosorbents. Therefore, it is still necessary to develop an innovative approach to solve these technical drawbacks.Developments in biomass immobilization systems, particularly those using immobilized bacteria or fungi, provide a promising solution to these problems. Several previous investigations have attempted to immobilize biomasses on natural polymers, such as Ca-alginate gels,6 chitosan,7 activated carbon,8 polymer fibers,9 and volcanic minerals,10 to increase the mechanical strength and resistance to chemical constituents, and to facilitate the separation of the biosorbents from the effluents. The soluble biopolymer poly-γ-glutamic acid (γ-PGA) is formed by glutamate polymerization, which is primarily produced via bacterial fermentation.11 γ-PGA produced by different bacterial species has different molecular configurations and molecular masses and consists of either L- or D-glutamic acid or a mixture of both enantiomers.12 γ-PGA has been recognized as a new biomaterial with many biotechnologically extraordinary properties, as it is biodegradable, edible, non-toxic, and non-immunogenic to humans.13,14 Moreover, the enrichment of carboxyl groups in its side chains and the flocculation capacity in aquatic systems have special appeal for the development of a biocomposite adsorbent that could be used for the remediation of heavy metal pollution.15,16
Glutamine-binding protein (GlnBP) is a periplasmic protein that is responsible for glutamine transport. In Escherichia coli, GlnBP consists of 226 amino acids (aa) that form two globular domains connected by two flexible hinges. Previous studies have shown that the glutamine ligand between the two globular domains is bound by hydrogen bonds and ionic interactions. The α-amino group of the glutamine ligand strongly interacts with Gly68, Thr70, and Asp157 of GlnBP via hydrogen bonds formed by the NH3+ group of the glutamine ligand, the carbonyl group of Gly68, the carbonyl group of Thr70, and the side chain of Asp157. The α-carboxyl group of the glutamine ligand interacts with Thr70, Arg75, and Gly119 of GlnBP via hydrogen bonds formed between the backbone N atoms of Thr70 and Gly119 and the ligand, as well as a salt bridge formed by the electron on the side chain of Arg75 and the COO− of the glutamine ligand. As expected, the glutamine ligand also binds to the cavity formed by Asp10, Gly68, Thr70, and Ala67 of GlnBP via hydrogen bonds and ionic interactions.17 In addition, GlnBP is thermostable, and glutamine can bind to GlnBP over a relatively broad range of pH values, temperatures, and ionic strengths.18
The display of a metal-binding protein (MBP) on the bacterial cell surface using cell surface display technology is an admirable strategy for removing heavy metals; it enhances the reaction rate, eliminates the mass transfer limitation, and minimizes the toxicity of heavy metals to living cells.19 The metallothioneins (MTs) are a family of cysteine-rich metal-binding proteins that are capable of taking up heavy metals from the environment. Microbial MTs mainly consist of the BmtA family of proteins and the Cu-binding protein MymT. Moreover, a cadmium-binding protein of the BmtA family, namely, SmtA from the cyanobacterium Synechococcus PCC7942, has the well-characterized ability to take up not only Cd2+ but also other metal ions, including Zn2+, Cu2+, Hg2+, etc.20,21 At a low pH (as low as pH 2.35 for half dissociation), the adsorbed metal ions can be desorbed from SmtA.21 Thus, by displaying SmtA on the bacterial cell surface and by assigning different pH conditions, it is possible to govern the adsorption/desorption cycles of metal ions and to remove and recover metal ions from polluted aquatic systems.
In the present study, we constructed a composite bioadsorbent with the polymer γ-PGA and the recombinant P. putida cells with SmtA surface-display to investigate the Cu2+ adsorption, the induced flocculation, and the desorption/recovery of Cu2+ of this material. Firstly, P. putida AB92019 was engineered to display the fusion protein (InaQ-N)2–GlnBP–(SmtA)2, which consisted of two tandemly repeated anchor InaQ-N proteins,22 GlnBP, and two repeats of SmtA. When exogenous γ-PGA was added, it bound to the GlnBP subunit of the fusion protein to form a triple composite biomaterial “Cell-γ-PGA-Cell surface SmtA” (MB546-PGA). Subsequently, the optimal adsorption conditions, the adsorption kinetics, and the maximum adsorption capacity of Cu2+ of this biomaterial were determined. The interactions between γ-PGA and P. putida cells, as well as the biosorbent system and Cu2+, were examined using scanning electron microscopy (SEM) and Fourier transform infrared (FTIR) spectroscopy, respectively. Finally, experiments were conducted to investigate the flocculation activity upon the induction of certain coagulants and the adsorption/desorption activities of the biomaterial towards Cu2+.
