DOI:
10.1039/C7RA01845B
(Paper)
RSC Adv., 2017,
7, 18724-18744
Phosphonium carbosilane dendrimers for biomedical applications – synthesis, characterization and cytotoxicity evaluation†
Received
14th February 2017
, Accepted 21st March 2017
First published on 28th March 2017
Abstract
We report the synthesis and cytotoxicity evaluation of a completely new class of cationic carbosilane dendrimers functionalized with several different phosphonium peripheral groups and an ammonium functionalised one as a reference. The carbosilane dendrimers with NMe3, PMe3, P(Et2)2(CH2)3OH, PBu3, P(C6H4-OMe)3 and P(Ph)3 peripheral substituents were synthesized, thoroughly characterized and modelled by computer simulations. The cytotoxicities of the dendrimers were investigated in vitro on three model cell lines (B14, BRL and NRK cells) by MTT and CV assay methods. Generally, the cytotoxicities of PMe3 carbosilane dendrimers were similar or slightly lower when compared with NMe3 dendrimers. The substitution of methyl groups in PMe3 carbosilane dendrimers with more hydrophobic and bulky alkyl substituents (PBu3 and P(Et2)2(CH2)3OH dendrimers) resulted in an increase of cytotoxicity. The P(C6H4-OMe)3 dendrimer showed exceptionally low cytotoxicity across all cell lines or assay methods used. Generally, phosphonium carbosilane dendrimers could represent a valuable alternative to ammonium ones in gene therapy applications due to comparable or lower cytotoxicities, the presence of positive charge for nucleic acid electrostatic binding and in the cases of P(C6H4-OMe)3 and P(Ph)3 dendrimers high potential of mitochondrial targeting.
1. Introduction
Recently, numerous types of drug delivery systems (DDS) have been studied for nonviral gene delivery to treat various genetic diseases such as cystic fibrosis, haemophilia, cancer or several types of infection diseases.1 Cationic polymers and lipids, able to interact by electrostatic interactions with negatively charged nucleic acids are used for the formation of dendriplexes and lipoplexes. Such nanoparticles stabilize DNA or RNA, protect them from degradation by nucleases, enable their cellular uptake by endocytosis and finally cause endosomal escape finished by release of genetic material into the cytoplasmic space or nucleus.2 Among the other polymeric systems, cationic dendrimers (DDMs) such as the commercially available poly(amidoamine) (PAMAM) dendrimers have been shown to be effective for in vitro cell transfection.3–6 Up to now, most of the dendrimer-based gene delivery systems were surface modified by nitrogen-containing cationic groups including ammonium cations.7
Despite their successful in vitro application, the toxicity of positively charged amine groups is the main obstacle for in vivo use of dendrimers and the other cationic vectors as gene delivery vehicles.7 Several different approaches were presented, which address this problem by surface engineering of dendrimers with low-toxic outer shell, shielding the positively charged groups present in the inner layer of dendrimer. Modification of dendrimers with poly(ethylene)glycol,7–10 carbohydrates,11–14 amino acids,15 acetyls16 or by attaching targeting ligands (peptides, proteins)17 was described and their resulting lower cytotoxicity compared with unmodified cationic dendrimers demonstrated. On the other hand, a compromise between the toxicity, ability to complex nucleic acids and transfection efficacy of such nanoparticles has to be always considered.
Apart from surface functional groups, the core and branching units of dendrimer can also contribute to the overall toxicity. New less toxic DDMs having biodegradable core or less cytotoxic branching units were therefore synthesized. As an example, the polyester,18 polyether imine,19 phosphate20 or triazine21,22 low-toxic DDMs were synthesized. Another promising DDS candidates of this group are carbosilane dendrimers (CS-DDMs).11,23–28 For example, low toxicity of the second generation ammonium-terminated CS-DDMs was proven by several complementary approaches.27 It was also shown that these cationic DDMs favourably interact with anti-HIV oligodeoxyribonucleotides and form dendriplex nanoparticles, suitable for cell transfection.23 The siRNA delivery into the brain by carbosilane dendrimers was also reported.28
Beside this, much less effort has been focused on investigations of dendrimer modifications by other types of positively charged groups, like e.g. phosphonium or arsenium cationic moieties. As was already demonstrated, such cationic groups are able to substitute primary amines or ammonium groups in lipid or polymer-based transfectant molecules with significantly lower cytotoxicity and higher transfection efficiency.29–31 As an example, phosphonium- and arsenium-containing lipids have shown ability to bind DNA and to mediate higher gene transfection with lower cytotoxicity in comparison with the ammonium-based analogs in vitro and in vivo32–34. Similarly, phosphonium-containing AB diblock copolymers, which efficiently complexed pDNA to generate core–shell nanoparticles were also reported. Resulted polyplexes demonstrated HepaRG cells specific transfection and low cytotoxicity.29 The same group of authors also systematically investigated the role of variable length of alkyl substituent attached to the phosphonium cation in DNA complexation and transfection efficiency of different cell lines.31 The synthesis of phosphonium polymeric material for nucleic acid transfection consisting of polyacrylate polymer backbone modified with phosphonium groups has been reported.30 Further on, in agreement with other studies,29 systematic investigation has shown important influence of different types of alkyl substituents on the phosphonium cations on transfection efficiency and toxicity of polyplexes.30
Despite the proved benefits in other types of transfection materials, there exist only few attempts to synthesize dendrimers with phosphonium surface groups and to use them for gene delivery applications.35–37 Most of the work done was based on surface group modification of PAMAM dendrimers. As an example, mitochondria-targeted dendrimer was synthesized via the acid-amine-coupling conjugation reaction between the acid group of (3-carboxypropyl)triphenylphosphonium bromide and the primary amines of the acetylated generation 5 poly(amidoamine) (PAMAM) dendrimer. Resulting triphenylphosphonium (TPP)-anchored dendrimer was efficiently taken up by the cells and demonstrated mitochondrial targeting with greater cell viability on fibroblast cells (NIH-3T3) as compared to unmodified PAMAM-NH2 dendrimer.37 In the similar way, an efficient HeLa and COS-7 cells transfection with EGFP and luciferase reporter gene expression plasmids has been shown. Transfection efficacy was improved in comparison with commercial transfectants Lipo2000 and unmodified dendrimers along with decreased cytotoxicity.36 Apart from those two literature examples, which focused solely on TPP surface modification of PAMAM dendrimers, no systematic comparative investigation of different types of phosphonium groups on dendrimer cytotoxicity and eventually, transfection efficacy has been presented.
Herein we present synthesis and characterization of completely new class of generation 1–3 carbosilane dendrimers surface modified with various types of phosphonium groups and one type of ammonium terminated CS-DDM for comparison. The core of carbosilane dendrimers was synthesized based on our previous work on synthesis of CS-DMMs functionalized with titanocene moieties.38–41 Ammonium and phosphonium groups were connected by quaternization of trimethyl amine or an appropriate phosphine by iodopropyl-terminated dendrimers. To manifest their potential in biomedical applications, we have performed comparative in vitro cytotoxicity studies on three model cell lines.
2. Results and discussion
2.1 Synthesis and characterization of dendrimers
The cationic dendrimers were prepared by the sequence of reactions as shown in Scheme 1. In the first step carbosilane dendrimers with 3-chloropropyl terminal groups were synthesized via hydrosilylation of allyl-terminated starting compounds by (3-chloropropyl)dimethylsilane. The reactions were performed in Schlenk tubes using pentane as solvent and the Karstedt catalyst. The progress of the reactions was monitored by 1H NMR spectroscopy. The most significant signals of the starting compounds are those of the terminal double bonds consisting of one doublet at ca. δ 1.8 and two multiplets at ca. δ 5.4 and 6.0 in 1H NMR spectra and three resonances at ca. δ 22.9, 128.3, and 143.1 in 13C NMR spectra. Disappearance of these signals indicates the completeness of reaction. No side products resulting from undesired α-hydrosilylation leading to branched alkyl spacers in the structure were detected. Siloxane, which was sometimes observed as the product of hydrolysis followed by condensation of (3-chloropropyl)dimethylsilane was removed by evaporation in high vacuum. The chloropropyl functionality was subsequently transformed to iodopropyl by Finkelstein reaction. The triplets of the methylenes connected to halogens are after the substitution shifted to higher field from 3.50 ppm for chloride to 3.18 ppm for iodide. As the starting uncharged dendrimers 1–6 were not easily ionisable, even using APCI ionization technique, a different approach was employed. The compounds were dissolved in chloroform, a few drops of methanol were added followed by addition of ammonium formate buffer. These solutions were then measured using ESI source in the positive mode resulting in observation of [M + NH4]+ ions. Different synthetic protocol based on hydrosilylation of bromoalkenes by Si–H terminated carbosilane dendrimers was employed by De la Mata et al. for preparation bromo-terminated carbosilane dendrimers with longer spacer.42
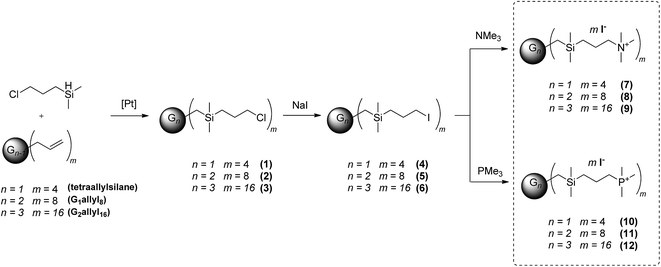 |
| | Scheme 1 Synthesis of trimethylonium salts terminated carbosilane dendrimers. | |
The last step of the synthesis consists in the formation of onium salts on the periphery of dendrimers. The quarternisation reaction with 3-iodopropyl-terminated dendrimers was carried out in acetonitrile with slight excess of trimethylamine or an appropriate phosphine. Insoluble starting hydrophobic dendrimers became gradually soluble in acetonitrile as soon as the number of onium groups created on the periphery was sufficient to transfer compounds into solution. After completion of the reaction, the excess of amine or phosphine was extracted in toluene. Depending on terminal groups, the products were isolated as white solids or liquids having variable viscosity.
In this manner we prepared a series of carbosilane dendrimers decorated by onium salts on the periphery. For ammonium–phosphonium comparative study we synthetized a series of methylated analogues which differ only in cationic centre i.e. nitrogen (7–9) or phosphorus (10–12) atom. As expected, due to the much higher nucleophilicity of phosphines toward carbon compared with amines, quaternization of amines requires longer reaction time to achieve fully substituted product. For ammonium derivatives (7–9) was recently published procedure based on quaternization of amino groups on periphery of dendrimers with an excess of MeI.43
The above described synthetic procedure was followed also in the synthesis of a new family of phosphonium-terminated dendrimers (Scheme 2). Representative groups having diverse environments of cationic centres were designed and characterized. The NMR spectroscopic and analytical data for all derivatives are consistent with their proposed structures. The presence of Si(CH2)3P unit in molecule was documented by a doublet of outer silicon atoms in 29Si {1H} NMR, typically with coupling constant 4JSiP ca. 2.5 Hz (Fig. 1). However chemical shifts in 29Si {1H} NMR showed no significant change with respect to the environment of phosphorus atom. 31P {1H} NMR was in this sense more sensitive. Signals occurred in the range from about 21 ppm for methoxyphenyl-terminated dendrimers to 43 ppm for dendrimers 13–15. In the case of all quarternary ammonium or phosphonium salts, the ions were easily observable via electrospray ionisation. Due to the ability of target dendrimer molecules to form multiply charged species, almost all the possible cations were observed in each case (for example, if the dendrimer structure is Dend.PnIn, all the possible [Dend.PnIn−i]i+ ions are formed) (see Tables S1 and S2†). This feature offers an opportunity to observe counteranions in the positive mode, which simplifies the monitoring of I−/Cl− ion exchange reaction leading to the compounds 19–21, 25–27 and 31–33.
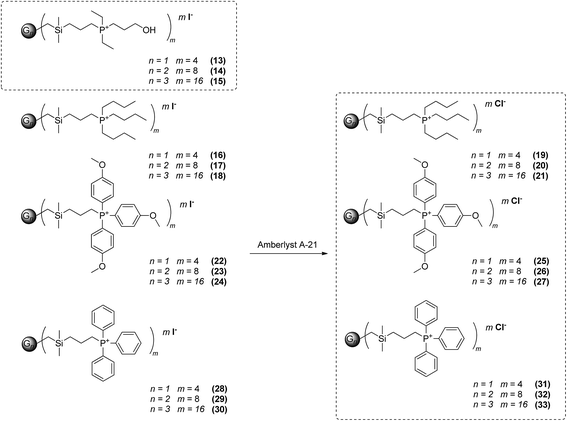 |
| | Scheme 2 Phosphonium carbosilane dendrimers synthesized in this work. | |
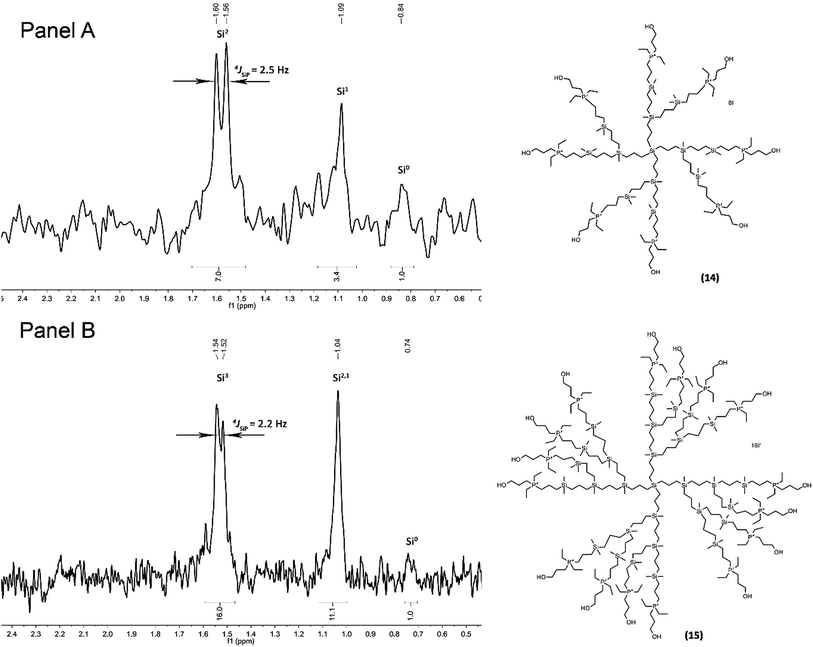 |
| | Fig. 1 Dendrimer structure and 29Si {1H} NMR. Panel A: compound 14; panel B: compound 15. | |
Solubility in aqueous solutions is a critical parameter for applicability of dendrimers in biological systems and their use in biomedical applications. Because of the inherent hydrophobicity of the interior of carbosilane dendrimers the appropriate solubility behaviour can be achieved by targeted design of peripheral groups. The solubility of onium salts is known to be strongly dependent on counter-anion,44 therefore ion exchange was suggested as a useful method to enhance solubility in water. Dendrimers containing butyl (16–18), methoxyphenyl (22–24) and phenyl (28–30) substituents showed only negligible solubility in water when the counter-anion was the iodide ion despite being well soluble in polar aprotic solvents such as DMSO, acetonitrile, but also in methanol. To enhance water solubility of poorly soluble compounds we exchanged the iodide anions for chloride using ion exchange resign with the result of increasing the solubility in water considerably. For example solubility of all dendrimers with butyl groups was increased from <5 mg mL−1 for derivatives with iodine to >100 mg mL−1 for those with chloride anion. Table S3† summarizes typical values of solubility of prepared compounds in water.
2.2 Computer modelling of dendrimers
To obtain detailed information about the shape, size, space distribution of peripheral groups etc. of all dendrimers in water, their computer simulation models were studied using molecular dynamics in explicit water. Models show that higher generation dendrimers maintain spherical shape (Fig. 2 panel A). Calculated radii of gyration (Rg) and “geometric radii” (Rmax) are shown in Table 1. The sizes of NMe3 and PMe3 dendrimers, which have the same structure except terminal (quaternized) atoms (N or P), are almost identical in all generations. The other structures are slightly bigger than those two structures P(C6H4-OMe)3 dendrimer being the biggest.