Results and discussions
Construction of recombinant P. putida MB546 and surface localization analysis of the fusion protein (InaQ-N)2–GlnBP–(SmtA)2
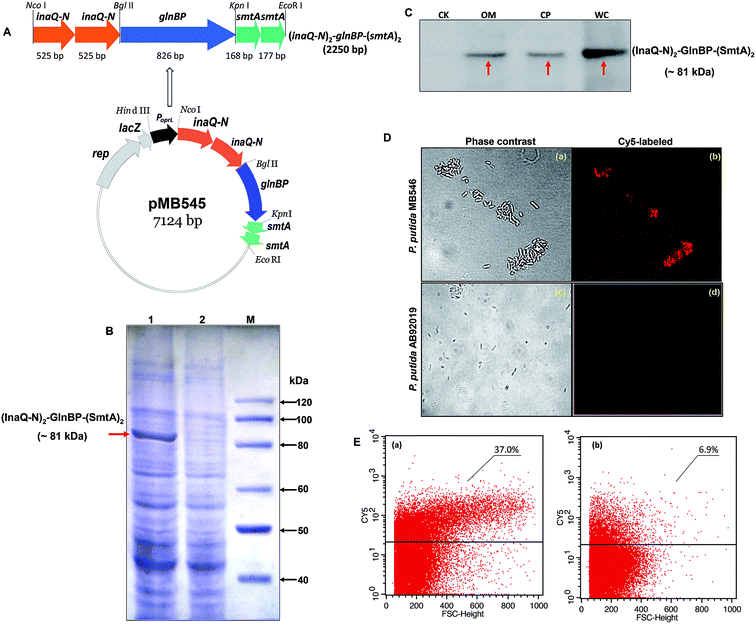
As shown in Fig. 1A, the recombinant pMB545 was constructed to express the fusion gene (inaQ-N)2–glnBP–(smtA)2 that is driven by an E. coli–Pseudomonas dual-active constitutive promoter PoprL.23 Two copies of the smtA gene were used to construct pMB545 to increase the adsorption capacity of SmtA for metal ions. Theoretically, the fusion protein (InaQ-N)2–GlnBP–(SmtA)2 has a molecular mass (Mr) of approximately 81 kDa. As shown in Fig. 1B, a band corresponding to a protein with a predicted Mr that was apparently equal to the Mr of (InaQ-N)2–GlnBP–(SmtA)2 was clearly present in the MB546 profile (lane 1, indicated by an arrow), but was not found in the profile of the parent strain AB92019, verifying the accurate expression of this fusion protein in the host cells. This result was further confirmed by Western blot analysis, in which the profile of the expressed proteins from the whole cell fraction of MB546 cells (Fig. 1C, lane WC) revealed clear bands (Fig. 1C, lane WC), whereas no bands were observed in the control AB92019 cells (Fig. 1C, lane CK).The Western blot profile of the outer membrane-complex fraction (OM) showed the presence of a protein band (Fig. 1C, lane OM) corresponding to the bands present in the WC fraction (Fig. 1C, lane WC) and the cytoplasmic fraction (CP) (Fig. 1C, lane CP), thereby confirming the surface localization of this fusion protein in principle. Immunofluorescence micrographs (Fig. 1D) and FACS analysis (Fig. 1E) also showed clear signs of the surface occurrence of (InaQ-N)2–GlnBP–(SmtA)2 on P. putida MB546 cells. In contrast, the control P. putida AB92019 cells were unable to react with externally added Cy5-labeled antiserum. Therefore, these assays confirmed the surface localization of (InaQ-N)2–GlnBP–(SmtA)2 on P. putida MB546 cells.
Preparation of the MB546-PGA biocomposite
We developed a biocomposite adsorbent that consisted of the engineered P. putida MB546 cells and γ-PGA, which is presumed to confer a joint adsorption capacity for metal ions. γ-PGA was designed to bind to the surface of recombinant bacterial cells through two types of linkages: (i) covalent binding through the formation of amide bonds between the activated side chain carboxyl groups of γ-PGA and the amino groups of aa residues on the MB546 cell surface, such as those from lysine of GlnBP. As shown in Fig. S1(a),† γ-PGA and EDC formed an unstable reactive O-acylisourea ester by NHS treatment; then, EDC was replaced with NHS to form a semi-stable amine-reactive NHS ester with γ-PGA, allowing the complete activation of γ-PGA24 and leading to the formation of stable amide bonds with the amino groups on the cell surface. As a result, the activated γ-PGA was bound to the bacterial surface. (ii) γ-PGA and GlnBP bind through ionic and hydrogen bonds. As shown in Fig. S1(b),† γ-PGA can insert into the cavity between the two domains of GlnBP and can then become stabilized by forming ionic interactions and hydrogen bonds with glutamine and other aa residues around the cavity.17The morphology of the MB546-PGA biocomposite that was or was not bound to the activated γ-PGA was examined using SEM. Both P. putida AB92019 cells (Fig. 2A) and AB92019 + PGA (Fig. 2B) were observed in natural dispersion states, without significant morphological differences. MB546 cells that were bound to the inactivated γ-PGA also appeared in a naturally dispersed state (Fig. 2C). However, the biocomposite MB546-PGA formed a certain amount of cell aggregates (Fig. 2D). These results indicated that the formation of “Cell-γ-PGA-Cell surface SmtA” aggregates in the biocomposite MB546-PG and γ-PGA served as the crucial coupler to mediate cellular aggregation.