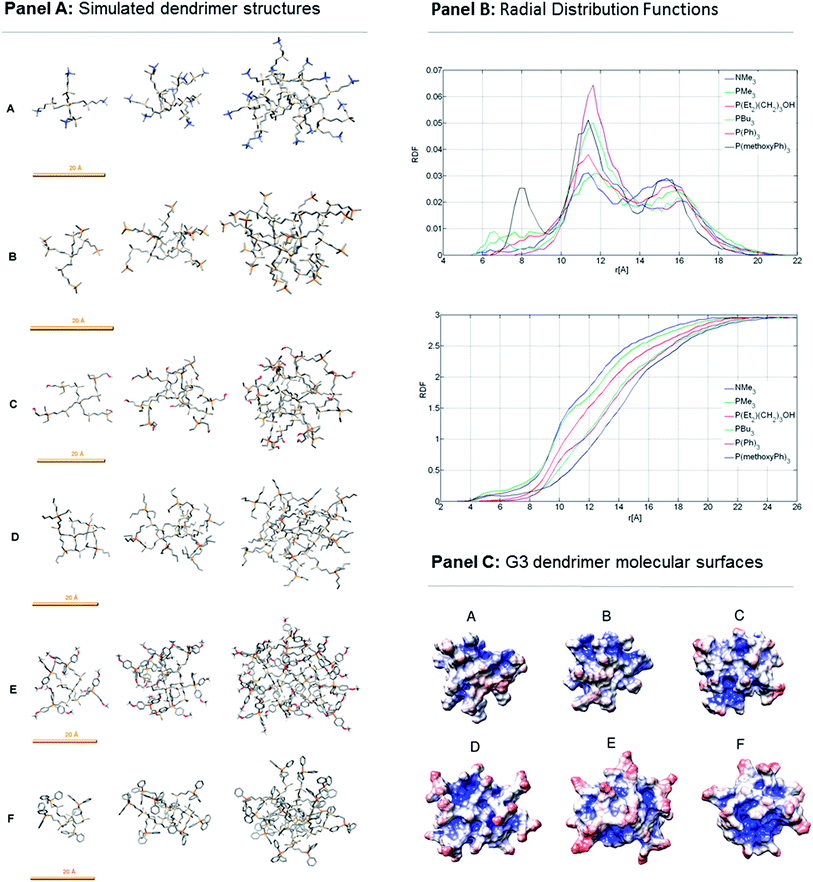 |
| | Fig. 2 Computer models of dendrimer structures and their characteristics. Panel A: Simulated dendrimer structures. The first (G1, left), second (G2, middle) and third (G3, right) generations are shown. (A) NMe3; (B) PMe3; (C) P(Et2)2(CH2)3OH; (D) PBu3; (E) P(C6H4-OMe)3; (F) P(Ph)3. Hydrogens are omitted (except those attached to oxygens or those belonging to terminal methyl groups attached to oxygens) for better clarity. Orange scale bar denotes length 20 Å. Colours: C – grey; O – red; H – white; Si – beige; N – blue; P – orange. Panel B: Radial distribution functions (relative density profiles, RDF) of terminal P(N) atoms with respect to central Si atom in case of 3rd generation dendrimers (upper graph); RDF of water with respect to central Si atom (just G3, lower graph). Panel C: simulated dendrimer structures (just G3) with molecular surface shown, which is coloured based on electrostatic potential. (A) NMe3; (B) PMe3; (C) P(Et2)2(CH2)3OH; (D) PBu3; (E) P(C6H4-OMe)3; (F) P(Ph)3. Red – lower potential (0 V and less); white – potential 0.063 V; blue – higher potential (0.127 V and higher). Influence of water, Na+ and Cl− is taken implicitly in an account (APBS calculation). | |
Table 1 The size characteristics of dendrimers. Rg – radius of gyration of the simulated dendrimer structures, Rmax – maximal distance of the dendrimer atoms from the dendrimer center of geometry (for spherical molecules estimate of their radius). In each cell of the table the average value and standard deviation is presented
| |
NMe3 |
PMe3 |
P(Et2)2(CH2)3OH |
PBu3 |
P(C6H4-OMe)3 |
P(Ph)3 |
| Rg [Å] |
| G3 |
10.80 ± 0.21 |
10.83 ± 0.20 |
11.94 ± 0.25 |
12.47 ± 0.21 |
13.10 ± 0.20 |
12.66 ± 0.17 |
| G2 |
8.80 ± 0.28 |
8.98 ± 0.36 |
9.86 ± 0.34 |
10.28 ± 0.40 |
10.23 ± 0.23 |
10.02 ± 0.28 |
| G1 |
6.98 ± 0.31 |
7.27 ± 0.30 |
7.91 ± 0.56 |
8.13 ± 0.41 |
8.04 ± 0.29 |
7.90 ± 0.42 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| Rmax [Å] |
| G3 |
19.77 ± 1.21 |
19.70 ± 0.90 |
21.58 ± 1.20 |
22.95 ± 1.16 |
24.70 ± 0.92 |
22.41 ± 0.96 |
| G2 |
16.37 ± 1.25 |
16.59 ± 1.23 |
17.82 ± 1.15 |
19.33 ± 1.38 |
19.53 ± 0.63 |
18.25 ± 1.11 |
| G1 |
12.36 ± 0.79 |
12.79 ± 0.74 |
13.77 ± 1.11 |
15.07 ± 1.16 |
15.52 ± 0.85 |
14.12 ± 0.84 |
Radial distribution functions of dendrimer atoms and water with respect to the central Si atom were calculated for the 3rd generation (G3) dendrimers. Density analysis of terminal P(N) atoms revealed more pronounced back-folding in case of PMe3 dendrimer (ca. 6–9 Å area from central Si atom) compared with NMe3 which has higher density in more distant area (ca. 13–16 Å). Otherwise the density profiles of those two dendrimers are quite similar. Furthermore, the density profiles have rather similar shape in all dendrimers showing one main peak around ca. 11.5 Å and one secondary peak at ca. 15.5 Å distance from the central Si atom. P(C6H4-OMe)3 dendrimer has one additional remarkable peak at 8 Å indicating back-folded branch/es (see Fig. 2 panel B, upper graph). Radial density profiles of water with respect to central Si atom are shown at Fig. 2 panel B, lower graph. Dendrimers with the most hydrated interior are NMe3 and PMe3. NMe3 is slightly more hydrated than PMe3 dendrimer. P(C6H4-OMe)3 dendrimer has the smallest water density in majority of its interior when compared with other dendrimer types.
Fig. 2 panel C shows molecular surfaces of dendrimers (G3) coloured according to their electrostatic potential. As presented in Table S4 (see ESI†) the average value of surface potential is very similar (around 0.14 V) for all structures but there are more significant differences in the range of electrostatic potential values (min, max) and standard deviation.
2.3 In vitro cytotoxicity evaluation
To determine the potential of biomedical applications of novel class of dendrimers presented, we have further focused on investigation of their cytotoxicity, as one of the most important parameters which influence and limit the practical use of the majority of drug delivery systems. Cytotoxicity profiles of carbosilane dendrimers terminated with different phosphonium and NMe3 functional groups were investigated in vitro on three model cell lines (B14, BRL and NRK cells) with the aim to reflect the potential tissue/cell type related variations. Two methods were used to determine the effect of dendrimers on cell viability. The first one was MTT assay, the method which is based on monitoring of NAD(P)H-dependent cellular oxidoreductase enzymes activity and is therefore related to inhibition of metabolic processes in mitochondria. The other method was the crystal violet (CV) staining assay. This method is based on crystal violet property to stain cell's DNA and is therefore connected to number of cells after cultivation experiment (monitors the inhibition of the cell growth).
With the aim to investigate the main parameters influencing the toxicity profiles of individual dendrimer types, cell viability was plotted in logarithmic concentration scale (Fig. 3) and fitted with four-parameter logistic function (4PL function, R2 > 0.95) using nonlinear regression analysis as described in Experimental section (see Section 4.5). Based on the calculated parameters of each curve, the IC50 values (concentration of dendrimers which inhibits the cell viability to 50% of the control sample) were obtained (Table 2) and used for further evaluation of dendrimer cytotoxicity. Toxicity of dendrimers was studied based on IC50 as variable of dendrimer type, generation, assay method and cell line.
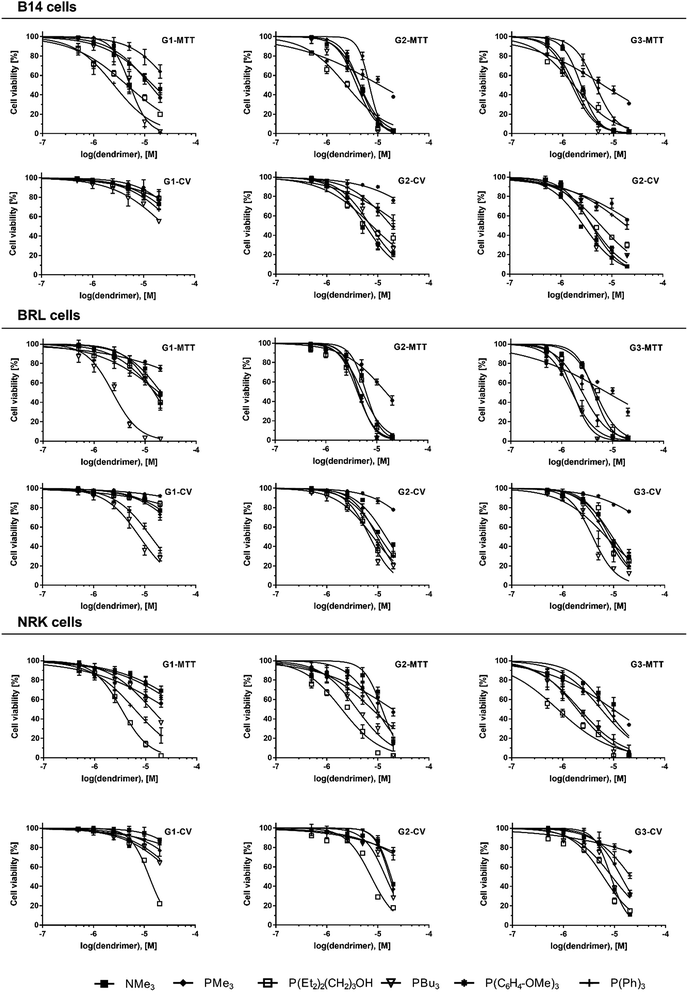 |
| | Fig. 3 Graphical presentation of data obtained from cell viability assays. Measured data from each experimental variant were fitted with four-parameter logistic function (R2 ≥ 0.95). Each graph compares the viability of cells (B14, BRL, NRK cells) exposed to concentration gradient of different dendrimer types (NMe3, PMe3, P(Et2)2(CH2)3OH, PBu3, P(C6H4-OMe)3, P(Ph)3) of the same generation (G1-3) and measured by the same method (MTT or CV). | |
Table 2 IC50 values of dendrimers (given in μM) and standard error of the mean (SEM) derived from the best fit (four-parameter logistic function, R2 ≥ 0.95) of experimental data obtained from cell viability measurements. Values are organized according to the (i) cell line used for toxicity evaluation (B14, BRL, NRK); (ii) toxicity assay method (MTT, CV); (iii) generation of dendrimer (G3, G2, G1); (iv) type of dendrimer (NMe3, PMe3, P(Et2)2(CH2)3OH, PBu3, P(C6H4-OMe)3, P(Ph)3)
| |
NMe3 |
PMe3 |
P(Et2)2(CH2)3OH |
PBu3 |
P(C6H4-OMe)3 |
P(Ph)3 |
| B14-MTT |
| G3 |
2.29 ± 0.041 |
4.43 ± 0.17 |
1.64 ± 0.11 |
1.69 ± 0.08 |
6.23 ± 0.37 |
1.51 ± 0.04 |
| G2 |
4.42 ± 0.11 |
6.78 ± 0.28 |
2.37 ± 0.23 |
3.83 ± 0.18 |
10.81 ± 0.96 |
4.41 ± 0.08 |
| G1 |
14.75 ± 1.36 |
33.18 ± 4.08 |
4.51 ± 0.27 |
5.15 ± 0.22 |
12.75 ± 0.51 |
2.50 ± 0.24 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| B14-CV |
| G3 |
2.81 ± 0.11 |
4.18 ± 0.23 |
6.14 ± 0.39 |
4.04 ± 0.33 |
31.94 ± 6.02 |
19.14 ± 1.73 |
| G2 |
5.70 ± 0.29 |
18.16 ± 1.02 |
7.60 ± 0.60 |
7.13 ± 0.45 |
110.7 ± 28.52 |
28.68 ± 3.34 |
| G1 |
58.7 ± 9.07 |
161.7 ± 32.01 |
153.7 ± 29.85 |
27.1 ± 2.83 |
66.7 ± 15.91 |
42.63 ± 3.47 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| BRL-MTT |
| G3 |
4.29 ± 0.14 |
2.54 ± 0.15 |
4.64 ± 0.24 |
1.68 ± 0.04 |
7.84 ± 0.90 |
1.59 ± 0.08 |
| G2 |
6.06 ± 0.17 |
4.35 ± 0.16 |
5.11 ± 0.32 |
4.39 ± 0.14 |
14.69 ± 0.99 |
4.11 ± 0.15 |
| G1 |
20.82 ± 1.18 |
15.16 ± 0.76 |
15.9 ± 1.31 |
2.34 ± 0.54 |
219.01 ± 38.74 |
19.14 ± 1.79 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| BRL-CV |
| G3 |
9.42 ± 0.36 |
7.42 ± 0.21 |
8.44 ± 0.57 |
3.87 ± 0.20 |
84.8 ± 12.79 |
7.11 ± 0.61 |
| G2 |
14.32 ± 0.71 |
9.68 ± 0.48 |
8.00 ± 0.44 |
6.91 ± 0.43 |
65.8 ± 7.35 |
10.74 ± 0.60 |
| G1 |
81.72 ± 20.93 |
62.06 ± 14.03 |
624.5 ± 83.75 |
7.98 ± 0.46 |
1891.00 ± 258.40 |
13.36 ± 0.41 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| NRK-MTT |
| G3 |
7.41 ± 0.89 |
5.68 ± 0.55 |
0.79 ± 0.08 |
1.92 ± 0.12 |
9.68 ± 0.67 |
2.17 ± 0.15 |
| G2 |
12.07 ± 0.93 |
9.15 ± 0.76 |
1.95 ± 0.14 |
3.90 ± 0.23 |
19.33 ± 2.56 |
7.67 ± 0.48 |
| G1 |
68.45 ± 15.13 |
53.31 ± 13.24 |
3.36 ± 0.13 |
13.04 ± 0.77 |
28.47 ± 3.65 |
6.09 ± 0.36 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| NRK-CV |
| G3 |
8.48 ± 0.25 |
13.44 ± 0.48 |
6.02 ± 0.43 |
8.93 ± 0.59 |
316.3 ± 64.12 |
20.73 ± 1.71 |
| G2 |
18.06 ± 0.68 |
16.91 ± 0.67 |
7.34 ± 0.43 |
14.08 ± 0.72 |
149.8 ± 41.52 |
86.93 ± 8.77 |
| G1 |
106.3 ± 24.26 |
77.54 ± 18.52 |
12.47 ± 0.35 |
42.19 ± 4.58 |
335.6 ± 83.51 |
48.01 ± 7.89 |
2.3.1 Influence of dendrimer peripheral groups on cytotoxicity. As is apparent from Fig. 4 (generation 3) and Fig. S70 and S71† (generation 2, 1) there is no simple relative order of toxicity of individual types of dendrimers which holds for all experimental variants and assay methods investigated. More likely it is possible to recognize some groups or individual types which differ from the rest of the dendrimers according to the specific conditions of experiment. Focusing first on the phosphonium and ammonium dendrimers with methyl groups (NMe3 – dendrimer 7–9 and PMe3 – dendrimer 10–12) quite similar cytotoxic effect with IC50 in the order of 2–50 μM (based on the cell type and generation) was observed by MTT method. This is in good agreement with previously published results27 of NMe3 2nd generation carbosilane dendrimers cytotoxicity with the IC50 measured by MTT assay in the range of 1–10 μM. In comparison with other types of dendrimers, those two dendrimers stay always close together in relative order of toxicity showing slightly less toxic effect (exception being P(C6H4-OMe)3 dendrimer 25–27 as discussed further) on mitochondria (MTT assay), but have very similar effect on inhibition of cell growth with other types of dendrimers (CV assay, IC50 = 2–160 μM based on cell type and dendrimer generation). PMe3 dendrimer was significantly less toxic when compared with NMe3 one on B14 cells. This applies to all generations measured by MTT and in case of G1 also to CV assay. Based on such observations, we may conclude that generally the cytotoxicity of PMe3 is similar or slightly lower when compared with NMe3 dendrimers based on the cell line tested. This is in agreement with the published data focused on comparative studies of phosphonium- and ammonium-modified lipid transfectants.32–34
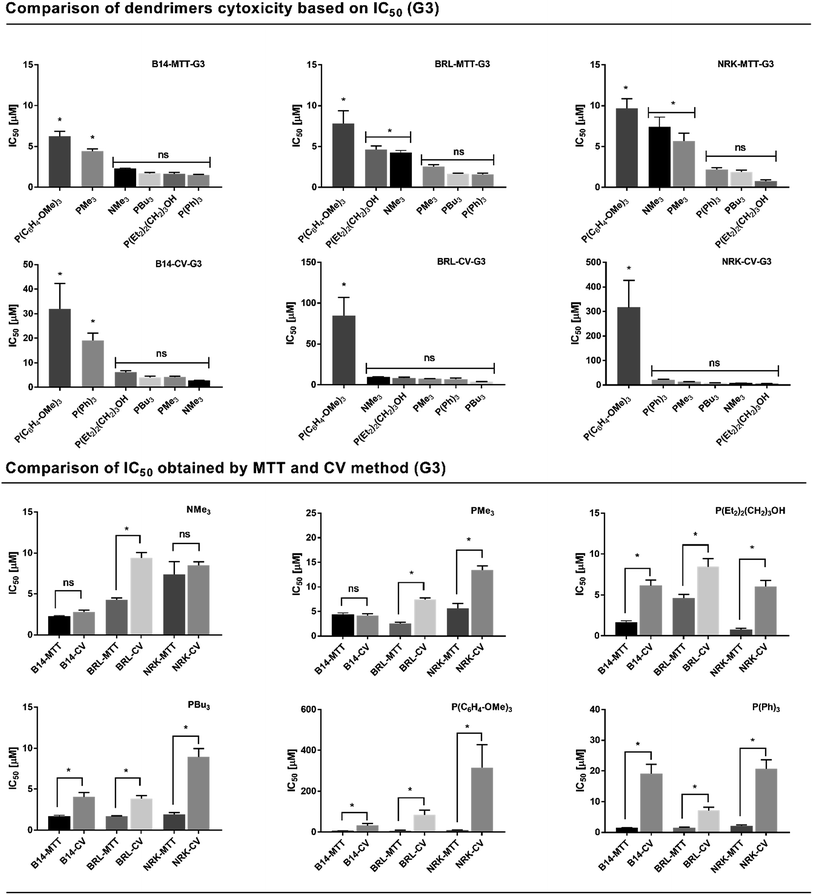 |
| | Fig. 4 Evaluation of G3 dendrimers cytotoxicity based on IC50 values. Upper panel: To evaluate and rank cytotoxic effect of each kind of dendrimer based on comparison of IC50 values summarized in Table 2, data of IC50 of all dendrimer types were plotted in a common column graph for each cell line (B14, BRL, NRK) and assay method used (MTT, CV). Each dendrimer (column) is placed in the order of decreasing mean value of IC50 (from left to right, increasing toxicity). With the aim to determine a statistical significance of difference between the IC50 values of all dendrimers, one-way ANOVA followed with Tukey–Kramer multiply comparison test was used to quantify the p values between each dendrimer. Values p < 0.05 were supposed to be statistically significant (*), values p ≥ 0.05 non-significant (ns). Columns were grouped in the graph based on their p values as compared to lowest IC50 mean value present and p values related to their nearest neighbour. Statistical difference between the grouped dendrimers is therefore p ≥ 0.05 (ns), whereas between groups is p < 0.05 (*). In this way, a relative order of dendrimer toxicity was constructed for each cell line and assay method used. Data are presented as mean ± SEM (n = 3). Lower panel: To compare the difference between the toxicity of G3 dendrimers measured by MTT and CV methods, a set of graphs representing IC50 values of the same dendrimer assayed by both methods on different cell lines were constructed. To determine a statistical significance of difference between the IC50 values measured by MTT and CV method for one cell line, unpaired t-test was used to quantify the p values. Values p < 0.05 were supposed to be statistically significant (*), values p ≥ 0.05 non-significant (ns). Data are presented as mean ± SEM (n = 3). | |
Significant differences in comparison with all other dendrimers were shown by P(C6H4-OMe)3 dendrimer 25–27. In fact, it's very low cytotoxicity was measured across all cell lines, assay methods used and generations (except some small differences of G1 on B14 cells). IC50 calculated for MTT assay were in the range of 6–219 μM, whereas for CV assay those values were shifted up to 66–1890 μM (based on the cell line and dendrimer generation). This exceptionally low cytotoxicity as compared to other dendrimer types in this study falls in the range reported for sugar or even PEG coated dendrimers.7,13,45 For example, Janaszewska et al.46 reported the IC50 value of maltose-modified open shell and dense shell poly(propylene imine) (PPI) dendrimers between 100–145 μM when studied on SKOV3 cell line, contrary to unmodified cationic PPI dendrimers with IC50 < 8 μM. Similarly, Wrobel et al.13 showed the significant haemolytic activity of maltose functionalized hyperbranched poly(ethylene imine)s starting at 100–300 μM concentration depending on density of maltose units on PEI polymer. It is not so straightforward to explain such exceptional properties of P(C6H4-OMe)3 dendrimer. We may hypothesize that it's relatively low cytotoxicity may be possibly related to delocalization of positive charge on phosphorus atoms caused by three electron-donating methoxy groups in para positions capable of conjugation with electrons of aromatic rings and with the phosphonium center. The delocalization of positive charge of outer shell together with larger concentration of charged groups in the interior of dendrimer, caused by significant backfolding of dendrimer branches (as discussed in chapter 3.2) could decrease the toxic interactions with the cell membrane and proteins in cytoplasmic space. Because still being positively charged and therefore able to complex siRNA we suggest that this type of dendrimer could be an interesting alternative to low-toxic pegylated or sugar coated dendrimers for receptor targeted siRNA delivery in vivo.
Interestingly, recognizable large differences in cell viability as measured by MTT and CV assay were in several cases, along with P(C6H4-OMe)3 dendrimer, also observed for P(Ph)3 dendrimer (31–33). This dendrimer is quite toxic when measured by MTT (IC50 < 2.5 μM at G3). On the contrary, the toxicity measured by CV was in some cases (e.g. B14 G3, NRK G2) significantly lower (IC50 > 19 μM for B14-G3 and 86 μM for NRK-G2) when compared with other types of dendrimers ranking it second (after P(C6H4-OMe)3 dendrimer) in line (decreasing order of toxicity, see e.g. Fig. 4, B14-CV-G3). Such results are quite interesting taking in mind a relatively high hydrophobicity of phenyl groups. Moreover, lower toxic influence on the growth of cells as observed in some cases and relatively high toxicity observed by MTT suggests possible preferential targeting of this dendrimer into mitochondria. This, in fact, supports the previously published results on triphenylphosphonium (TPP) modified PAMAM dendrimers.37 Fluorescently labeled PAMAM dendrimers, surface functionalized with TPP were selectively targeted into the mitochondria and showed a decreased overall toxicity to cells as compared to unmodified dendrimer. In fact, it is well known that triphenylphosphonium (TPP), a cation with sufficient lipophilicity and delocalized positive charge, is an efficient mitochondriotropic ligand which promotes accumulation of various TPP conjugated molecules in the mitochondria.47 Thus, despite having no direct evidence, we suggest that P(C6H4-OMe)3 and P(Ph)3 dendrimers may preferentially accumulate in mitochondria causing relatively large differences in toxicity as observed by MTT and CV assay. If this holds true than those two dendrimers could represent a unique drug delivery systems (DDSs) for mitochondrial targeting with relatively low toxicity as compared to other types of comparable systems. Such hypothesis will be verified by further investigations.
The last group of dendrimers which are characterized by quite similar cytotoxic effects are PBu3 and P(Et2)2(CH2)3OH dendrimers (19–21, 13–15). Both are ranked among the most cytotoxic dendrimers in both – MTT and CV assays (see Table 2) with some exception of P(Et2)2(CH2)3OH observed at G1 (B14, BRL – CV assay). The reason for the higher cytotoxicity observed could be the presence of hydrophobic alkyl substituents on phosphonium groups. Interestingly, there is no significant positive effect of –OH groups present on P(Et2)2(CH2)3OH dendrimer on decreasing of cytotoxic effect.
Based on these results, we may conclude that the replacement of methyl substituents on phosphonium group with more hydrophobic and bulky alkyl groups (P(Et2)2(CH2)3OH, PBu3) leads to the increase of overall cytotoxicity towards the mitochondria and strong inhibition of cell growth. Substitution of methyl substituents with phenyl and p-tolyl groups leads to significant differences in toxicity as observed by MTT and CV method, with higher observed toxicity of dendrimers towards the mitochondria. This effect is extremely pronounced in the case of P(C6H4-OMe)3 dendrimers which are more than order of magnitude less toxic when measured by CV method as compared to MTT. Such effects are more visible at higher (G2, G3) generations of dendrimers, where the larger amount of phosphonium groups on dendrimer is present. Similar results were demonstrated by Ornelas-Megiatto et al.30 in their study of cytotoxicity of alkyl-functionalized polyphosphonium polymers. The nature of the alkyl substituents of the phosphonium cations was also shown to have important influence on the transfection efficiency where the phosphonium groups of the analogous composition as ammonium ones achieved a much better transfection efficiency and lower cytotoxicity. Therefore, we expect that a similar positive effect could be observed with phosphonium carbosilane dendrimers in dependence on the type of substituent used.
2.3.2 Influence of dendrimer generation on cytotoxicity. Dendrimer generation is another important parameter, which influence their overall cytotoxicity. As is apparent from Fig. 5, dendrimer cytotoxicity significantly increases between G1 to G2 generation regardless on assay method and cell line used. On the contrary, much smaller differences were observed between G2 and G3 dendrimers. The differences between IC50 of G2 and G3 were statistically significant only in few cases (see Fig. 5). The toxicity of dendrimer is always a function of toxicity of core and outer shell. First generation dendrimers, where a hydrophobic core is more open and the number of ammonium/phosphonium groups is low are much less toxic than G2 and G3 dendrimers where the number of positively charged groups increases and creates an outer shell of dendrimer. Thus, the contribution of hydrophobic core to overall cytotoxicity of dendrimers is negligible.
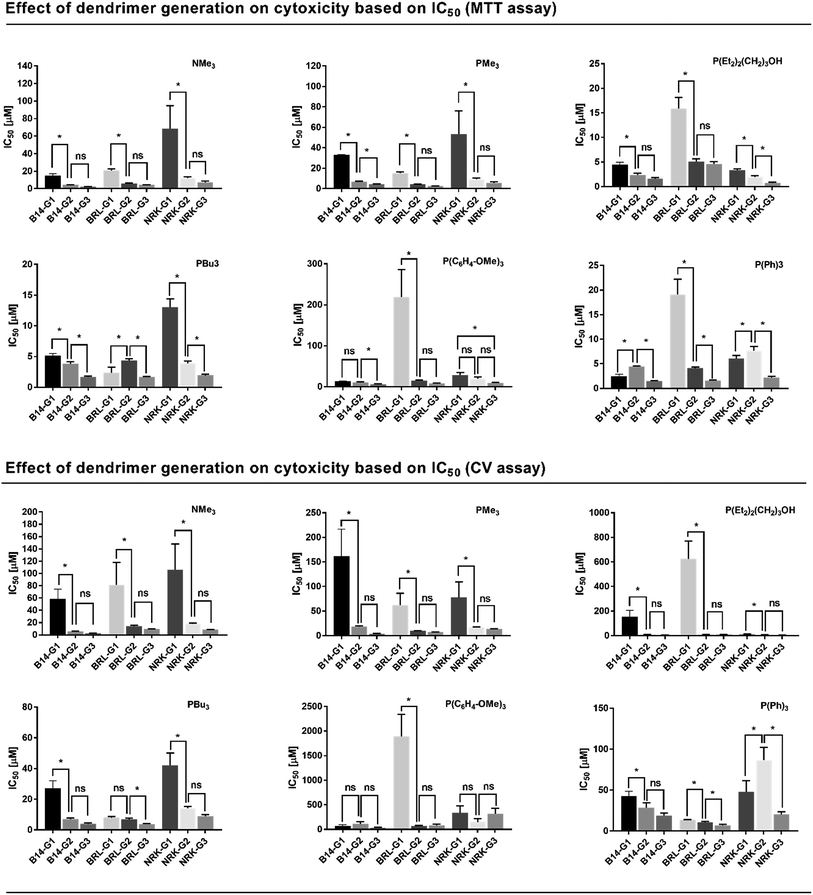 |
| | Fig. 5 Effect of dendrimer generation on cytotoxicity based on IC50. Upper panel: Data obtained by MTT assay. Lower panel: Data obtained by CV assay. Each graph represents data analyzed for one type of dendrimer and for three generations (G1–G3) grouped and compared each to other for each cell line. To determine a statistical significance of difference between the IC50 values of all three generations of dendrimers on one cell line, one-way ANOVA followed with Tukey–Kramer multiply comparison test was used to quantify the p values between each generation. Values p < 0.05 were supposed to be statistically significant (*), values p ≥ 0.05 non-significant (ns). Data are presented as mean ± SEM (n = 3). | |
2.3.3 Influence of assay method. MTT and CV assay were selected for cytotoxicity screening since they measure the effect of compound (dendrimer) based on different principles. The MTT is related to specific enzymatic activity connected with mitochondria, CV monitors the growth of cell population. As is apparent from Fig. 4, S70 and S71,† MTT assay is more sensitive to presence of dendrimer than CV assay. Statistically significant differences in IC50 of MTT and CV (IC50 MTT < IC50 CV) were observed in the most of the experimental variations regardless on dendrimer type, generation and cell line. Moderate differences between IC50 of MTT and CV (differences within an order of magnitude) were present mainly in case of NMe3, PMe3, P(Et2)2(CH2)3OH and PBu3 dendrimers. Contrary to this, much higher differences (IC50 MTT ≪ IC50 CV) were apparent in case of P(C6H4-OMe)3 and P(Ph)3 dendrimers as was already discussed above. Similar discrepancies between the IC50 of various inhibitors measured by MTT and CV method have been already discussed elsewhere.48 The reason of such observations could be the preferential interaction of toxic compound with mitochondrial metabolism or provoking a cell stress which onset is faster than reduction of cell population growth by necrosis or apoptosis.48,49 Higher differences observed in case of P(C6H4-OMe)3 and P(Ph)3 dendrimers could be connected to their preferential targeting and therefore also higher influence on the metabolism of mitochondria, as suggested from other published works.37 Our experiments also support the conclusions discussed elsewhere,48 that to evaluate reliably the cytotoxicity of novel types of nanoparticles, several assay methods must be performed simultaneously, since they can give a different or sometimes even misleading results.
3. Conclusions
The carbosilane dendrimers with NMe3, PMe3, P(Et2)2(CH2)3OH, PBu3, P(C6H4-OMe)3 and P(Ph)3 periphery substituents were synthesized, thoroughly characterized and simulated by computer modelling approaches. Dendrimers containing butyl, methoxyphenyl and phenyl substituents showed only negligible solubility in water when the counter-anion was the iodide ion. By exchange of the iodide anions for chloride using ion exchange resign the solubility in water was considerably increased to >100 mg mL−1.
Computer simulations show that higher generation dendrimers maintain spherical shape and their dimensions are comparable with only negligible differences. Radial distribution function revealed interesting properties of P(C6H4-OMe)3 dendrimer with apparent backfolding of peripheral groups and the smallest water density in the interior of dendrimer in comparison with other dendrimer types.
Generally, the cytotoxicity of PMe3 carbosilane dendrimers was similar or slightly lower as compared to NMe3 dendrimers which correlates well with the published works focused on phosphonium- and ammonium-modified lipid transfectants.32–34 The substitution of methyl groups of PMe3 carbosilane dendrimers with more hydrophobic and bulky alkyl substituents (PBu3 and P(Et2)2(CH2)3OH dendrimers) resulted in increase of cytotoxicity as measured by MTT and CV method regardless of cell line tested. Significant differences in comparison with all other dendrimers were shown for P(C6H4-OMe)3 dendrimer and partially also in the case of P(Ph)3 dendrimer. P(C6H4-OMe)3 dendrimer is of very low cytotoxicity across all cell lines or assay methods used and can be even compared to low cytotoxic sugar or PEG coated dendrimers.7,13,45 Recognizably large differences in cell viability as was measured by MTT and CV assay were in several cases, along with P(C6H4-OMe)3 dendrimer, observed also for P(Ph)3 dendrimer. Based on such observations, we suggest that P(C6H4-OMe)3 and P(Ph)3 dendrimers may preferentially accumulate in mitochondria. Therefore, both dendrimers could represent unique drug delivery systems (DDS) for mitochondrial targeting with relatively low cytotoxicity as compared to other types of DDS systems.
4. Experimental section
4.1 General considerations
Unless otherwise stated, reagents were used as received from commercial sources. Literature procedures were followed in the preparation of (3 chloropropyl)dimethylsilane50 and allyl-terminated carbosilane dendrimers.51 Karstedt catalyst (obtained as a 2–3% solution in xylenes), trimethylphosphine (1.0 M solution in THF), triphenylphosphine and AMBERLYST A-21 were purchased from Sigma Aldrich, trimethylamine (ca. 25% solution in isopropyl alcohol), tris(p-methoxyphenyl)phosphine and tributylphosphine from TCI chemicals. Diethyl-(3-hydroxypropyl)-phosphine was taken from laboratory supplies. The 1H (299.9 or 499.9 MHz), 13C {1H} (75.4 or 125.7 MHz) and 29Si {1H} (59.6 or 99.3 MHz), 31P {1H} (121.4 or 202.4 MHz) NMR spectra were measured on a Varian Mercury 300 or Varian Innova 500 spectrometer at 25 °C. The 1H and 13C NMR spectra were referenced to the line of the solvent (δ/ppm; δH/δC: CDCl3, 7.26/76.99; DMSO-d6, 2.51/39.52; CD3CN, 1.94/1.32). The 29Si and 31P NMR spectra were referenced to external standards.
HRMS spectra were measured using Bruker MicrOTOF-QIII apparatus. Due to the structure of target compounds, the electrospray ionisation source in positive mode was used with the parameters adjusted as follows: the capillary voltage was 4200 V, the end plate offset was −500 V. The collision cell RF ranged from 350 Vpp to 1000 Vpp, based on the size of the measured molecule. Nitrogen was used as the nebulizer gas (at the pressure of 1.6 bar), just as the drying gas (heated to 180 °C, with the flow of 8 L min−1). The scans of MS spectra were carried out in the mass range of m/z 80–1550 for the smaller and 200–6000 for the bigger dendrimers. For HRMS, the calibration on Na-formate or CsPFHA clusters was employed. To support ionisation in solutions of uncharged dendrimers, ammonium formate buffer (5 mL MeOH, 1.25 mL HCOOH, 3.9 g HCOO−NH4+, filled up to 25 mL with water) was used. The samples were delivered into the ESI source by direct infusion, using a syringe pump (Kd-Scientific, KDS-100-CE, 0.5 mL Hamilton syringe, flow 180 mL min−1) coupled to the MicrOTOF-QIII mass spectrometer.