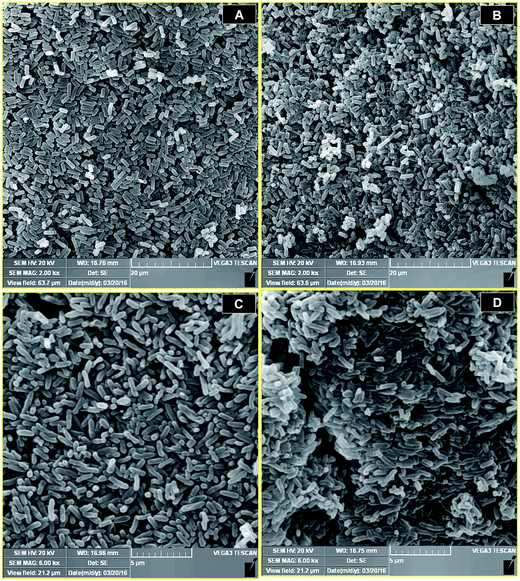 | ||
| Fig. 2 Representative SEM micrograph of P. putida AB92019 cells (A), P. putida AB92019 cells+PGA (B), P. putida MB546 cells (C), and the MB546-PGA biocomposite (D). | ||
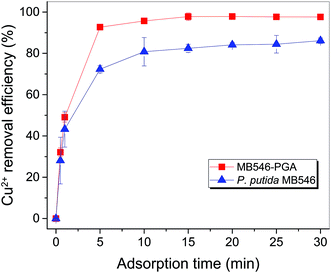
Cu2+ adsorption of MB546-PGA
Fig. S2A† showed that at pH 5.5, 30 °C, an initial Cu2+ concentration of 32 mg L−1 and a MB546-PGA loading equivalent to 0.6 mg mL−1 dry biomass, the Cu2+ adsorption reaction reached the equilibrium state in 15 min, and Cu2+ removal efficiency reached 90% of the maximum value in 5 min, reflecting the fast adsorption reaction conducted by MB546-PGA. Fig. S2B and S2C† showed that the high Cu2+ removal efficiencies (>96%) were maintained within a range of 20–60 °C and pH 3.5–5.5, respectively. Fig. S2D† shows that Cu2+ removal efficiencies were relatively stable at high levels (>96%) across an MB546-PGA loading range of 0.06 mg mL−1 to 1.8 mg mL−1.Based on the results describe above, an optimized immobilization condition for Cu2+ biosorption by MB546-PGA can be defined as follows: a pH of 5.5, 30 °C, an initial Cu2+ concentration of 32 mg L−1, an adsorption time of 30 min, and an MB546-PGA loading amount equivalent to 0.6 mg mL−1 dry MB546 cells. Fig. 3 shows that these two materials exhibited consistent adsorption patterns for Cu2+ over the time course under the optimized adsorption conditions. The Cu2+ removal efficiency of MB546-PGA was higher than that of MB546 alone, with an ultimate increase in Cu2+ removal efficiency by approximately 13% in 30 min. Two-way ANOVA analysis was performed at 0.05 significance level to evaluate whether the difference of Cu2+ removal efficiency between MB546 and MB546-PGA was significant or not. The ANOVA result (Table S1†) showed that the P values of the factor A (MB546) and factor B (MB546-PGA) were all less than 0.01, indicating that the Cu2+ removal efficiency of MB546 and MB546-PGA is very significant.
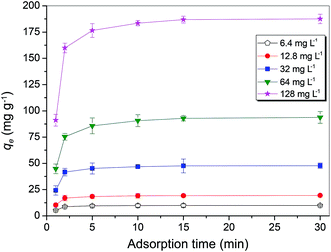
Kinetics of biosorption
The optimum biosorption conditions were further applied to determine the Cu2+ adsorption capacity of MB546-PGA towards different initial Cu2+ concentrations of 6.4 mg L−1, 12.8 mg L−1, 32.0 mg L−1, 64.0 mg L−1, and 128.0 mg L−1. Fig. 4 shows that the biosorption of Cu2+ was rapid, with an approximately maximum adsorption equilibrium capacity obtained within the first 5 min towards low initial concentration of Cu2+ (6.4–32 mg L−1), or approximately 90% of the maximum adsorption capacity towards a high initial concentration of Cu2+ (64–128 mg L−1). The adsorption capacity maintained a stable or slightly increasing trend until the final adsorption equilibrium was obtained at 30 min. Pseudo first-order and second-order equations were used to model the sorption data over the time course. The pseudo-first-order equation based on the adsorption equilibrium is represented as follows:25
ln(qe − qt) = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) qe − k1t qe − k1t
| (1) |
The pseudo-second-order equation can be generally presented as follows:25
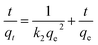 | (2) |
The values of ln(qe − qt) and 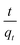 were calculated. Since ln(qe − qt) and t in eqn (1), and
were calculated. Since ln(qe − qt) and t in eqn (1), and 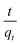 and t in eqn (2) are linearly related, the time course sorption profiles were linearly fitted using eqn (1) and (2), and the data are shown in Fig. S3A and S3B.† The fitting parameters of the pseudo-first-order kinetics and pseudo-second-order kinetics were shown in Tables S2 and S3.† Each value of Prob > F of these two models was less than 0.01, indicating that both models were well fitted. Comparison of R2 of each models indicated pseudo-second-order kinetics model (R2 = 0.98–0.99) was able to describe the sorption kinetics better. The pseudo-second-order equation show that as the initial Cu2+ concentration increased, the adsorption capacity qe increased, and the pseudo-second-order kinetic constant k2 was generally decreased.
and t in eqn (2) are linearly related, the time course sorption profiles were linearly fitted using eqn (1) and (2), and the data are shown in Fig. S3A and S3B.† The fitting parameters of the pseudo-first-order kinetics and pseudo-second-order kinetics were shown in Tables S2 and S3.† Each value of Prob > F of these two models was less than 0.01, indicating that both models were well fitted. Comparison of R2 of each models indicated pseudo-second-order kinetics model (R2 = 0.98–0.99) was able to describe the sorption kinetics better. The pseudo-second-order equation show that as the initial Cu2+ concentration increased, the adsorption capacity qe increased, and the pseudo-second-order kinetic constant k2 was generally decreased.