4.2 Synthesis of dendrimers
4.2.1 Dendrimer (1). Two drops of Karstedt catalyst solution were added to a solution of tetraallylsilane (1.00 g, 5.2 mmol) in pentane (2 mL). After the mixture was stirred for 10 min, (3-chloropropyl)dimethylsilane (2.91 g, 21.3 mmol) was added and the mixture was refluxed for 3 h and then stirred at room temperature overnight. The solvent and the excess of silane were removed on the rotary evaporator and finally under high vacuum. Yield: 3.80 g (5.15 mmol, 99%) of dendrimer 1 as colourless liquid. Data for 1: NMR (CDCl3): 1H (499.99 MHz): δ −0.02 (s, 24H, Si1CH3), 0.54–0.61 (m, 24H, Si0CH2, CH2Si1CH2), 1.28–1.35 (m, 8H, Si0CH2CH2), 1.72–1.78 (m, 8H, CH2CH2Cl), 3.50 (t, 3JHH = 7.0 Hz, 8H, CH2Cl). 13C {1H} (125.70 MHz): δ −3.4 (Si1CH3), 13.0 (CH2CH2CH2Cl), 17.5 (Si0CH2), 18.5 (Si0CH2CH2), 20.1 (Si0CH2CH2CH2), 27.7 (CH2CH2Cl), 48.0 (CH2Cl). 29Si {1H} (59.60 MHz): δ 0.58 (Si0), 2.05 (Si1). HRMS (C32H72Cl4Si5): (ESI+) m/z: [M + NH4]+ calcd for C32H72Cl4Si5NH4: 756.3551 found: 756.3559, [M + Na]+ calcd for C32H72Cl4Si5Na: 761.3104, found: 761.3104.
4.2.2 Dendrimer (2). The compound was synthesized according to the procedure for 1 from G1allyl8 (1.3 g, 1.86 mmol) and (3-chloropropyl)dimethylsilane (2.24 g, 16.4 mmol). Yield: 3.30 g (1.84 mmol, 99%) of a yellow oil. Data for 2: NMR (CDCl3): 1H (499.99 MHz): δ −0.07 (s, 12H, Si1CH3), −0.02 (s, 48H, Si2CH3), 0.53–0.60 (m, 64H, Si0CH2, CH2Si1CH2, CH2Si2CH2), 1.28–1.34 (m, 24H, CH2CH2Si1CH2CH2), 1.72–1.78 (m, 16H, CH2CH2Cl), 3.50 (t, 3JHH = 7.0 Hz, 16H, ClCH2). 13C {1H} (125.70 MHz): δ −5.0 (Si1CH3), −3.4 (Si2CH3), 13.0 (CH2CH2CH2Cl), 17.7 (Si0CH2), 18.4 (Si1CH2CH2CH2Si2), 18.6 (Si0CH2CH2), 18.8 (Si1CH2CH2CH2Si2), 19.2 (Si0CH2CH2CH2), 19.9 (Si1CH2CH2CH2Si2), 27.7 (CH2CH2CH2Cl), 48.0 (CH2Cl). 29Si {1H} (59.60 MHz): δ 0.49 (Si0), 0.97 (Si1); 2.06 (Si2). HRMS (C80H180Cl8Si13): (ESI+) m/z: [M + NH4]+ calcd for C80H180Cl8Si13NH4: 1808.8888 found: 1808.8838.
4.2.3 Dendrimer (3). The compound was synthesized according to the procedure for 1 from G2allyl16 (1.5 g, 0.88 mmol) and (3-chloropropyl)dimethylsilane (2.11 g, 15.46 mmol). Yield: 3.36 g (0.87 mmol, 99%) of a yellow oil. Data for 3: NMR (CDCl3): 1H (499.99 MHz): δ −0.08 (s, 36H, Si1,2CH3), −0.02 (s, 96H, Si3CH3), 0.53–0.60 (m, 144H, Si0CH2, CH2Si1CH2, CH2Si2CH2, CH2Si3CH2), 1.28–1.33 (m, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.71–1.7 (m, 32H, CH2CH2Cl), 3.49 (t, 3JHH = 7.0 Hz, 32H, ClCH2). 13C {1H} (125.70 MHz): δ −5.0 (Si1CH3), −4.9 (Si2CH3), −3.4 (Si3CH3), 13.0 (CH2CH2CH2Cl), 17.7 (Si0CH2), 18.4 (Si2CH2CH2CH2Si3), 18.52 (Si1CH2CH2CH2Si2), 18.55 (Si0CH2CH2CH2Si1), 18.8 (Si2CH2CH2CH2Si3), 18.96, 19.03 (Si1CH2CH2CH2Si2), 19.3 (Si0CH2CH2CH2), 19.9 (Si2CH2CH2CH2Si3), 27.7 (CH2CH2Cl), 48.0 (CH2Cl). 29Si {1H} (59.60 MHz): δ 0.38 (Si0), 0.90 (Si1), 0.98 (Si2), 2.05 (Si3).
4.2.4 Dendrimer (4). Mixture of chlorine terminated dendrimers 1 (2.00 g, 2.71 mmol) and sodium iodide (8.11 g, 54.2 mmol) in 40 mL of butan-2-one was heated to reflux for 2 days. The reaction mixture was then cooled to room temperature. The product was extracted to diethyl ether (2 × 30 mL), the extract filtered through silica gel and dried over anhydrous MgSO4. The volatiles were removed on the rotary evaporator and finally under high vacuum. Yield: 2.51 g (2.2 mmol, 84%) of a yellow oil. Data for 4: NMR (CDCl3): 1H (499.99 MHz): δ −0.02 (s, 24H, Si1CH3), 0.53–0.60 (m, 24H, Si0CH2, CH2Si1CH2), 1.27–1.32 (m, 8H, Si0CH2CH2), 1.77–1.84 (m, 8H, CH2CH2I), 3.18 (t, 3JHH = 7.3 Hz, 8H, CH2I). 13C {1H} (125.70 MHz): δ −3.3 (Si1CH3), 11.5 (CH2I), 17.4 (Si0CH2), 17.5 (CH2CH2CH2I), 18.5 (Si0CH2CH2), 20.1 (Si0CH2CH2CH2), 28.9 (CH2CH2I). 29Si {1H} (99.31 MHz): δ 0.56 (Si0), 1.47 (Si1). HRMS (C32H72I4Si5): (ESI+) m/z: [M + NH4]+ calcd for C32H72I4Si5NH4: 1122.0997, found: 1122.1000, [M + Na]+ calcd for C32H72I4Si5Na: 1127.0551, found: 1127.0553.
4.2.5 Dendrimer (5). The compound was synthesized according to the procedure for dendrimer 4 from dendrimer 2 (1 g, 0.56 mmol). Yield: 1.14 g (0.45 mmol, 81%) of a yellow oil. Data for 5: NMR (CDCl3): 1H (499.99 MHz): δ −0.07 (s, 12H, Si1CH3), −0.02 (s, 48H, Si2CH3), 0.53–0.60 (m, 64H, Si0CH2, CH2Si1CH2, CH2Si2CH2), 1.28–1.33 (m, 24H, CH2CH2Si1CH2CH2), 1.78–1.84 (m, 16H, CH2CH2I), 3.18 (t, 3JHH = 7.3 Hz, 16H, ICH2). 13C {1H} (125.70 MHz): δ −4.9 (Si1CH3), −3.2 (Si2CH3), 11.4 (CH2I), 17.5 (CH2CH2CH2I), 17.7 (Si0CH2), 18.4 (Si1CH2CH2CH2Si2), 18.6 (Si0CH2CH2CH2), 18.8 (Si1CH2CH2CH2Si2), 19.2 (Si0CH2CH2CH2), 19.9 (Si1CH2CH2CH2Si2), 28.9 (CH2CH2I). 29Si {1H} (59.60 MHz): δ 0.50 (Si0), 0.99 (Si1); 1.50 (Si2). HRMS (C80H180I8Si13): (ESI+) m/z: [M + NH4]+ calcd for C80H180I8Si13NH4: 2540.3799 found: 2540.3789.
4.2.6 Dendrimer (6). The compound was synthesized according to the procedure for dendrimer 4 from dendrimer 3 (1 g, 0.26 mmol). Yield: 1.18 g (0.22 mmol, 86%) of a yellow oil. Data for 6: NMR (CDCl3): 1H (499.99 MHz): δ −0.06 (s, 36H, Si1,2CH3), −0.02 (s, 96H, Si3CH3), 0.54–0.61 (m, 144H, CH2CH2CH2Si1CH2, CH2Si2CH2, CH2Si3CH2), 1.29–1.34 (m, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.78–1.85 (m, 32H, CH2CH2I), 3.18 (t, 3JHH = 7.2 Hz, 32H, ICH2). 13C {1H} (125.70 MHz): δ −4.9 (Si1CH3), −4.8 (Si2CH3), −3.2 (Si3CH3), 11.4 (CH2I), 17.5 (CH2CH2CH2I), 17.8 (Si0CH2), 18.4 (Si2CH2CH2CH2Si3), 18.56 (Si1CH2CH2CH2Si2), 18.59 (Si0CH2CH2), 18.8 (Si2CH2CH2CH2Si3), 19.02, 19.07 (Si1CH2CH2CH2Si2), 19.4 (Si0CH2CH2CH2), 20.0 (Si2CH2CH2CH2Si3), 29.0 (CH2CH2I). 29Si {1H} (99.31 MHz): δ 0.35 (Si0), 0.88 (Si1), 0.97 (Si2), 1.47 (Si3).
4.2.7 Dendrimer (7). To a solution of dendrimer 4 (1.2 g, 1.09 mmol) in acetonitrile (15 mL) the commercial solution of trimethylamine (3 M in isopropyl amine) was added (4.3 mL, 13.03 mmol) at 0 °C. The reaction mixture was then stirred at room temperature for 48 hours. The solvents and the excess of phosphine were removed on the rotary evaporator and finally under high vacuum to give product as a white powder. Yield: 1.44 g (1.07 mmol, 99%) data for 7: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.02 (s, 24H, Si1CH3), 0.35–0.39 (m, 8H, CH2CH2CH2N), 0.52–0.60 (m, 16H, Si0CH2CH2CH2), 1.26–1.34 (m, 8H, Si0CH2CH2), 1.59–1.66 (m, 8H, CH2CH2N), 3.09 (s, 36H, CH3N), 3.28–3.33 (m, 8H, CH2N). 13C {1H} (125.70 MHz): δ −3.4 (Si1CH3), 11.0 (CH2CH2CH2N), 16.8 (Si0CH2), 16.9 (CH2CH2N), 18.0 (Si0CH2CH2), 19.3 (Si0CH2CH2CH2), 52.1 (CH3N), 67.7 (CH2N). 29Si {1H} (99.31 MHz): δ 0.74 (Si0), 2.42 (Si1). HRMS (C44H108N4Si5I4): (ESI+) m/z: [M − I−]+ calcd for C44H108N4Si5I3: 1213.4549 found: 1213.4536, [M − 2I−]2+ calcd for C44H108N4Si5I2: 543.2749 found: 543.2749, [M − 3I−]3+ calcd for C44H108N4Si5I: 319.8816 found: 319.8822.
4.2.8 Dendrimer (8). The compound was synthesized according to the procedure for dendrimer 7 from dendrimer 5 (1.1 g, 0.44 mmol). Yield: 1.29 g (0.43 mmol, 99%) of a white powder. Data for 8: NMR (DMSO-d6): 1H (299.99 MHz): δ −0.09 (s, 12H, Si1CH3), −0.00 (s, 48H, Si2CH3), 0.37–0.40 (m, 16H, CH2CH2CH2N), 0.52–0.59 (m, 48H, Si0CH2, CH2Si1CH2CH2CH2), 1.29–1.33 (m, 24H, CH2CH2Si1CH2CH2), 1.62–1.64 (m, 16H, CH2CH2N), 3.04–3.09 (m, 72H, CH3N), 3.26–3.31 (m, 16H, NCH2). 13C {1H} (75.44 MHz): δ −4.9 (Si1CH3), −3.4 (Si2CH3), 11.2 (CH2CH2CH2N), 17.0 (CH2CH2N), 17.1 (Si0CH2), 18.0 (Si1CH2CH2CH2Si2), 18.3 (Si0CH2CH2CH2Si1CH2), 18.5 (Si0CH2CH2CH2), 19.3 (Si1CH2CH2CH2Si2), 52.2 (NCH3), 67.9 (CH2N). 29Si {1H} (59.60 MHz): δ 0.74 (Si0), 0.98 (Si1); 2.34 (Si2). HRMS (C104H252I8N8Si13): (ESI+) m/z: [M − 2I−]2+ calcd for C104H252I6N8Si13: 1370.5623 found: 1370.5612, [M − 3I−]3+ calcd for C104H252I5N8Si13: 871.4065 found: 871.4065, [M − 4I−]4+ calcd for C104H252I4N8Si13: 621.8286 found: 621.8289, [M − 5I−]5+ calcd for C104H252I3N8Si13: 472.0819 found: 472.0813, [M − 6I−]6+ calcd for C104H252I2N8Si13: 372.2507 found: 372.2527, [M − 7I−]7+ calcd for C104H252IN8Si13: 300.9428 found: 300.9423.
4.2.9 Dendrimer (9). The compound was synthesized according to the procedure for dendrimer 7 from dendrimer 6 (1 g, 0.19 mmol). Yield: 1.14 g (0.18 mmol, 97%) of a white powder. Data for 9: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.11 (s, 12H, Si1CH3), −0.09 (s, 24H, Si2CH3), 0.00 (s, 96H, Si3CH3), 0.37–0.40 (m, 32H, CH2CH2CH2N), 0.51–0.59 (m, 112H, Si0CH2, CH2Si1CH2, CH2Si2CH2CH2CH2), 1.28–1.33 (m, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.61–1.66 (m, 32H, CH2CH2N), 3.08 (br s, 144H, NCH3), 3.22–3.28 (m, 32H, NCH2). 13C {1H} (125.70 MHz): δ −4.9 (Si1,2CH3), −3.4 (Si3CH3), 11.2 (CH2CH2CH2N), 17.0 (CH2CH2N), 18.0 (Si2CH2CH2CH2Si3), 18.2 (Si1CH2CH2CH2Si2CH2), 18.4 (Si1CH2CH2CH2Si2), 19.4, (Si2CH2CH2CH2Si3), 52.2 (CH3N), 67.9 (CH2N), (Si0CH2CH2CH2 were overlap or not detected). 29Si {1H} (99.31 MHz): δ (Si0 was not detected), 1.06 (Si1,2), 2.42 (Si3). HRMS (C224H540N16Si29I16): (ESI+) m/z: [M − 4I−]4+ calcd for C224H540N16Si29I12: 1448.8655 found: 1448.8647, [M − 5I−]5+ calcd for C224H540N16Si29I11: 1133.7114 found: 1133.7114, [M − 6I−]6+ calcd for C224H540N16Si29I10: 923.6086 found: 923.6073. Anal. calc. C224H540I16N16Si29 (6303.74 g mol−1): C, 42.68; H, 8.63; exp.: C, 42.97; H, 8.89.
4.2.10 Dendrimer (10). To a solution of dendrimer 4 (1.2 g, 1.09 mmol) in acetonitrile (15 mL) the commercial solution of trimethylphosphine (1 M in THF) was added (8.7 mL, 8.67 mmol) at 0 °C. The reaction mixture was then stirred at room temperature for 48 hours. The solvents and the excess of phosphine were removed on the rotary evaporator and finally under high vacuum to give product as a white powder. Yield: 1.52 g (1.07 mmol, 99%). Data for 10: NMR (DMSO-d6): 1H (299.99 MHz): δ −0.01 (s, 24H, Si1CH3), 0.52–0.62 (m, 24H, Si0CH2, CH2Si1CH2), 1.26–1.36 (m, 8H, Si0CH2CH2), 1.45–1.54 (m, 8H, PCH2CH2), 1.82 (d, 2JHP = 14.8 Hz, 48H, PCH3), 2.14–2.24 (m, 8H, PCH2). 13C {1H} (75.44 MHz): δ −3.4 (s, Si1CH3), 7.3 (d, 1JPC = 53.7 Hz, PCH3), 15.8 (d, 2JPC = 4.5 Hz, PCH2CH2), 16.7 (d, 3JPC = 14.1 Hz, PCH2CH2CH2), 16.9 (s, Si0CH2), 18.1 (s, Si0CH2CH2), 19.4 (s, Si0CH2CH2CH2), 25.9 (d, 1JPC = 49.6 Hz, CH2P). 29Si {1H} (59.60 MHz): δ 0.74 (Si0), 1.66 (d, 4JSiP = 2.4 Hz, Si1). 31P {1H} (121.44 MHz): δ 26.61. HRMS (C44H108I4P4Si5): (ESI+) m/z: [M − I−]+ calcd for C44H108I3P4Si5: 1281.3376 found: 1281.3373, [M − 2I−]2+ calcd for C44H108I2P4Si5: 557.2163, found: 557.2169, [M − 3I−]3+ calcd for C44H108IP4Si5: 342.5092, found: 342.5118.