Biosorption isotherms
The Langmuir model (Fig. S3C†) and the Freundlich model (Fig. S3D†) were used to analyze the biosorption isotherm data for Cu2+ adsorption by MB546-PGA at different initial Cu2+ concentrations. The Langmuir model assumes a monolayer sorption of a solute from a liquid solution and can be expressed as:
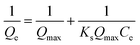 | (3) |
The Freundlich model is based upon sorption on a heterogeneous surface, which can be expressed as:
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qe = ln Qe = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kf + 1/n(ln Kf + 1/n(ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce) Ce)
| (4) |
The values of Qe−1, Ce−1, ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qe and ln
Qe and ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce were calculated. Qe−1 and Ce−1 in eqn (3), and ln
Ce were calculated. Qe−1 and Ce−1 in eqn (3), and ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qe and ln
Qe and ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce in eqn (4) are linearly related, and linear fitting of these parameters was adopted using the software Origin Pro 8 SRO v8.0724 (B724). The fitting parameters of the Freundlich isotherm and Langmuir isotherm models were calculated by fitting these equations. The results (Tables S4 and S5†) showed that the adsorption activity of MB546-PGA was more consistent with the Freundlich model (R2 > 0.99; P-value = 6.85 × 10−5). Under optimized conditions, the MB546-PGA biosorbent obtained a maximum Cu2+ adsorption capacity of 145.99 mg g−1 through the Langmuir model and other isotherm model calculations.
Ce in eqn (4) are linearly related, and linear fitting of these parameters was adopted using the software Origin Pro 8 SRO v8.0724 (B724). The fitting parameters of the Freundlich isotherm and Langmuir isotherm models were calculated by fitting these equations. The results (Tables S4 and S5†) showed that the adsorption activity of MB546-PGA was more consistent with the Freundlich model (R2 > 0.99; P-value = 6.85 × 10−5). Under optimized conditions, the MB546-PGA biosorbent obtained a maximum Cu2+ adsorption capacity of 145.99 mg g−1 through the Langmuir model and other isotherm model calculations.
Flocculation induction by MB546-PGA
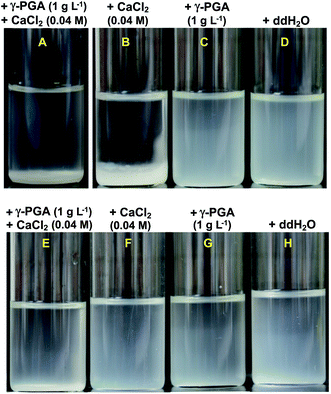
Effective flocculation can definitely confer a desirable potential for the recovery of adsorbed heavy metal ions and for the enhanced overall removal capacity of metal ions. As shown in Fig. 5A and B, the combination of 0.04 M CaCl2 and 1 g L−1 γ-PGA to MB546-PGA suspension or adding 0.04 M CaCl2 alone induced flocculation; however, γ-PGA alone failed to induce flocculation (Fig. 5C), indicating that 0.04 M CaCl2 induced flocculation. Interestingly, neither the CaCl2/γ-PGA combination nor CaCl2 or γ-PGA alone was able to induce flocculation in the control MB546 cell suspensions (Fig. 5E and F), suggesting that the distinctive “Cell-γ-PGA-Cell” linkages of MB546-PGA caused the formation of aggregates and that γ-PGA pivotally bound to cells and contributed to the flocculation upon CaCl2 induction.The Cu2+ adsorption/desorption cycles
Fig. S4† shows that in the first round, MB546-PGA exhibited a Cu2+ adsorption capacity of 100 mg g−1 and an approximately half dissociation of Cu2+ at pH 2.5. However, in the second round, the Cu2+ adsorption of MB546-PGA significantly decreased to approximately 20% of the capacity in the first round, indicating that the low pH condition caused the remarkable ineffectiveness of this biomaterial for Cu2+ adsorption.FTIR spectroscopic analysis
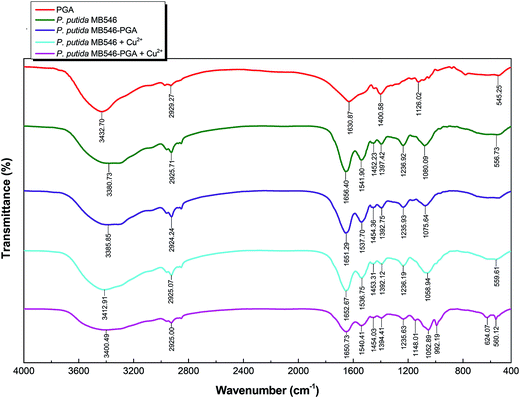
FTIR spectroscopic analyses of P. putida MB546 cells and MB546-PGA before and after Cu2+ adsorption were performed to verify the chemical groups that were involved in Cu2+ and γ-PGA binding. As shown in Fig. 6, the peaks in the 3300–2800 region were attributed to the vibrations of alkyl groups, such as CH3, CH2, and CH.10,26 The peaks at 1656.40 and 1651.39 cm−1 were produced by the stretching vibration of carbonyl groups in MB546 and MB546-PGA, respectively, in the absence of Cu2+ binding. The peaks at 1652.67 and 1650.73 cm−1 represent the stretching vibration of carbonyl groups in Cu2+-bound MB546 and Cu2+-bound MB546-PGA, respectively. The peaks at 1537.70 and 1541.90 cm−1 were attributed to the stretching vibration of the NH groups in MB546-PGA and unbound MB546, respectively, whereas the peaks at 1536.75 and 1540.41 cm−1 were attributed to the stretching vibration of the NH groups in Cu2+-bound MB546 and Cu2+-bound MB546-PGA, respectively. In addition, the peaks at 3385.85 and 3400.49 cm−1 were attributed to the stretching vibration of the hydroxyl groups in MB546-PGA prior to Cu2+ adsorption. These results showed that the vibrational peaks of the carbonyl and amino groups were shifted to low frequencies after γ-PGA bound to the MB546 cells, which suggested that the carbonyl and amino groups on the surface of the recombinant P. putida MB546 cells might be involved in the binding of γ-PGA to the cells. After Cu2+ was adsorbed by MB546-PGA, the vibrational peaks of the amino and hydroxyl groups were shifted to high frequencies, which indicated that the amino and hydroxyl groups of MB546-PGA might be involved in Cu2+ adsorption.The aim of this study was to construct a composite biosorbent with high adsorption and inducible flocculation capacities for the biosorption of Cu2+. The engineered P. putida cells that displayed SmtA and γ-PGA on their surface appear to be particularly valuable for this purpose because of their high Cu2+-binding activities and, for γ-PGA, the