4.2.11 Dendrimer (11). The compound was synthesized according to the procedure for dendrimer 10 from dendrimer 5 (1 g, 0.40 mmol). Yield: 1.22 g (0.39 mmol, 98%) of a white powder. Data for 11: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.09 (s, 12H, Si1CH3), −0.02 (s, 48H, Si2CH3), 0.52–0.60 (m, 64H, Si0CH2, CH2Si1CH2, CH2Si2CH2), 1.27–1.33 (m, 24H, CH2CH2Si1CH2CH2), 1.47–1.52 (m, 16H, CH2CH2P), 1.83 (d, 1JHP = 14.8 Hz, 72H, CH3P), 2.17–2.23 (m, 16H, PCH2). 13C {1H} (125.70 MHz): δ −4.9 (Si1CH3), −3.4 (Si2CH3), 7.4 (d, 1JCP = 53.6 Hz, PCH3), 15.8 (d, 2JCP = 4.4 Hz, CH2CH2P), 16.6 (d, 3JCP = 13.9 Hz, CH2CH2CH2P), 17.1 (Si0CH2), 17.9 (Si1CH2CH2CH2Si2), 18.1 (Si0CH2CH2CH2Si0CH2), 18.4 (Si0CH2CH2CH2), 19.3 (Si1CH2CH2CH2Si2), 25.9 (d, 1JCP = 49.7 Hz, CH2P). 29Si {1H} inept (99.31 MHz): δ 0.82 (Si0), 1.07 (Si1), 1.64 (d, 4JSiP = 2.3 Hz, Si2). 31P {1H} (202.37 MHz): δ 26.59. HRMS (C104H252I8P8Si13): (ESI+) m/z: [M − 2I−]2+ calcd for C104H252I6P8Si13: 1437.4438 found: 1437.4433, [M − 3I−]3+ calcd for C104H252I5P8Si13: 915.9941 found: 915.9941, [M − 4I−]4+ calcd for C104H252I4P8Si13: 655.2694 found: 655.2692, [M − 5I−]5+ calcd for C104H252I3P8Si13: 498.8345 found: 498.8340, [M − 6I−]6+ calcd for C104H252I2P8Si13: 394.5446 found: 394.5444, [M − 7I−]7+ calcd for C104H252IP8Si13: 320.0517 found: 320.0511.
4.2.12 Dendrimer (12). The compound was synthesized according to the procedure for dendrimer 10 from dendrimer 6 (1 g, 0.19 mmol). Yield: 1.20 g (0.18 mmol, 98%) of a white powder. Data for 12: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.11 (s, 12H, Si1CH3), −0.09 (s, 24H, Si2CH3), −0.02 (s, 96H, Si3CH3), 0.52–0.60 (m, 144H, Si0CH2, CH2Si1CH2, CH2Si2CH2, CH2Si3CH2), 1.27–1.33 (m, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.46–1.53 (m, 32H, CH2CH2P), 1.88 (d, 1JPH = 14.8 Hz, 144H, PCH3), 2.22–2.28 (d, 32H, PCH2). 13C {1H} (125.70 MHz): δ −4.84 (Si2CH3), −4.81 (Si1CH3), −3.3 (Si3CH3), 7.5 (d, 1JPC = 53.7 Hz,CH3P), 15.9 (d, 2JCP = 3.9 Hz, CH2CH2P), 16.7 (d, 3JCP = 14.0 Hz, CH2CH2CH2P), 18.0 (Si2CH2CH2CH2Si3), 18.2 (Si1CH2CH2CH2Si2), 18.3 (Si2CH2CH2CH2Si3), 18.4 (Si1CH2CH2CH2Si2), 19.5, (Si2CH2CH2CH2Si3), 25.9 (d, 1JCP = 49.4 Hz, CH2P). (Si0CH2CH2CH2 were overlap or not detected). 29Si {1H} (99.31 MHz): δ (Si0 was not detected), 1.05 (Si1,2), 1.62 (d, 4JSiP = 2.2 Hz, Si3). 31P {1H} (202.37 MHz): δ 26.59. HRMS (C224H540I16P16Si29): (ESI+) m/z: [M − 3I−]3+ calcd for C224H540I13P16Si29: 2064.6327 found: 2064.6333, [M − 4I−]4+ calcd for C224H540I12P16Si29: 1516.7483 found: 1516.7482, [M − 5I−]5+ calcd for C224H540I11P16Si29: 1188.0176 found: 1188.0170, [M − 6I−]6+ calcd for C224H540I10P16Si29: 968.8639 found: 968.8644, [M − 7I−]7+ calcd for C224H540I9P16Si29: 812.3254 found: 812.3267, [M − 8I−]8+ calcd for C224H540I8P16Si29: 694.9216 found: 694.9223.
4.2.13 Dendrimers (13–18, 22–24, 28–30). Dendrimers 13–18, 22–24, 28–30 were synthetized according to similar procedure using iododerivate 4 for first 5 for second and 6 for third generation. Appropriate phosphine was used in slight excess. In all cases yields are in range between 97–100%. Dendrimers 14–15 were prepared as colourless liquids, 16–33 as white powders.
4.2.14 Dendrimer (13). To a solution of dendrimer 4 (1.2 g, 1.09 mmol) in acetonitrile (15 mL) the diethyl-(3-hydroxypropyl)-phosphin was added (8.7 mL, 8.67 mmol). The reaction mixture was then stirred at 70 °C for 48 hours. The solvents were removed on the rotary evaporator and to a crude product toluene (2 mL) was added and mixture was stirred at 80 °C for 6 h followed by filtration at room temperature and drying of product at vacuum. The product was obtained as a colourless liquid. Yield: 1.52 g (1.07 mmol, 99%). Data for 13: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.02 (s, 24H, Si1CH3), 0.53–0.59 (m, 16H, Si0CH2CH2CH2), 0.60–0.64 (m, 8H, Si1CH2CH2CH2P), 1.13 (dt, 3JHP = 18.2 Hz, 3JHH = 7.7 Hz, 24H, CH3), 1.27–1.34 (m, 8H, Si0CH2CH2), 1.44–1.52 (m, 8H, Si1CH2CH2CH2P), 1.60–1.66 (m, 8H, CH2CH2OH), 2.19–2.29 (m, 32H, CH2P), 3.48 (q, 3JHH = 5.6 Hz, 8H, CH2OH), 4.77 (t, 3JHH = 5.3 Hz, 4H, OH). 13C {1H} (125.70 MHz): δ −3.4 (s, Si1CH3), 5.3 (d, 2JCP = 5.5 Hz, PCH2CH3), 11.1 (d, 1JCP = 48.6 Hz, CH3CH2P), 14.0 (d, 1JCP = 48.9 Hz, PCH2CH2CH2OH), 15.9 (d, 2JCP = 4.6 Hz, Si1CH2CH2CH2P), 16.92 (s, Si0CH2), 19.94 (d, 3JCP = 12.8 Hz, Si1CH2CH2CH2P), 18.1 (Si0CH2CH2), 19.4 (s, Si0CH2CH2CH2), 20.7 (d, 1JCP = 44.7 Hz, Si1CH2CH2CH2P), 24.0 (d, 2JCP = 4.3 Hz, OHCH2CH2CH2P), 60.4 (d, 3JCP = 15.4 Hz, OHCH2). 29Si {1H} (99.31 MHz): δ 0.71 (s, Si0), 1.58 (d, 4JSiP = 2.5 Hz, Si1). 31P {1H} (121.41 MHz): δ 42.63. HRMS (C60H140O4Si5I4): (ESI+) m/z: [M − I−]+ calcd for C60H140O4Si5I3: 1569.5677 found: 1569.5668, [M − 2I−]2+ calcd for C60H140O4Si5I2: 721.3313 found: 721.3316, [M − 3I−]3+ calcd for C60H140O4Si5I: 438.5858 found: 438.5856, [M − 4I−]4+ calcd for C60H140O4Si5: 297.2131 found: 297.2135. Anal. calc. C60H140I4O4P4Si5 (1697.69 g mol−1): C, 42.45; H, 8.31; exp.: C, 43.26; H, 8.73.
4.2.15 Dendrimer (14). Data for 14: NMR (DMSO-d6): 1H (299.99 MHz): δ −0.09 (s, 12H, Si1CH3), −0.02 (s, 48H, Si2CH3), 0.50–0.65 (m, 64H, Si0CH2, CH2Si1CH2CH2CH2Si2CH2), 1.13 (dt, 3JHP = 18.0 Hz, 3JHH = 7.6 Hz, 48H, CH3), 1.25–1.34 (m, 24H, Si0CH2CH2CH2Si1CH2CH2), 1.43–1.53 (m, 16H, Si2CH2CH2CH2P), 1.60–1.67 (m, 16H, PCH2CH2CH2OH), 2.19–2.32 (m, 64H, CH2P), 3.48 (q, 3JHH = 5.4 Hz, 16H, CH2OH), 4.78 (t, 3JHH = 5.4 Hz, 8H, OH). 13C {1H} (125.70 MHz): δ −4.9 (s, Si1CH3), −3.4 (s, Si2CH3), 5.3 (d, 2JCP = 5.3 Hz, PCH2CH3), 11.1 (d, 1JCP = 48.5 Hz, CH3CH2P), 14.1 (d, 1JCP = 49.4 Hz, PCH2CH2CH2OH), 15.9 (d, 2JCP = 4.3 Hz, Si2CH2CH2CH2P), 16.90 (d, 3JCP = 13.0 Hz, Si2CH2CH2CH2P), 17.1 (s, Si0CH2), 18.0 (s, Si1CH2CH2CH2Si2), 18.3 (s, Si1CH2CH2CH2Si2), 18.5 (s, Si0CH2CH2), 19.4 (s, Si1CH2CH2CH2Si2), 20.8 (d, 1JCP = 43.4 Hz, Si2CH2CH2CH2P), 24.1 (d, 2JCP = 4.3 Hz, OHCH2CH2), 60.4 (d, 3JCP = 15.5 Hz, OHCH2), (Si0CH2CH2CH2 was not detected). 29Si {1H} (59.60 MHz): δ 0.84 (s, Si0), 1.08 (s, Si1), 2.48 (d, 4JSiP = 2.5 Hz, Si2). 31P {1H} (121.44 MHz): δ 42.61. HRMS (C136H316O8Si13I8): (ESI+) m/z: [M − 2I−]2+ calcd for C136H316O8Si13I6: 1726.6756 found: 1726.6756, [M − 3I−]3+ calcd for C136H316O8Si13I5: 1108.8154 found: 1108.8152, [M − 4I−]4+ calcd for C136H316O8Si13I4: 799.8853 found: 799.8849, [M − 5I−]5+ calcd for C136H316O8Si13I3: 614.5272 found: 614.5272, [M − 6I−]6+ calcd for C136H316O8Si13I2: 490.9551 found: 490.9552, [M − 7I−]7+ calcd for C136H316O8Si13I: 402.6894 found: 402.6888.
4.2.16 Dendrimer (15). Data for 15: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.10 (s, 36H, Si1,2CH3), −0.03 (s, 96H, Si3CH3), 0.51–0.57 (m, 112H, Si0CH2, CH2Si1CH2, CH2Si2CH2CH2CH2Si3), 0.61–0.64 (m, 32H, Si3CH2CH2CH2P), 1.13 (dt, 3JHP = 15.4 Hz, 3JHH = 7.4 Hz, 96H, CH3), 1.26–1.33 (m, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.46–1.52 (m, 32H, Si3CH2CH2CH2P), 1.60–1.68 (m, 32H, CH2CH2OH), 2.22–2.31 (m, 128H, CH2P), 3.47–3.50 (m, 32H, CH2OH), 4.77 (br s, 16H, OH). 13C {1H} (125.70 MHz): δ −4.9 (s, Si1,2CH3), −3.4 (s, Si3CH3), 5.4 (d, 2JCP = 4.4 Hz, PCH2CH3), 11.2 (d, 1JCP = 48.8 Hz, CH3CH2P), 14.2 (d, 1JCP = 47.9 Hz, PCH2CH2CH2OH), 16.0 (d, 2JCP = 4.4 Hz, Si3CH2CH2CH2P), 16.90 (d, 3JCP = 12.7 Hz, Si3CH2CH2CH2P), 18.0 (s, Si2CH2CH2CH2Si3), 18.2 (s, Si1CH2CH2CH2Si2), 18.3 (s, Si2CH2CH2CH2Si3), 18.4 (s, Si1CH2CH2CH2Si2), 19.5 (s, Si2CH2CH2CH2Si3), 20.9 (d, 1JCP = 43.3 Hz, Si3CH2CH2CH2P), 24.1 (d, 2JCP = 4.3 Hz, OHCH2CH2CH2P), 60.4 (d, 3JCP = 15.5 Hz, OHCH2), (Si0CH2CH2CH2 were overlap or not detected). 29Si {1H} (99.31 MHz): δ 0.74 (s, Si0), 1.04 (s, Si1,2), 1.53 (d, 4JSiP = 2.2 Hz, Si3). 31P {1H} (121.44 MHz): δ 42.57. HRMS (C288H668O16P16Si29I16): (ESI+) m/z: [M − 3I−]3+ calcd for C288H668O16P16Si29I13: 2449.2736 found: 2449.2686, [M − 4I−]4+ calcd for C288H668O16P16Si29I12: 1805.2290 found: 1805.2277, [M − 5I−]5+ calcd for C288H668O16P16Si29I11: 1418.8022 found: 1418.8014, [M − 6I−]6+ calcd for C288H668O16P16Si29I10: 1161.1843 found: 1161.1835, [M − 7I−]7+ calcd for C288H668O16P16Si29I9: 977.1715 found: 977.1711, [M − 8I−]8+ calcd for C288H668O16P16Si29I8: 839.1620 found: 839.1614, [M − 9I−]9+ calcd for C288H668O16P16Si29I7: 731.8212 found: 731.8220, [M − 10I−]10+ calcd for C288H668O16P16Si29I6: 645.9486 found: 645.9441, [M − 11I−]11+ calcd for C288H668O16P16Si29I5: 575.6891 found: 575.6874.
4.2.17 Dendrimer (16). Data for 16: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.02 (s, 24H, Si1CH3), 0.53–0.57 (m, 16H, Si0CH2CH2CH2), 0.59–0.63 (m, 8H, Si1CH2CH2CH2P), 0.92 (t, 3JHH = 7.1 Hz, 36H, CH3), 1.27–1.33 (m, 8H, Si0CH2CH2CH2), 1.39–1.49 (m, 56H, Si1CH2CH2CH2PCH2CH2CH2), 2.17–2.23 (m, 32H, CH2P). 13C {1H} (125.70 MHz): δ −35 (s, Si1CH3), 13.3 (s, CH2CH3), 15.9 (d, 2JCP = 4.5 Hz, Si1CH2CH2CH2P), 16.8 (d, 3JCP = 13.0 Hz, Si1CH2CH2CH2P), 17.0 (s, Si0CH2), 17.3 (d, 1JCP = 47.3 Hz, PCH2CH2CH2CH3), 18.1 (s, Si0CH2CH2), 19.4 (s, Si0CH2CH2CH2), 21.3 (d, 1JCP = 44.7 Hz, Si1CH2CH2CH2P), 22.7 (d, 2JCP = 4.3 Hz, CH3CH2CH2CH2P), 23.3 (d, 3JCP = 15.4 Hz, CH3CH2). 29Si {1H} (99.31 MHz): δ 0.67 (s, Si0), 1.61 (d, 4JSiP = 2.5 Hz, Si1). 31P {1H} (202.35 MHz): δ 32.75. HRMS (C80H180P4Si5I4): (ESI+) m/z: [M − I−]+ calcd for C80H180P4Si5I3: 1786.9036 found: 1786.9038, [M − 2I−]2+ calcd for C80H180P4Si5I2: 829.9992 found: 829.9985, [M − 3I−]3+ calcd for C80H180P4Si5I: 511.0311 found: 511.0317, [M − 4I−]4+ C80H180P4Si5: 351.5461 found: 351.5461.
4.2.18 Dendrimer (17). Data for 17: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.09 (s, 12H, Si1CH3), −0.03 (s, 48H, Si2CH3), 0.51–0.56 (m, 48H, Si0CH2, CH2Si1CH2CH2CH2), 0.60–0.63 (m, 16H, Si2CH2CH2CH2P), 0.92 (t, 3JHH = 7.1 Hz, 72H, CH3), 1.28–1.32 (m, 24H, CH2CH2Si1CH2CH2), 1.39–1.47 (m, 112H, Si2CH2CH2CH2P, CH2CH2CH3), 2.19–2.25 (m, 64H, CH2P). 13C {1H} (125.70 MHz): δ −4.9 (s, Si1CH3), −3.4 (s, Si2CH3), 13.3 (s, CH2CH3), 16.0 (d, 2JCP = 3.8 Hz, Si2CH2CH2CH2P), 16.8 (d, 3JCP = 12.8 Hz, Si2CH2CH2CH2P), 17.1 (s, Si0CH2), 17.4 (d, 1JCP = 47.4 Hz, PCH2CH2CH2CH3), 18.0 (s, Si1CH2CH2CH2Si2), 18.2 (s, Si0CH2CH2), 18.3 (s, Si1CH2CH2CH2Si2), 18.5 (s, Si0CH2CH2CH2), 19.4 (s, Si1CH2CH2CH2Si2), 21.4 (d, 1JCP = 44.3 Hz, Si2CH2CH2CH2P), 22.7 (d, 2JCP = 4.4 Hz, CH3CH2CH2CH2P), 23.3 (d, 3JCP = 15.5 Hz, CH3CH2). 29Si (inept) {1H} (99.31 MHz): δ 0.79 (s, Si0), 1.03 (s, Si1), 1.56 (d, 4JSiP = 2.5 Hz, Si2). 31P {1H} (202.37 MHz): δ 32.71. HRMS (C176H396I8P8Si13): (ESI+) m/z: [M − 2I−]2+ calcd for C176H396I6P8Si13: 1943.5098 found: 1943.5091, [M − 3I−]3+ calcd for C176H396I5P8Si13: 1253.3715 found: 1253.3708, [M − 4I−]4+ calcd for C176H396I4P8Si13: 908.3024 found: 908.3029, [M − 5I−]5+ calcd for C176H396I3P8Si13: 701.2609 found: 701.2603, [M − 6I−]6+ calcd for C176H396I2P8Si13: 563.2332 found: 563.2317, [M − 7I−]7+ calcd for C176H396IP8Si13: 464.6420 found: 464.6411.
4.2.19 Dendrimer (18). Data for 18: NMR (CD3CN): 1H (499.99 MHz): δ −0.07 (s, 12H, Si1CH3), −0.06 (s, 24H, Si2CH3), 0.01 (s, 96H, Si3CH3), 0.55–0.62 (m, 112H, Si0CH2, CH2Si1CH2, CH2Si2CH2CH2CH2Si3), 0.66–0.70 (m, 32H, Si3CH2CH2CH2P), 0.96 (t, 3JHH = 7.1 Hz, 144H, CH3), 1.31–1.39 (m, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.44–1.56 (m, 224H, Si3CH2CH2CH2P, CH2CH2CH3), 2.20–2.28 (m, 128H, CH2P). 13C {1H} (125.70 MHz): δ −4.1 (s, Si1,2CH3), −2.8 (s, Si3CH3), 13.8 (s,CH2CH3), 17.6 (d, 2JCP = 4.9 Hz, Si3CH2CH2CH2P), 18.1 (d, 3JCP = 13.2 Hz, Si3CH2CH2CH2P), 18.5 (br s, Si0CH2), 19.3 (d, 1JCP = 47.5 Hz, PCH2CH2CH2CH3), (Si0CH2CH2 overlap), 19.4 (s, Si2CH2CH2CH2Si3), 19.5 (s, Si1CH2CH2CH2Si2), 19.6 (s, Si2CH2CH2CH2Si3), 19.8 (s, Si1CH2CH2CH2Si2), 20.0 (s, Si0CH2CH2CH2), 20.7 (s, Si2CH2CH2CH2Si3), 23.3 (d, 1JCP = 44.8 Hz, Si3CH2CH2CH2P), 24.2 (d, 2JCP = 4.5 Hz, CH3CH2CH2CH2P), 24.6 (d, 3JCP = 15.4 Hz, CH3CH2). 29Si {1H} (59.60 MHz): δ 0.72 (s, Si0), 0.99 (s, Si1,2), 1.55 (d, 4JSiP = 2.5 Hz, Si3). 31P {1H} (202.37 MHz): δ 33.55. HRMS (C368H828I16P16Si29): (ESI+) m/z: [M − 4I−]4+ calcd for C368H828I12P16Si29: 2021.8131 found: 2021.8132, [M − 5I−]5+ calcd for C368H828I11P16Si29: 1592.0695 found: 1592.0695, [M − 6I−]6+ calcd for C368H828I10P16Si29: 1305.5737 found: 1305.5729, [M − 7I−]7+ C368H828I9P16Si29: 1100.9338 found: 1100.9317, [M − 8I−]8+ C368H828I8P16Si29: 947.4540 found: 947.4557, [M − 9I−]9+ C368H828I7P16Si29: 828.0807 found: 828.0775, [M − 10I−]10+ C368H828I6P16Si29: 732.5822 found: 732.5795.
4.2.20 Dendrimer (22). Data for 22: NMR (DMSO-d6): 1H (299.99 MHz): δ −0.16 (s, 24H, Si1CH3), 0.40–0.45 (m, 16H, Si0CH2CH2CH2), 0.65–0.70 (m, 8H, CH2CH2CH2P), 1.16–1.19 (m, 8H, Si0CH2CH2CH2), 1.46–1.52 (m, 8H, CH2CH2P), 3.35–3.44 (m, 8H, CH2P), 3.87 (s, 36H, OCH3), 7.25–7.29 (m, 24H CH), 7.63–7.70 (m, 24H, CH). 13C {1H} (75.44 MHz): δ −3.6 (s, Si1CH3), 16.6 (d, 3JCP = 13.9 Hz, CH2CH2CH2P), 16.7 (s, Si0CH2), 16.9 (d, 2JCP = 3.9 Hz, CH2CH2P), 18.0 (s, Si0CH2CH2), 19.2 (s, Si0CH2CH2CH2), 24.6 (d, 1JCP = 49.1 Hz, PCH2), 55.9 (s, OCH3), 109.5 (d, 1JCP = 93.1 Hz, CP), 115.8 (d, 3JCP = 13.5 Hz, CHCHCP), 135.4 (d, 2JCP = 11.6 Hz, CHCP), 163.9 (d, 4JCP = 2.9 Hz, Cipso). 29Si {1H} (59.60 MHz): δ 0.70 (s, Si0), 1.53 (d, 4JSiP = 2.7 Hz, Si1). 31P {1H} (121.44 MHz): δ 26.47. HRMS (C80H180Cl4P4Si5): (ESI+) m/z: [M − Cl−]+ calcd for C80H180Cl3P4Si5: 1512.0940 found: 1512.0932, [M − 2Cl−]2+ calcd for C80H180Cl2P4Si5: 738.5621 found: 738.5627, [M − 3Cl−]3+ calcd for C80H180ClP4Si5: 480.3858 found: 480.3859, [M − 4Cl−]4+ calcd for C80H180P4Si5: 351.5466 found: 351.5471.
4.2.21 Dendrimer (23). Data for 23: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.21 (s, 12H, Si1CH3), −0.18 (48H, Si2CH3), 0.39–0.44 (m, 48H, Si0CH2, CH2Si1CH2CH2CH2), 0.65–0.68 (m, 16H, CH2CH2CH2P), 1.14–1.22 (m, 24H, CH2CH2Si1CH2CH2), 1.44–1.50 (m, 16H, CH2CH2P), 3.38–3.42 (m, 16H, CH2P), 3.85 (s, 72H, OCH3), 7.24–7.27 (m, 48H CH), 7.63–7.67 (m, 48H, CH). 13C {1H} (125.70 MHz): δ −5.1 (s, Si1CH3), −3.6 (s, Si2CH3), 16.5 (d, 3JCP = 13.4 Hz, CH2CH2CH2P), 16.9 (d, 2JCP = 4.4 Hz, CH2CH2P), 17.0 (s, Si0CH2), 17.9 (s, Si1CH2CH2CH2Si2), 18.1 (s, Si1CH2CH2CH2Si2), (Si0CH2CH2 overlap), 18.4 (s, Si0CH2CH2CH2), 19.2 (s, Si1CH2CH2CH2Si2), 24.6 (d, 1JCP = 49.2 Hz, CH2P), 55.9 (s, OCH3), 109.5 (d, 1JCP = 93.1 Hz, CP), 115.8 (d, 3JCP = 13.5 Hz, CHCHCP), 135.4 (d, 2JCP = 11.6 Hz, CHCP), 163.9 (d, 4JCP = 3.2 Hz, Cipso). 29Si {1H} (99.31 MHz): δ 0.7 (s, Si0), 0.95 (s, Si1), 1.43 (d, 4JSiP = 2.9 Hz, Si2). 31P {1H} (202.37 MHz): δ 20.59. HRMS (C176H396Cl8P8Si13): (ESI+) m/z: [M − 3Cl−]3+ calcd for C176H396Cl5P8Si13: 1100.8111 found: 1100.8111, [M − 4Cl−]4+ calcd for C176H396Cl4P8Si13: 816.8661 found: 816.8653, [M − 5Cl−]5+ calcd for C176H396Cl3P8Si13: 646.2991 found: 646.2989, [M − 6Cl−]6+ calcd for C176H396Cl2P8Si13: 532.7545 found: 532.7547, [M − 7Cl−]7+ calcd for C176H396ClP8Si13: 451.5083 found: 451.5086, [M − 8Cl−]8+ calcd for C176H396P8Si13: 390.6987 found: 390.6983. Anal. calc. C248H348I8O24P8Si13 (5341.54 g mol−1): C, 55.76; H, 6.57; exp.: C, 56.22; H, 6.78.
4.2.22 Dendrimer (24). Data for 24: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.20 (br s, 132H, Si1,2,3CH3), 0.40 (br s, 112H, Si0CH2, CH2Si1CH2, CH2Si2CH2CH2CH2Si3), 0.68 (br s, 32H, CH2CH2CH2P), 1.17 (br s, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.47 (br s, 32H, CH2CH2P), 3.42 (br s, 32H, CH2P), 3.83 (s, 144H, OCH3), 7.23–7.25 (m, 96H, CH), 7.64–7.68 (m, 96H, CH). 13C {1H} (125.70 MHz): δ −5.1 (s, Si1,2CH3), −3.7 (s, Si3CH3), 16.5 (d, 3JCP = 14.0 Hz, CH2CH2CH2P), 16.9 (d, 2JCP = 4.0 Hz, CH2CH2P); 17.9 (s, Si2CH2CH2CH2Si3), 18.1 (s, Si1CH2CH2CH2Si2), 18.3 (s, Si2CH2CH2CH2Si3), 18.6 (s, Si0CH2CH2CH2), 19.2 (s, Si2CH2CH2CH2Si3), 24.7 (d, 1JCP = 49.6 Hz, CH2P), 55.9 (s, OCH3), 109.5 (d, 1JCP = 93.2 Hz, CP), 115.8 (d, 3JCP = 13.4 Hz, CHCHCP), 135.4 (d, 2JCP = 11.7 Hz, CHCP), 163.9 (d, 4JCP = 3.1 Hz, Cipso), (Si0CH2CH2 were not detected or overlap). 29Si {1H} (inept) (99.31 MHz): δ Si0 was not detected, 0.92 (s, Si1,2), 1.39 (d, 4JSiP = 2.9 Hz, Si3). 31P {1H} (202.37 MHz): δ 20.57. HRMS (C368H828Cl16P16Si29): (ESI+) m/z: [M − 2Cl−]2+ calcd for C368H828Cl14P16Si29: 3529.9769 found: 3529.9799, [M − 3Cl−]3+ calcd for C368H828Cl13P16Si29: 2341.6616 found: 2341.6592, [M − 4Cl−]4+ calcd for C368H828Cl12P16Si29: 1747.2541 found: 1747.2537, [M − 5Cl−]5+ calcd for C368H828Cl11P16Si29: 1390.8095 found: 1390.8097, [M − 6Cl−]6+ calcd for C368H828Cl10P16Si29: 1153.0131 found: 1153.0127, [M − 7Cl−]7+ calcd for C368H828Cl9P16Si29: 983.3014 found: 983.2992, [M − 8Cl−]8+ calcd for C368H828Cl8P16Si29: 855.7677 found: 855.7692, [M − 9Cl−]9+ calcd for C368H828Cl7P16Si29: 756.9080 found: 756.9035, [M − 10Cl−]10+ calcd for C368H828Cl6P16Si29: 677.6204 found: 677.6217.
4.2.23 Dendrimer (28). Data for 28: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.17 (s, 24H, Si1CH3); 0.38–0.43 (m, 16H, Si0CH2CH2CH2), 0.68–0.72 (m, 8H, Si1CH2CH2CH2P), 1.16–1.18 (m, 8H, Si0CH2CH2), 1.50–1.52 (m, 8H, CH2CH2P), 3.61–3.64 (m, 8H, CH2P), 7.74–7.90 (m, 60H, CH Ph). 13C {1H} (125.70 MHz): δ −3.6 (s, Si1CH3), 16.6 (d, 3JCP = 13.6 Hz, CH2CH2CH2P), 16.7 (s, Si0CH2), 17.0 (d, 2JCP = 4.6 Hz, CH2CH2P), 18.0 (s, Si0CH2CH2), 19.2 (s, Si0CH2CH2CH2), 23.5 (d, 1JCP = 46.0 Hz, PCH2), 118.5 (d, 1JCP = 85.4 Hz, CP), 130.2 (d, 3JCP = 12.4 Hz, CHCHCP), 133.5 (d, 2JCP = 10.0 Hz, CHCP), 134.9 (d, 4JCP = 3.2 Hz, CHCHCHCP). 29Si {1H} (99.31 MHz): δ 0.69 (s, Si0), 1.55 (d, 4JSiP = 2.7 Hz, Si1). 31P {1H} (202.37 MHz): δ 22.88. HRMS: C104H132P4Si5I4 (ESI+): m/z calc. for [C104H132P4Si5I3]+ calc. 2026.5281 found 2026.5244; [C104H132P4Si5I2]2+ calc. 949.8115 found 949.8118; [C104H132P4Si5I]3+ calc. 590.9060 found 590.9052 [C104H132P4Si5]4+ calc. 411.4532 found 411.4533.
4.2.24 Dendrimer (29). Data for 29: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.20 (br s, 60H, Si1CH3, Si2CH3), 0.38–0.45 (m, 48H, CH2CH2CH2Si1CH2CH2CH2), 0.66–0.70 (m, 16H, CH2CH2CH2P), 1.15–1.21 (m, 24H, CH2CH2Si1CH2CH2), 1.47–1.52 (m, 16H, CH2CH2P), 3.56–3.62 (m, 16H, CH2P), 7.72–7.89 (m, 120H, CH Ph). 13C {1H} (125.70 MHz): δ −5.0 (s, Si1CH3), −3.6 (s, Si2CH3), 16.5 (d, 3JCP = 13.6 Hz, CH2CH2CH2P), 16.97 (d, 2JCP = 4.8 Hz, CH2CH2P), 17.04 (s, Si0CH2), 17.9 (s, Si1CH2CH2CH2Si2), 18.1 (s, Si1CH2CH2CH2Si2), 18.2 (s, Si0CH2CH2), 18.4 (s, Si0CH2CH2CH2), 19.2 (s, Si1CH2CH2CH2Si2), 23.5 (d, 1JCP = 46.2 Hz, CH2P), 118.5 (d, 1JCP = 85.2 Hz, CP), 130.2 (d, 3JCP = 12.4 Hz, CHCHCP), 133.5 (d, 2JCP = 10.1 Hz, CHCP), 134.9 (d, 4JCP = 3.1 Hz, CHCHCHCP). 29Si {1H} (99.31 MHz): δ 0.74 (s, Si0), 1.00 (s, Si1), 1.52 (d, 4JSiP = 2.9 Hz, Si2). 31P {1H} (202.37 MHz): δ 22.91. HRMS: C224H300P8Si13I8 (ESI+): m/z calc. for [C224H300P8Si13I6]2+ calc. 1943.5098 found 1943.5098; [C224H300P8Si13I5]3+ calc. 1253.3715 found 1253.3708; [C224H300P8Si13I4]4+ calc. 908.3024 found 908.3029; [C224H300P8Si13I3]5+ calc. 701.2609 found 701.2603; [C224H300P8Si13I2]6+ calc. 563.2332 found 563.2317; [C224H300P8Si13I]7+ calc. 464.6420 found 464.6420.
4.2.25 Dendrimer (30). Data for 30: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.22 (br s, 132H, Si1,2,3CH3), 0.38–0.46 (m, 112H, CH2CH2CH2Si1CH2CH2CH2Si2CH2CH2CH2), 0.67–0.71 (m, 32H, CH2CH2CH2P), 1.14–1.24 (m, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.45–1.52 (m, 32H, CH2CH2P), 3.57–3.66 (m, 32H, CH2P), 7.73–7.86 (m, 240H, CH Ph). 13C {1H} (125.70 MHz): δ −5.0 (s, Si1,2CH3), −3.6 (s, Si3CH3), 16.5 (d, 3JCP = 13.4 Hz, CH2CH2CH2P), 17.0 (d, 2JCP = 4.0 Hz, CH2CH2P), 17.9 (s, Si2CH2CH2CH2Si3), (Si0CH2CH2CH2 were not detected or overlap), 18.07 (s, Si1CH2CH2CH2Si2), 18.12 (s, Si2CH2CH2CH2Si3), 18.3 (s, Si1CH2CH2CH2Si2), 19.2 (s, Si2CH2CH2CH2Si3), 23.5 (d, 1JCP = 46.1 Hz, CH2P), 118.5 (d, 1JCP = 85.2 Hz, CP), 130.2 (d, 3JCP = 12.3 Hz, CHCHCP), 133.6 (d, 2JCP = 10.1 Hz, CHCP), 134.9 (d, 4JCP = 2.9 Hz, CHCHCHCP). 29Si {1H} (59.60 MHz): δ Si0 was not detected, 0.97 (s, Si1, Si2), 1.47. (d, 4JSiP = 3.0 Hz, Si3). 31P {1H} (202.37 MHz): δ 22.87. HRMS: C464H636P16Si29I16 (ESI+): m/z calc. for [C464H636P16Si29I13]3+ 3057.8860 found 3057.8863; [C464H636P16Si29I12]4+ calc. 2261.6882 found 2261.6880; [C464H636P16Si29I11]5+ calc. 1783.9696 found 1783.9639; [C464H636P16Si29I10]6+ calc. 1465.4905 found 1465.4898; [C464H636P16Si29I9]7+ calc.1238.0054 found 1238.0038; [C464H636P16Si29I8]8+ calc. 1067.3916 found 1067.3900; [C464H636P16Si29I7]9+ calc. 934.6920 found 934.6933; [C464H636P16Si29I6]10+ 828.5323 found 828.5305; [C464H636P16Si29I5]11+ calc. 741.6744 found 741.6765; [C464H636P16Si29I4]12+ calc. 669.2927 found 669.2976; [C464H636P16Si29I3]13+ calc. 608.0467 found 608.0451.Dendrimers 19–21, 25–27, 31–33 were prepared according to similar procedure. A respective iodo derivative was converted to chloro derivative by ion exchange on AMBERLYST A-21 with methanol as the mobile phase. In all cases yields are in range between 98–100%.