distinctive cell-coupling and inducible flocculation capabilities. The introduction of the glutamine-binding protein GlnBP to the fusion protein that was anchored on cell surface could be considered pivotal for the functionality of this system. Theoretically, γ-PGA could covalently bind to GlnBP by its Gln that acts as a ligand through an “enzyme–substrate” interaction mode. Previous investigations have revealed a cavity between the two domains of the GlnBP 3D structure that is presumed to bind Gln in E. coli.27 However, because GlnBP is trapped in the middle of the “triple” configuration of the fusion protein, it remains uncertain whether this structural configuration would affect the spatial structure of GlnBP and ultimately affect the formation of its cavity. Thus, we employed a type of EDC/NHS activation linkage method for γ-PGA to ensure that γ-PGA could effectively bind to GlnBP. The side chain carboxyl groups of γ-PGA were activated by EDC to form an unstable reactive o-acylisourea ester, which would react with NHS to form a semi-stable amine-reactive NHS ester. The latter would react with the amino groups of various amino acids, such as lysine of GlnBP anchored on the cell surface, to form stable amide bonds, which ultimately allowed γ-PGA to bind to GlnBP. SEM observations, FTIR characterization, and the successful flocculation performance indicated that γ-PGA successfully bound to GlnBP. It is worth noting that γ-PGA itself serves a good adsorbent for heavy metals.28,29 Therefore, γ-PGA is definitely multifunctional in MB546-PGA in terms of cell coupling, flocculation initiation, and enhanced metal ion adsorption.
A bacterial adsorbent with an easy flocculation operation not only facilitates the recovery of metals but also extends the scope of potential applications, such as applications in still polluted or waste waters. Currently, the flocculation of MB546-PGA following Cu2+ adsorption was successfully conducted by adding the environmentally compatible and economically available CaCl2 in laboratory trials; however, the feasibility of this system must be validated in the naturally occurring real polluted water bodies where various metals and pollutants coexist and could cause interference in this system. The development of a system based on MB546-PGA that could be applied to real metal-polluted waters will be our next primary research goal.
Conclusions
This study reports a new biocomposite adsorbent that is capable of adsorbing Cu2+ at a high capacity and that induced flocculation following Cu2+ adsorption in aqueous solutions. Using cell surface display technology, a recombinant P. putida stain was constructed that enabled the expression of the fusion protein with two tandemly repeated InaQ-N anchors, the glutamine-binding protein GlnBP and two cyanobacterial metallothionein SmtA repeats on the cell surface. Through γ-PGA-mediated covalent linkages, a triple biocomposite MB546-PGA consisting of “Cell-γ-PGA-Cell surface SmtA” was formed. The Cu2+ adsorption activity could be performed over a pH range of 3.5–5.5 and a relatively broad temperature range of 20–60 °C, and the activity rapidly reached equilibrium within 30 min. The maximum adsorption capacity for Cu2+ reached 145.99 mg g−1. This biocomposite exhibited easy and efficient flocculation induced by CaCl2. Infrared analysis indicated that the hydroxyl and amino groups of MB546-PGA were involved in the adsorption of Cu2+, whereas the carbonyl and amino groups of γ-PGA were bound to P. putida cells and were involved in Cu2+ uptake.Experimental section
Chemicals, bacterial strains, plasmids, and culture conditions
Industrial-grade γ-PGA (Na+ type) was purchased from SaiTaisi Biotechnology Co., Ltd. (Nanjing, China) and was used to construct the composite adsorbent. According to the manufacturer's quality inspection report, this γ-PGA product was a water-soluble homo-polypeptide polymerized by amide bonds on the γ-positions of mixed L-glutamic acid and D-glutamic acid monomers, with a polymerization degree ranging between 1000–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, a Mr of ≥1000 kDa, pH 5.0–7.0, and a total purity of ≥92%. N-Hydroxysuccinimide (NHS) and 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDC) were purchased from Adamas Reagent, Ltd. (Shanghai, China); 2-(N-morpholino) ethanesulfonic acid (MES) was purchased from Hefei New Source Biological Technology Co., Ltd. (Hefei, China). The wild-type P. putida Migula CCTCC (China Center for Type Culture Collection) AB92012 was used as the host strain for the construction of the surface display system. E. coli DH5α (TaKaRa Bio Inc.) was used as a host strain for constructing the recombinant plasmids. The genome of Bacillus thuringiensis BMB171 (ref. 30) (Microbial Genetic Stock Center, Wuhan, China) was the source of the glnBP gene that encodes the glutamine-binding protein.
000, a Mr of ≥1000 kDa, pH 5.0–7.0, and a total purity of ≥92%. N-Hydroxysuccinimide (NHS) and 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDC) were purchased from Adamas Reagent, Ltd. (Shanghai, China); 2-(N-morpholino) ethanesulfonic acid (MES) was purchased from Hefei New Source Biological Technology Co., Ltd. (Hefei, China). The wild-type P. putida Migula CCTCC (China Center for Type Culture Collection) AB92012 was used as the host strain for the construction of the surface display system. E. coli DH5α (TaKaRa Bio Inc.) was used as a host strain for constructing the recombinant plasmids. The genome of Bacillus thuringiensis BMB171 (ref. 30) (Microbial Genetic Stock Center, Wuhan, China) was the source of the glnBP gene that encodes the glutamine-binding protein.
All plasmids used in this study are shown in Table S6.† All strains were routinely cultured in lysogeny broth (LB) medium according to standard cultivation procedures.31 Recombinant E. coli cells were grown at 37 °C in LB broth containing 100 μg mL−1 (final concentration) ampicillin (Amp), whereas recombinant P. putida cells were grown at 28 °C in LB broth containing 500 μg mL−1 (final concentration) carbenicillin (Cb).
Construction of the recombinant plasmids
All polymerase chain reaction (PCR) primers used to construct the various recombinant plasmids are listed in Table S6.† Total bacterial and plasmid DNA were prepared according to standard methods.32 The construction of all recombinant plasmids is schematically illustrated in Fig. S5.† Briefly, an 846-bp glnBP gene was amplified by PCR from the B. thuringiensis BMB171 genome with the primers glnBP-F (BglII site underlined) and glnBP1-R (EcoRI site underlined). The amplified fragment was digested with BglII and EcoRI, followed by ligation into the BglII/EcoRI site of a previously digested pMB109 plasmid, yielding the recombinant pMB541 plasmid harboring the fusion gene (inaQ-N)2–glnBP. Using inaQ-N-F (NcoI site underlined) and glnBP2-R (PstI site underlined) as the primers, an 1894-bp fusion gene (inaQ-N)2–glnBP fragment was amplified by PCR from the pMB541 plasmid. The resulting fragment was digested with NcoI and PstI and was inserted into NcoI/PstI sites of the pTrcHis B E. coli expression vector to yield the 6185-bp recombinant pMB542 plasmid. PCR amplification was performed using primers smtA-F (KpnI site underlined) and smtA-R (EcoRI site underlined) to obtain two tandemly aligned smtA repeats from the pYN2S plasmid. After digesting this fragment with KpnI/EcoRI, it was ligated into the KpnI/EcoRI site of pMB542 to yield a 6525-bp recombinant pMB544 plasmid. Finally, pMB544 was digested with NcoI/EcoRI, and the 2252 bp digested fragment was inserted into the NcoI/EcoRI sites of the Pseudomonas expression vector pYNP to yield a 7124-bp recombinant plasmid pMB545, which harbored the fusion gene (inaQ-N)2–glnBP–(smtA)2.E. coli were transformed using standard protocols.31 Recombinant E. coli strains harboring the recombinant pMB541, pMB542, pMB544, and pMB545 plasmids were named E. coli MB541, MB542, MB544, and MB545, respectively. P. putida AB92019 were transformed by electroporation using a previously described protocol.33 Recombinant P. putida AB92019 harboring the recombinant plasmid pMB545 was designated as P. putida MB546.
Cell fractionation and preparation of cell surface proteins
P. putida MB546 cells were grown at 28 °C for 24 h and the cells were collected to prepare the fractionated samples following a previously described method.34 Equal volumes of each fractionated sample were subjected to SDS-PAGE and Western blot assays.SDS-PAGE and Western blot analyses
P. putida MB546 cells and fractionated samples, as described above, were subjected to SDS-PAGE analysis on a 10% separation gel using a previously described method.35 For the fractionated samples, Western blot analyses were further performed to verify the expression of the (InaQ-N)2–GlnBP–(SmtA)2 fusion protein using polyclonal InaQ-N antiserum as the primary antibody and horseradish peroxidase-conjugated goat anti-mice IgG (Chemicon, USA) as the secondary antibody, according to previously described procedures.36Immunofluorescence microscopy and FACS analysis
Immunofluorescence microscopy and FACS analysis of the recombinant P. putida MB546 cells were performed according to a previously described method,37 except that the polyclonal InaQ-N antiserum was used as the primary antibody. FACS measurements were recorded as the percentage of total fluorescence-labeled cells relative to the total Cy5 fluorescence.SEM
Morphological observations of P. putida MB546 cells and the biocomposite adsorbent MB546-PGA were conducted using a JSM-6390/LV SEM (NTC, Japan). The sample preparation procedures, including glutaraldehyde fixing, ethanol dehydration, freeze drying, and gold-plating the specimens, were performed according to the manufacturer's instructions.Construction of MB546-PGA by conjugating MB546 and γ-PGA
The biocomposite MB546-PGA was conjugated with the activated carboxyl side chains of γ-PGA and the surface-active groups of MB546 cells. A single LB-plate colony of a P. putida MB546 culture was incubated in LB broth and grown at 28 °C with shaking at 150 rpm for 12 h. The inoculum (1%, v/v) was inoculated into a 500 mL Erlenmeyer flask loaded with 200 mL of LB broth, and the culture was grown at 28 °C with shaking at 200 rpm for 24 h. The cells were collected by centrifugation (8000 rpm for 10 min), washed twice with PBS (pH 7.0), and resuspended to an OD600 of 1.5 with ddH2O, which was subsequently used for conjugation with γ-PGA. Each 50.0 mL aliquot of the resuspended cells was desiccated at 180 °C for 2 h to weigh the net biomass.The conjugation of MB546 cells and γ-PGA was performed using the following procedures. Firstly, an appropriate amount of γ-PGA was dissolved in the activation buffer (0.1 M MES and 0.5 M NaCl, pH 6.0) to a final concentration of 1 mg mL−1 (i.e., Solution I), and 0.4 mg of EDC (final concentration of 2 mM) and 0.6 mg of NHS (final concentration 5 mM) that had previously been equilibrated to room temperature (RT) were sequentially added to Solution I. The mixed solution was incubated at RT with shaking at 60 rpm for 15 min, and 1.4 μL of β-mercaptoethanol (final concentration of 20 mM) were added to stop the reaction to yield activated Solution I. Secondly, an appropriate amount of the resuspended P. putida MB546 cells, which was equivalent to 6 mg mL−1 dry weight, was added to the coupling buffer (100 mM phosphate and 150 mM NaCl, pH 7.2) to yield Solution II. Finally, equal volumes of activated Solution I and Solution II were mixed, incubated at RT with shaking at 60 rpm for 2 h, and centrifuged at 8000 rpm for 1 min to obtain the biocomposite MB546-PGA. This biomaterial was washed three times with ddH2O, then diluted to a unit concentration equivalent to 6 mg mL−1 dry weight of MB546 cells with ddH2O, and stored at 4 °C until use. For use as the control, P. putida AB92019 cells were treated in parallel to prepare the composite material named AB92019 + PGA.