4.2.26 Dendrimer (19). Data for 19: NMR (CD3CN): 1H (499.99 MHz): δ 0.00 (s, 24H, Si1CH3), 0.56–0.61 (m, 16H, Si0CH2CH2CH2), 0.64–0.67 (m, 8H, Si1CH2CH2CH2P), 0.95 (t, 3JHH = 7.1 Hz, 36H, CH3), 1.31–1.38 (m, 8H, Si0CH2CH2CH2), 1.42–1.55 (m, 56H, Si1CH2CH2CH2PCH2CH2CH2), 2.14–2.21 (m, 32H, CH2P). 13C {1H} (125.70 MHz): δ −3.2 (s, Si1CH3), 13.7 (d, 4JCP = 0.9 Hz, CH2CH3), 17.4 (d, 2JCP = 4.7 Hz, Si1CH2CH2CH2P), 18.0 (d, 3JCP = 12.9 Hz, Si1CH2CH2CH2P), 18.2 (s, Si0CH2), 19.1 (d, 1JCP = 47.8 Hz, PCH2CH2CH2CH3), 19.4 (s, Si0CH2CH2), 20.6 (s, Si0CH2CH2CH2), 23.0 (d, 1JCP = 44.6 Hz, Si1CH2CH2CH2P), 24.0 (d, 2JCP = 4.3 Hz, CH3CH2CH2CH2P), 24.5 (d, 3JCP = 15.3 Hz, CH3CH2). 29Si {1H} (99.31 MHz): δ 0.60 (s, Si0), 1.58 (d, 4JSiP = 2.4 Hz, Si1). 31P {1H} (121.41 MHz): δ 33.60. HRMS (C116H156P4Si5I4O12): (ESI+) m/z: [M − I−]+calcd for C116H156P4Si5I3O12: 2386.6549 found: 2386.6554, [M − 2I−]2+ calcd for C116H156P4Si5I2O12: 1129.8749 found: 1129.8755, [M − 3I−]3+ calcd for C116H156P4Si5I3O12: 710.9483 found: 710.9478, [M − 4I−]4+ calcd for C116H156P4Si5O12: 501.4849 found: 501.4853.
4.2.27 Dendrimer (20). Data for 20: NMR (CD3CN): 1H (499.99 MHz): δ −0.07 (s, 12H, Si1CH3), −0.01 (s, 48H, Si2CH3), 0.53–0.60 (m, 48H, Si0CH2, CH2Si1CH2CH2CH2), 0.64–0.67 (m, 16H, Si2CH2CH2CH2P), 0.94 (t, 3JHH = 7.1 Hz, 72H, CH3), 1.31–1.36 (m, 24H, CH2CH2Si1CH2CH2), 1.42–1.54 (m, 112H, Si2CH2CH2CH2P, CH2CH2CH3), 2.17–2.27 (m, 64H, CH2P). 13C {1H} (125.70 MHz): δ −4.4 (s, Si1CH3), −3.1 (s, Si2CH3), 13.7 (s, CH2CH3), 17.4 (d, 2JCP = 4.7 Hz, Si2CH2CH2CH2P), 18.0 (d, 3JCP = 12.9 Hz, Si2CH2CH2CH2P), 18.42 (s, Si0CH2), 19.1 (d, 1JCP = 47.6 Hz, PCH2CH2CH2CH3), 19.3 (s, Si1CH2CH2CH2Si2), 19.6 (s, Si1CH2CH2CH2Si2, Si0CH2CH2), 19.8 (s, Si0CH2CH2CH2), 20.6 (s, Si1CH2CH2CH2Si2), 23.1 (d, 1JCP = 44.5 Hz, Si2CH2CH2CH2P), 24.1 (d, 2JCP = 4.6 Hz, CH3CH2CH2CH2P), 24.6 (d, 3JCP = 15.7 Hz, CH3CH2). 29Si {1H} (59.60 MHz): δ 0.74 (s, Si0), 0.98 (s, Si1), 1.54 (d, 4JSiP = 2.5 Hz, Si2). 31P {1H} (202.37 MHz): δ 32.47. HRMS (C248H348P8O24Si13I8): (ESI+) m/z: [M − 3I−]3+ calcd for C248H348P8O24Si13I5: 1653.5399 found: 1653.5380, [M − 4I−]4+ calcd for C248H348P8O24Si13I4: 1208.4287 found: 1208.4270, [M − 5I−]5+ calcd for C248H348P8O24Si13I3: 941.3619 found: 941.3607, [M − 6I−]6+ calcd for C248H348P8O24Si13I2: 763.3174 found: 763.3156, [M − 7I−]7+ calcd for C248H348P8O24Si13I: 636.1428 found: 636.1405, [M − 8I−]8+ calcd for C248H348P8O24Si13: 540.7618 found: 540.7615.
4.2.28 Dendrimer (21). Data for 21: NMR (CD3CN): 1H (499.99 MHz): δ −0.08 (s, 12H, Si1CH3), −0.06 (s, 24H, Si2CH3), 0.00 (s, 96H, Si3CH3), 0.54–0.59 (m, 112H, Si0CH2, CH2Si1CH2, CH2Si2CH2CH2CH2Si3), 0.64–0.68 (m, 32H, Si3CH2CH2CH2P), 0.95 (t, 3JHH = 7.1 Hz, 144H, CH3), 1.32–1.37 (m, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.44–1.55 (m, 224H, Si3CH2CH2CH2P, CH2CH2CH3), 2.21–2.30 (m, 128H, CH2P). 13C {1H} (125.70 MHz): δ −4.2 (s, Si1,2CH3), −2.9 (s, Si3CH3), 13.8 (s,CH2CH3), 17.5 (d, 2JCP = 4.2 Hz, Si3CH2CH2CH2P), 18.1 (d, 3JCP = 13.0 Hz, Si3CH2CH2CH2P), 18.6 (s, Si0CH2), 19.2 (d, 1JCP = 47.8 Hz, PCH2CH2CH2CH3), (s, Si0CH2CH2 overlap), 19.3 (s, Si2CH2CH2CH2Si3), 19.5 (s, Si1CH2CH2CH2Si2), 19.6 (s, Si2CH2CH2CH2Si3), 19.8 (s, Si1CH2CH2CH2Si2), 20.0 (s, Si0CH2CH2CH2), 20.7 (s, Si2CH2CH2CH2Si3), 23.2 (d, 1JCP = 44.9 Hz, Si3CH2CH2CH2P), 24.2 (d, 2JCP = 4.4 Hz, CH3CH2CH2CH2P), 24.6 (d, 3JCP = 15.5 Hz, CH3CH2). 29Si {1H} (59.60 MHz): δ (Si0 was not detected), 1.01 (s, Si1,2), 1.53 (d, 4JSiP = 1.8 Hz, Si3). 31P {1H} (202.37 MHz): δ 32.43. HRMS (C512H732P16Si29I16O48): (ESI+) m/z: [M − 4I−]4+ calcd for C512H732P16Si29I12O48: 2621.8153 found: 2621.8113, [M − 5I−]5+ calcd for C512H732P16Si29I11O48: 2072.0712 found: 2072.0691, [M − 6I−]6+ calcd for C512H732P16Si29I10O48: 1705.5750 found: 1705.5777, [M − 7I−]7+ calcd for C512H732P16Si29I9O48: 1443.7923 found: 1443.7899, [M − 8I]8+ calcd for C512H732P16Si29I8O48: 1247.4551 found: 1247.4601, [M − 9I−]9+ calcd for C512H732P16Si29I7O48: 1094.7484 found: 1094.7459, [M − 10I−]10+ calcd for C512H732P16Si29I6O48: 972.5831 found: 972.5753, [M − 11I−]11+ calcd for C512H732P16Si29I5O48: 872.6296 found: 872.6241, [M − 12I−]12+ calcd for C512H732P16Si29I4O48: 789.3351 found: 789.3337, [M − 13I−]13+ calcd for C512H732P16Si29I3O48: 718.8555 found: 718.8503.
4.2.29 Dendrimer (25). Data for 25: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.16 (s, 24H, Si1CH3), 0.39–0.43 (m, 16H, Si0CH2CH2CH2), 0.64–0.67 (m, 8H, CH2CH2CH2P), 1.13–1.21 (m, 8H, Si0CH2CH2), 1.45–1.50 (m, 8H, CH2CH2P), 3.37–3.42 (m, 8H, CH2P), 3.86 (s, 36H, OCH3), 7.26–7.28 (m, 24H, CH), 7.63–7.68 (m, 24H, CH). 13C {1H} (125.70 MHz): δ −3.7 (s, Si1CH3), 16.6 (d, 3JCP = 13.8 Hz, CH2CH2CH2P), 16.8 (s, Si0CH2), 16.9 (d, 2JCP = 4.1 Hz, CH2CH2P), 18.0 (s, Si0CH2CH2), 19.2 (s, Si0CH2CH2CH2), 24.5 (d, 1JCP = 49.1 Hz, PCH2), 55.9 (s, OCH3), 109.6 (d, 1JCP = 93.0 Hz, CP), 115.8 (d, 3JCP = 13.5 Hz, CHCHCP), 135.4 (d, 2JCP = 11.7 Hz, CHCP), 163.9 (d, 4JCP = 3.0 Hz, Cipso). 29Si {1H} (99.31 MHz): δ 0.65 (s, Si0), 1.49 (d, 4JSiP = 2.8 Hz, Si1). 31P {1H} (202.37 MHz): δ 20.63. HRMS (C116H156P4Si5Cl4O12): (ESI+) m/z: [M − Cl−]+ calcd for C116H156P4Si5Cl3O12: 2112.8474 found: 2112.8485, [M − 2Cl−]2+ calcd for C116H156P4Si5Cl2O12: 1038.4390 found: 1038.4388, [M − 3Cl−]3+ calcd for C116H156P4Si5ClO12: 680.6365 found: 680.6366, [M − 4Cl−]4+ calcd for C116H156P4Si5O12: 501.4849 found: 501.4846.
4.2.30 Dendrimer (26). Data for 26: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.22 (s, 12H, Si1CH3), −0.19 (48H, Si2CH3), 0.39–0.42 (m, 48H, Si0CH2, CH2Si1CH2CH2CH2), 0.65–0.68 (m, 16H, CH2CH2CH2P), 1.18 (br s, 24H, CH2CH2Si1CH2CH2), 1.46 (br s, 16H, CH2CH2P), 3.41–3.50 (m, 16H, CH2P), 3.85 (s, 72H, OCH3), 7.24–7.27 (m, 48H CH), 7.66–7.69 (m, 48H, CH). 13C {1H} (125.70 MHz): δ −5.1 (s, Si1CH3), −3.6 (s, Si2CH3), 16.5 (d, 3JCP = 13.7 Hz, CH2CH2CH2P), 16.9 (d, 2JCP = 4.4 Hz, CH2CH2P), 17.1 (s, Si0CH2), 17.9 (s, Si1CH2CH2CH2Si2), 18.1 (s, Si1CH2CH2CH2Si2), 18.2 (s, Si0CH2CH2), 18.4 (s, Si0CH2CH2CH2), 19.2 (s, Si1CH2CH2CH2Si2), 24.5 (d, 1JCP = 49.1 Hz, CH2P), 55.9 (s, OCH3), 109.6 (d, 1JCP = 93.1 Hz, CP), 115.8 (d, 3JCP = 13.6 Hz, CHCHCP), 135.4 (d, 2JCP = 11.6 Hz, CHCP), 163.9 (d, 4JCP = 3.1 Hz, Cipso). 29Si {1H} (99.31 MHz): δ (Si0 was not detected), 0.97 (s, Si1), 1.44 (d, 4JSiP = 2.9 Hz, Si2). 31P {1H} (202.37 MHz): δ 20.64. HRMS (C248H348P8Si13Cl8O24): (ESI+) m/z: [M − 2Cl−]2+ calcd for C248H348P8Si13Cl6O24: 2269.4534 found: 2269.4517, [M − 3Cl−]3+ calcd for C248H348P8Si13Cl5O24: 1500.9793 found: 1500.9760, [M − 4Cl−]4+ calcd for C248H348P8Si13Cl4O24: 116.9923 found: 116.9917, [M − 5Cl−]5+ calcd for C248H348P8Si13Cl3O24: 886.4001 found: 886.3995, [M − 6Cl−]6+ calcd for C248H348P8Si13Cl2O24: 732.8386 found: 732.8341, [M − 7Cl−]7+ calcd for C248H348P8Si13ClO24: 623.0086 found: 623.0069, [M − 8Cl−]8+ calcd for C248H348P8Si13O24: 540.7618 found: 540.7604.
4.2.31 Dendrimer (27). Data for 27: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.23 (br s, 132H, Si1,2,3CH3), 0.39 (br s, 112H, Si0CH2, CH2Si1CH2, CH2Si2CH2CH2CH2Si3), 0.67 (br s, 32H, CH2CH2CH2P), 1.48 (br s, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.48 (br s, 32H, CH2CH2P), 3.51 (br s, 32H, CH2P), 3.83 (s, 144H, OCH3), 7.22–7.24 (m, 96H, CH), 7.66–7.70 (m, 96H, CH). 13C {1H} (125.70 MHz): δ −5.1 (s, Si1,2CH3), −3.7 (s, Si3CH3), 16.4 (d, 3JCP = 13.4 Hz, CH2CH2CH2P), 16.9 (d, 2JCP = 4.2 Hz, CH2CH2P); 17.9 (s, Si2CH2CH2CH2Si3); 18.1 (br s, Si1CH2CH2CH2Si2), 18.3 (s, Si2CH2CH2CH2Si3), 19.2 (s, Si2CH2CH2CH2Si3), 24.6 (d, 1JCP = 49.3 Hz, CH2P), 55.9 (s, OCH3), 109.6 (d, 1JCP = 85.2 Hz, CP), 115.8 (d, 3JCP = 13.2 Hz, CHCHCP), 135.4 (d, 2JCP = 11.6 Hz, CHCP), 163.9 (s, Cipso), (Si0CH2CH2CH2 were not detected or overlap). 29Si {1H} (inept) (99.31 MHz): δ Si0 was not detected, 0.91 (s, Si1,2), 1.36 (d, 4JSiP = 2.9 Hz, Si3). 31P {1H} (202.37 MHz): δ 20.64. HRMS (C512H732P16Si29Cl16O48): (ESI+) m/z: [M − 3Cl−]3+ calcd for C512H732P16Si29Cl13O48: 3141.9983 found: 3142.0013, [M − 4Cl−]4+ calcd for C512H732P16Si29Cl12O48: 2347.5066 found: 2347.5082, [M − 5Cl−]5+ calcd for C512H732P16Si29Cl11O48: 1871.0115 found: 1871.0099, [M − 6Cl−]6+ calcd for C512H732P16Si29Cl10O48: 1553.1815 found: 1553.1836, [M − 7Cl−]7+ calcd for C512H732P16Si29Cl9O48: 1326.3029 found: 1326.3036, [M − 8Cl−]8+ calcd for C512H732P16Si29Cl8O48: 1156.0189 found: 1156.0149, [M − 9Cl−]9+ calcd for C512H732P16Si29Cl7O48: 1023.6869 found: 1023.6872, [M − 10Cl−]10+ calcd for C512H732P16Si29Cl6O48: 917.7214 found: 917.7234, [M − 11Cl−]11+ calcd for C512H732P16Si29Cl5O48: 831.1132 found: 831.1105, [M − 12Cl−]12+ calcd for C512H732P16Si29Cl4O48: 758.8564 found: 758.8504, [M − 13Cl−]13+ calcd for C512H732P16Si29Cl3O48: 697.7929 found: 697.7889.
4.2.32 Dendrimer (31). Data for 31: NMR (DMSO-d6): 1H (400.13 MHz): δ −0.18 (s, 24H, Si1CH3), 0.38–0.43 (m, 16H, Si0CH2CH2CH2), 0.66–0.70 (m, 8H, CH2CH2CH2P), 1.11–1.20 (m, 8H, Si0CH2CH2), 1.45–1.55 (m, 8H, CH2CH2P), 3.60–3.67 (m, 8H, CH2P), 7.73–7.90 (m, 60H, CH Ph). 13C {1H} (100.62 MHz): δ −3.7 (s, Si1CH3), 16.6 (d, 3JCP = 13.6 Hz, CH2CH2CH2P), 16.8 (s, Si0CH2), 17.0 (d, 2JCP = 4.6 Hz, CH2CH2P), 18.0 (s, Si0CH2CH2), 19.2 (s, Si0CH2CH2CH2), 23.4 (d, 1JCP = 46.3 Hz, PCH2), 118.6 (d, 1JCP = 85.3 Hz, CP), 130.2 (d, 3JCP = 12.4 Hz, CHCHCP), 133.6 (d, 2JCP = 10.1 Hz, CHCP), 134.9 (d, 4JCP = 2.6 Hz, CHCHCHCP). 29Si {1H} (79.49 MHz): δ 0.68 (s, Si0), 1.56 (d, 4JSiP = 2.9 Hz, Si1). 31P {1H} (161.98 MHz): δ 22.99. C104H132P4Si5Cl4 (ESI+): m/z calc. for [C104H132P4Si5Cl3]+ calc. 1751.7194 found 1751.7174; [C104H132P4Si5Cl2]2+ calc. 858.3754 found 858.3751; [C104H132P4Si5Cl]3+ calc. 560.5941 found 560.5945 [C104H132P4Si5]4+ calc. 411.4532 found 411.4533.