Biosorption experiments
Unless otherwise specified, the Cu2+ adsorption through MB546-PGA was conducted at an initial Cu2+ concentration of 32 mg L−1 in a shaker-flask at 60 rpm. The experiments were conducted by varying the adsorption time (0–30 min), temperature (20–60 °C), pH (3.5–5.5), and MB546-PGA loading amount (equivalent to 0.06–1.8 mg mL−1 dry weight of MB546 cells) to investigate the optimal biosorption conditions. After the biosorption reaction, the supernatants were collected via centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 s. The residual Cu2+ contents of the supernatants were determined using an atomic absorption spectrophotometer (HITACHI 180-80, Japan). The residual Cu2+ contents were used to calculate the absolute Cu2+ removal efficiency using the following equation:
000 rpm for 30 s. The residual Cu2+ contents of the supernatants were determined using an atomic absorption spectrophotometer (HITACHI 180-80, Japan). The residual Cu2+ contents were used to calculate the absolute Cu2+ removal efficiency using the following equation:
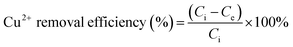 | (5) |
Kinetics and isotherm analysis
The biosorption experiments were performed with different initial Cu2+ concentrations ranging from 6.4 mg L−1 to 128.0 mg L−1 under the optimized reaction conditions (i.e., pH 5.5, 30 °C and 30 min). After the biosorption reaction equilibrium was reached, the residual Cu2+ content in the corresponding supernatant from each initial Cu2+ concentration was measured using the method described above. The equilibrium of the Cu2+ adsorption capacity (qe, mg g−1) was plotted, and the adsorption isotherm curves were established. Adsorption capacity was calculated using the following equation:
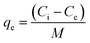 | (6) |
Desorption experiments
A Cu2+ solution with an initial concentration of 64 mg L−1 was used for the adsorption/desorption experiments with MB546-PGA at an equivalent loading dry weight of 0.6 mg mL−1. The adsorption experiments were conducted under the reaction conditions of pH 5.5 and 30 °C and were agitated at 60 rpm for 30 min. The biosorbents were harvested, washed three times with sterile ddH2O, and resuspended in a pH 2.5 solution. After shaking at 60 rpm for 30 min, the biosorbents were harvested and washed with a large amount of sterile deionized water before being used for a second round of adsorption/desorption experiments. The residual or redissolved Cu2+ contents in the corresponding solutions were measured using the method described above. The desorption experiments were performed in triplicate, and the adsorption capacity was calculated according to eqn (6), whereas the Cu2+ desorption capacity (qd, mg g−1) was calculated using the following equation:
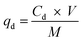 | (7) |
Flocculation experiments
The flocculation activity of MB546-PGA induced by ddH2O, γ-PGA and CaCl2 was recorded by measuring the OD600 of supernatant following the flocculation reaction. Each 20 mL MB546-PGA suspension (with a biomass concentration equivalent to 0.6 mg mL−1 dry weight of MB546 cells) was mixed with ddH2O, 1 g L−1 (final concentration) of γ-PGA, 0.04 M (final concentration) CaCl2, or a combination of γ-PGA and CaCl2. After stirring briefly, the mixed solutions were placed at RT for 30 min to monitor the occurrence of flocculation. For the control experiments, the flocculation induced by the unconjugated P. putida MB546 and P. putida AB92019 cells was examined in parallel.FTIR
The infrared spectra of MB546-PGA before and after Cu2+ adsorption, as well as the free or γ-PGA-bound P. putida MB546 cells, were analyzed using an FTIR spectrometer (Spectrum One, Perkin-Elmer) within the 400 to 4000 cm−1 range. The samples were generally prepared using previously described procedures.26Data analysis
All data were averaged from triplicate assays. The statistical analyses were performed using SPSS 17.0 statistical software. Statistical significance was defined as a P-value less than 0.05.Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31270158 and 31570123), the National Key Research and Development Program of China (Program No. 2016YFD0800702), and the Fundamental Research Funds for the Central Universities (Grant number 2662015PY189).Notes and references
- M. Bilal, J. A. Shah, T. Ashfaq, S. M. Gardazi, A. A. Tahir, A. Pervez, H. Haroon and Q. Mahmood, J. Hazard. Mater., 2013, 263, 322–333 CrossRef CAS PubMed.
- J. Y. Tseng, C. Y. Chang, C. F. Chang, Y. H. Chen, C. C. Chang, D. R. Ji, C. Y. Chiu and P. C. Chiang, J. Hazard. Mater., 2009, 171, 370–377 CrossRef CAS PubMed.
- B. Dhir, Environ. Sci. Pollut. Res., 2014, 21, 1614–1627 CrossRef CAS PubMed.
- J. Wang and C. Chen, Biotechnol. Adv., 2009, 27, 195–226 CrossRef CAS PubMed.
- R. Gupta and H. Mohapatra, Indian J. Exp. Biol., 2003, 41, 945–966 CAS.
- Y. A. Yahaya, M. Mat Don and S. Bhatia, J. Hazard. Mater., 2009, 161, 189–195 CrossRef CAS PubMed.
- J. Wang and C. Chen, Bioresour. Technol., 2014, 160, 129–141 CrossRef CAS PubMed.
- S. M. Lee, Lalhmunsiama, S. I. Choi and D. Tiwari, Environ. Sci. Pollut. Res., 2013, 20, 7464–7477 CrossRef CAS PubMed.
- K. Tsekova, D. Todorova, V. Dencheva and S. Ganeva, Bioresour. Technol., 2010, 101, 1727–1731 CrossRef CAS PubMed.
- H. Ni, Z. Xiong, T. Ye, Z. Zhang, X. Ma and L. Li, Chem. Eng. J., 2012, 204–206, 264–271 CrossRef CAS.
- J. M. Buescher and A. Margaritis, Crit. Rev. Biotechnol., 2007, 27, 1–19 CrossRef CAS PubMed.
- A. Ogunleye, A. Bhat, V. U. Irorere, D. Hill, C. Williams and I. Radecka, Microbiology, 2015, 161, 1–17 CrossRef CAS PubMed.
- Z. Luo, Y. Guo, J. Liu, H. Qiu, M. Zhao, W. Zou and S. Li, Biotechnol. Biofuels, 2016, 9, 134 CrossRef PubMed.
- B. Manocha and A. Margaritis, Crit. Rev. Biotechnol., 2008, 28, 83–99 CrossRef CAS PubMed.
- I. B. Bajaj and R. S. Singhal, Food Bioprocess Technol., 2009, 4, 745–752 CrossRef.
- S. W. Chang, J. F. Shaw, K. H. Yang, S. F. Chang and C. J. Shieh, Bioresour. Technol., 2008, 99, 2800–2805 CrossRef CAS PubMed.
- Y. J. Sun, J. Rose, B. C. Wang and C. D. Hsiao, J. Mol. Biol., 1998, 278, 219–229 CrossRef CAS PubMed.
- F. A. Quiocho, Philos. Trans. R. Soc., B, 1990, 326, 341–351 CrossRef CAS.
- K. Kuroda and M. Ueda, Curr. Opin. Biotechnol., 2011, 22, 427–433 CrossRef CAS PubMed.
- M. Vasak and D. W. Hasler, Curr. Opin. Chem. Biol., 2000, 4, 177–183 CrossRef CAS PubMed.
- J. Shi, W. P. Lindsay, J. W. Huckle, A. P. Morby and N. J. Robinson, FEBS Lett., 1992, 303, 159–163 CrossRef CAS PubMed.
- Q. Li, Q. Yan, J. Chen, Y. He, J. Wang, H. Zhang, Z. Yu and L. Li, Int. J. Biol. Sci., 2012, 8, 1097–1108 CrossRef CAS PubMed.
- T. Ye, C. Yao, Q. Li, H. Zhang and L. Li, Chin. J. Appl. Environ. Biol., 2008, 14, 528–533 CAS.
- Z. Grabarek and J. Gergely, Anal. Biochem., 1990, 185, 131–135 CrossRef CAS PubMed.
- M. Iqbal, A. Saeed and S. I. Zafar, J. Hazard. Mater., 2007, 148, 47–55 CrossRef CAS PubMed.
- K. Akhtar, A. M. Khalid, M. W. Akhtar and M. A. Ghauri, Bioresour. Technol., 2009, 100, 4551–4558 CrossRef CAS PubMed.
- C. D. Hsiao, Y. J. Sun, J. Rose and B. C. Wang, J. Mol. Biol., 1996, 262, 225–242 CrossRef CAS PubMed.
- B. S. Inbaraj, C. P. Chiu, G. H. Ho, J. Yang and B. H. Chen, J. Hazard. Mater., 2006, 137, 226–234 CrossRef CAS PubMed.
- J. Yao, H. Xu, J. Wang, M. Jiang and P. Ouyang, J. Biomater. Sci., Polym. Ed., 2007, 18, 193–204 CrossRef CAS PubMed.
- J. He, X. Shao, H. Zheng, M. Li, J. Wang, Q. Zhang, L. Li, Z. Liu, M. Sun, S. Wang and Z. Yu, J. Bacteriol., 2010, 192, 4074–4075 CrossRef CAS PubMed.
- J. Sambrook and D. W. Russell, Molecular cloning: a laboratory manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., New York, 3rd edn, 2001 Search PubMed.
- F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith and K. Struhl, Current protocols in molecular biology, John Wiley & Sons, Inc., New York, 5th edn, 2002 Search PubMed.
- K. Iwasaki, H. Uchiyama, O. Yagi, T. Kurabayashi, K. Ishizuka and Y. Takamura, Biosci., Biotechnol., Biochem., 1994, 58, 851–854 CrossRef CAS PubMed.
- L. Li, D. G. Kang and H. J. Cha, Biotechnol. Bioeng., 2004, 85, 214–221 CrossRef CAS PubMed.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
- Q. Li, H. Ni, S. Meng, Y. He, Z. Yu and L. Li, J. Microbiol. Biotechnol., 2011, 21, 1330–1335 CrossRef CAS PubMed.
- Q. Li, Z. Yu, X. Shao, J. He and L. Li, FEMS Microbiol. Lett., 2009, 299, 44–52 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra01546a |
| This journal is © The Royal Society of Chemistry 2017 |