4.2.33 Dendrimer (32). Data for 32: NMR (DMSO-d6): 1H (499.99 MHz): δ −0.21 (br s, 60H, Si1CH3, Si2CH3), 0.38–0.49 (m, 48H, CH2CH2CH2Si1CH2CH2CH2), 0.70–0.76 (m, 16H, CH2CH2CH2P), 1.12–1.27 (m, 24H, CH2CH2Si1CH2CH2), 1.43–1.58 (m, 16H, CH2CH2P), 3.61–3.76 (m, 16H, CH2P), 7.71–7.89 (m, 120H, CH Ph). 13C {1H} (125.70 MHz): δ −5.0 (s, Si1CH3), −3.7 (s, Si2CH3), 16.5 (d, 3JCP = 13.4 Hz, CH2CH2CH2P), 17.0 (d, 2JCP = 4.7 Hz, CH2CH2P), 17.1 (s, Si0CH2), 17.9 (s, Si1CH2CH2CH2Si2), 18.1 (s, Si1CH2CH2CH2Si2), 18.2 (s, Si0CH2CH2), 18.4 (s, Si0CH2CH2CH2), 19.2 (s, Si1CH2CH2CH2Si2), 23.4 (d, 1JCP = 46.0 Hz, CH2P), 118.6 (d, 1JCP = 85.3 Hz, CP), 130.2 (d, 3JCP = 12.4 Hz, CHCHCP), 133.6 (d, 2JCP = 10.2 Hz, CHCP), 134.8 (d, 4JCP = 3.2 Hz, CHCHCHCP). 29Si {1H} (79.49 MHz): δ 0.70 (s, Si0), 0.97 (s, Si1), 1.48 (d, 4JSiP = 3.0 Hz, Si2). 31P {1H} (161.98 MHz): δ 22.98. HRMS: (ESI+): m/z calc. for C224H300P8Si13Cl8 (ESI+): [C224H300P8Si13Cl6]2+ calc. 1908.8261 found 1908.8280; [C224H300P8Si13Cl5]3+ calc.1260.8945 found 1260.8946; [C224H300P8Si13Cl4]4+ calc. 936.6787 found 936.6796; [C224H300P8Si13Cl3]5+ calc. 742.3492 found 742.3495; [C224H300P8Si13Cl2]6+ calc. 612.6295 found 612.6282; [C224H300P8Si13Cl]7+ calc. 520.1155 found 520.1142; [C224H300P8Si13]8+ calc. 450.6049 found 450.6046. Anal. calc. C224H300Cl8P8Si13 (3889.30 g mol−1): C, 69.17; H, 7.17; exp.: C, 69.82; H, 7.31.
4.2.34 Dendrimer (33). Data for 33: NMR (DMSO-d6): 1H (400.13 MHz): δ −0.23 (br s, 132H, Si1,2,3CH3), 0.38–0.46 (m, 112H, CH2CH2CH2Si1CH2CH2CH2Si2CH2CH2CH2), 0.64–0.72 (m, 32H, CH2CH2CH2P), 1.10–1.24 (m, 56H, CH2CH2Si1CH2CH2CH2Si2CH2CH2), 1.42–1.55 (m, 32H, CH2CH2P), 3.67–3.81 (m, 32H, CH2P), 7.69–7.85 (m, 240H, CH Ph). 13C {1H} (100.62 MHz): δ −5.0 (s, Si1,2CH3), −3.7 (s, Si3CH3), 16.4 (d, 3JCP = 13.8 Hz, CH2CH2CH2P), 17.0 (d, 2JCP = 4.5 Hz, CH2CH2P), 17.9 (s, Si2CH2CH2CH2Si3), (Si0CH2CH2CH2 were not detected or overlap), 18.07 (s, Si1CH2CH2CH2Si2), 18.12 (s, Si2CH2CH2CH2Si3), 18.3 (s, Si1CH2CH2CH2Si2), 19.2 (s, Si2CH2CH2CH2Si3), 23.4 (d, 1JCP = 45.9 Hz, CH2P), 118.7 (d, 1JCP = 85.2 Hz, CP), 130.1 (d, 3JCP = 12.4 Hz, CHCHCP), 133.6 (d, 2JCP = 10.1 Hz, CHCP), 134.8 (d, 4JCP = 2.0 Hz, CHCHCHCP). 29Si {1H} (79.49 MHz): δ Si0 was not detected, 0.94 (s, Si1, Si2), 1.42 (d, 4JSiP = 3.0 Hz, Si3). 31P {1H} (161.98 MHz): δ 23.02. HRMS: (ESI+): m/z calcd for C464H636P16Si29Cl16 (ESI+): [C464H636P16Si29Cl13]3+ calc. 2661.4953 found 2661.4946; [C464H636P16Si29Cl12]4+ calc. 1987.1293 found 1987.1277; [C464H636P16Si29Cl11]5+ calc. 1582.7097 found 1582.7090; [C464H636P16Si29Cl10]6+ calc. 1312.9299 found 1312.9299; [C464H636P16Si29Cl9]7+ calc. 1120.3730 found 1120.3722; [C464H636P16Si29Cl8]8+ calc. 975.8303 found 975.8358; [C464H636P16Si29Cl7]9+ calc. 863.5192 found 863.5160; [C464H636P16Si29Cl6]10+ calc. 773.6705 found 773.6698; [C464H636P16Si29Cl5]11+ calc. 700.0669 found 700.0637; [C464H636P16Si29Cl4]12+ calc. 638.8139 found 638.8103; [C464H636P16Si29Cl3]13+ calc. 586.9075 found 586.9050.
4.3 Computer modelling of dendrimers
3D computer models of dendrimer structures were created using dendrimer builder, as implemented in the Materials Studio software package from BIOVIA (formerly Accelrys). The RESP technique52 was used for calculation of dendrimer atoms partial charges. For this charge parametrisation the R.E.D.-IV tools53 was used. The necessary QM calculations (QM structure minimisations, molecular electrostatic potential (MEP) calculations) were done using GAMESS.54,55 The default, HF/6-31G*, level of theory was used for all charge-related QM calculations and the MEP potential was fitted on Connolly molecular surface. GAFF force field (Generalized Amber Force Field), was used for parametrization of dendrimers.56 Missing force field parameters were fitted by minimizing the differences between QM and force field based relative energies of properly chosen molecular fragments. QM energies were calculated at MP2/HF/6-31G** level of theory using GAMESS and fitting was accomplished using paramfit routine from AMBER14 software.57 Slightly adjusted van der Waals parameters for Si atoms from MM3 force field were used in this study.58 Dendrimer models were solvated in explicit water (TIP3P model) with the proper number of Na+ and Cl− ions to preserve neutrality of the system and to ensure the physiological ionic strength (0.15 M).59 First the systems were minimized (5000 steps with 2 kcal (mol Å2)−1 restraint + 5000 without restraint), heated (200 ps NVT) to 294 K and equilibrated using 70 ns long molecular dynamics simulations (NPT, T = 294 K, P = 0.1 MPa). The first 0.5 ns with restrained solute. Hydrogens were constrained with the SHAKE algorithm to allow 2 fs time step60 and Langevin thermostat with collision frequency 2 ps−1 was used for all MD runs.61 The pressure relaxation time for weak-coupling barostat was 2 ps. Particle mesh Ewald method (PME) was used to treat long range electrostatic interactions under periodic conditions with a direct space cutoff of 10 Angstroms. The same cutoff was used for van der Waals interactions. The pmemd.cuda module from Amber14 package was used for all simulation steps.62 Radial distribution function calculations were done using last 20 ns of MD trajectory (2000 frames analyzed). In case of size characteristics calculations (Rg, Rmax) last 40 ns was used (80 frames analyzed). The cpptraj module from Amber14 was used for last mentioned analysis.63 Adaptive Poisson Boltzmann Solver (APBS) was used for electrostatic potential calculation.64 Chimera software was used for all visualizations.65
4.4 In vitro toxicity evaluation
4.4.1 Cell culture. B14 cells (Cricetulus griseus, ATCC, CCL-14.1, Sigma-Aldrich Inc.) were grown in High glucose DMEM medium with 10% (v/v) fetal bovine serum (FBS) and 4 mM glutamine, BRL 3A (Rattus norvegicus, ATCC, CRL-1442, Sigma-Aldrich Inc.) in Ham's F12 medium with 10% (v/v) FBS and 2 mM glutamine, and NRK 52E (Rattus norvegicus, ATCC, CRL-1571, Sigma-Aldrich Inc.) in high glucose DMEM with 2 mM glutamine, 10% (v/v) new born calf serum (NBCS) and 1% (v/v) nonessential amino acid solution (NEAA), all media were supplemented with 0.1% (w/v) penicillin and 0.1% (w/v) streptomycin. The cells were maintained in culture flasks in a 37 °C humidified atmosphere of 5% CO2/95% air (incubator) and passaged every 2–3 days. Cells were harvested and used in experiments after obtaining 80–90% confluence. The number of viable cells was determined by trypan blue exclusion on a haemocytometer. Then cells were suspended in media in a concentration of 1.0 × 105 cells per mL and plated in flat bottom 96-well plates. Plates with cells were incubated 24 h at a 37 °C in a humidified atmosphere of 5% CO2 to allow adherence of the cells before the administration of dendrimers.
4.4.2 MTT assay. Cytotoxicity of dendrimers was assayed with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium-bromide). Briefly, after 24 h incubation when cells attached on 96-well plates they were treated with concentration range from 0.5 to 20 μM carbosilane dendrimers. After 24 h incubation, a solution of MTT in PBS was added to each well. Four hours later the medium was removed and the formazan precipitate dissolved in DMSO for absorbance measurement at 580 nm and reference 700 nm. Viability is given graphically as a percent of the control values (without dendrimers).
4.4.3 Crystal violet staining assay (CVS). The cytotoxicity effect was also evaluated by crystal violet assay. Cells were seeded in 96-well plates and were grown in 100 μl of appropriate growth medium for 24 h. Then cells were treated with carbosilane dendrimers as it has been done in a MMT assay. After 24 h incubation cells were washed by phosphate buffer and 50 μl of 0.05% crystal violet solution in 20% methanol was added. Cells had been incubated with staining solution for 30 min and after incubation double washed with distilled water and left to dry. After drying process the crystal violet was washed from dry cells by methanol and the absorbance was measured at 570 nm and reference 700 nm. Viability is given graphically as a percent of the control values (without dendrimers).
4.5 IC50 calculations
The IC50 values (concentration of dendrimers which inhibit the cell viability to 50% of the control sample) were calculated from the best fit of experimental data with four-parameter logistic function (4PL function, R2 > 0.95) using nonlinear regression analysis in Graph Pad Prism software (GraphPad Software, Inc. USA, version 7):| | |
Y = 100/(1 + (XHillSlope)/(IC50HillSlope))
| (1) |
where X = log of dendrimer concentration, Y = growth inhibition value normalized to control and the HillSlope represent unitless factor.
4.6 Statistical analysis
Measured data are presented as the mean value ± standard deviation (error bars, S.D.). Each experimental variant was conducted in at least three independent runs. The statistical analysis was performed with GraphPad Prism 7 (GraphPad Software, Inc, USA) software using an unpaired t-test or one-way ANOVA followed with Tukey–Kramer multiply comparison test. p < 0.05 was considered statistically significant.
Acknowledgements
The authors acknowledge the project No. 15-05903S of the Czech Science Foundation and project of the Internal Grant Agency of UJEP. The authors also acknowledge the assistance provided by the Research Infrastructure NanoEnviCz, supported by the Ministry of Education, Youth and Sports of the Czech Republic under Project No. LM2015073.
References
- J. M. Oliveira, A. J. Salgado, N. Sousa, J. F. Mano and R. L. Reis, Prog. Polym. Sci., 2010, 35, 1163–1194 CrossRef CAS.
- V. Gajbhiye, V. K. Palanirajan, R. K. Tekade and N. K. Jain, J. Pharm. Pharmacol., 2009, 61, 989–1003 CrossRef CAS PubMed.
- C. Dufès, I. F. Uchegbu and A. G. Schätzlein, Adv. Drug Delivery Rev., 2005, 57, 2177–2202 CrossRef PubMed.
- D. Shcharbin, E. Pedziwiatr, J. Blasiak and M. Bryszewska, J. Controlled Release, 2010, 141, 110–127 CrossRef CAS PubMed.
- A. J. Salgado, J. M. Oliveira, R. P. Pirraco, V. H. Pereira, J. S. Fraga, A. P. Marques, N. M. Neves, J. F. Mano, R. L. Reis and N. Sousa, Macromol. Biosci., 2010, 10, 1130–1140 CrossRef CAS PubMed.
- A. M. Caminade, C. O. Turrin and J. P. Majoral, Chemistry, 2008, 14, 7422–7432 CrossRef CAS PubMed.
- K. Jain, P. Kesharwani, U. Gupta and N. K. Jain, Int. J. Pharm., 2010, 394, 122–142 CrossRef CAS PubMed.
- J. Maly, H. Lampova, A. Semeradtova, M. Stofik and L. Kovacik, Nanotechnology, 2009, 20, 385101 CrossRef CAS PubMed.
- S. Zhu, M. Hong, L. Zhang, G. Tang, Y. Jiang and Y. Pei, Pharm. Res., 2010, 27, 161–174 CrossRef CAS PubMed.
- Y. Zhao, S. Liu, Y. Li, W. Jiang, Y. Chang, S. Pan, X. Fang, Y. A. Wang and J. Wang, J. Colloid Interface Sci., 2010, 350, 44–50 CrossRef CAS PubMed.
- K. Hatano, K. Matsuoka and D. Terunuma, Chem. Soc. Rev., 2013, 42(11), 4574–4598 RSC.
- D. Wrobel, D. Appelhans, M. Signorelli, B. Wiesner, D. Fessas, U. Scheler, B. Voit and J. Maly, Biochim. Biophys. Acta, Biomembr., 2015, 1848, 1490–1501 CrossRef CAS PubMed.
- D. Wrobel, A. Janaszewska, D. Appelhans, B. Voit, M. Bryszewska and J. Maly, Int. J. Pharm., 2015, 496, 475–488 CrossRef CAS PubMed.
- J. Maly, E. Pedziwiatr-Werbicka, M. Maly, A. Semeradtova, D. Appelhans, A. Danani, M. Zaborski, B. Klajnert and M. Bryszewska, Curr. Med. Chem., 2012, 19, 4708–4719 CrossRef CAS PubMed.
- H. B. Agashe, T. Dutta, M. Garg and N. K. Jain, J. Pharm. Pharmacol., 2006, 58, 1491–1498 CrossRef CAS PubMed.
- S. Nimesh, A. Aggarwal, P. Kumar, Y. Singh, K. C. Gupta and R. Chandra, Int. J. Pharm., 2007, 337, 265–274 CrossRef CAS PubMed.
- J. Maly, O. Stanek, J. Frolik, M. Maly, F. Ennen, D. Appelhans, A. Semeradtova, D. Wrobel, M. Stofik, T. Knapova, M. Kuchar, L. C. Stastna, J. Cermak, P. Sebo and P. Maly, Macromol. Biosci., 2016, 16, 553–566 CrossRef CAS PubMed.
- Y. Hirayama, T. Nakamura, S. Uehara, Y. Sakamoto, K. Yamaguchi, Y. Sei and M. Iwamura, Org. Lett., 2005, 7, 525–528 CrossRef CAS PubMed.
- T. R. Krishna, S. Jain, U. S. Tatu and N. Jayaraman, Tetrahedron, 2005, 61, 4281–4288 CrossRef CAS.
- D. M. Domański, M. Bryszewska and G. Salamończyk, Biomacromolecules, 2004, 5, 2007–2012 CrossRef PubMed.
- L.-L. Lai, C.-H. Lee, L.-Y. Wang, K.-L. Cheng and H.-F. Hsu, J. Org. Chem., 2007, 73, 485–490 CrossRef PubMed.
- J. Lim and E. E. Simanek, Adv. Drug Delivery Rev., 2012, 64, 826–835 CrossRef CAS PubMed.
- E. Pedziwiatr-Werbicka, D. Shcharbin, J. Maly, M. Maly, M. Zaborski, B. Gabara, P. Ortega, F. J. de la Mata, R. Gomez, M. A. Munoz-Fernandez, B. Klajnert and M. Bryszewska, J. Biomed. Nanotechnol., 2012, 8, 57–73 CrossRef CAS PubMed.
- R. Gomez, F. J. de la Mata, J. L. Jimenez-Fuentes, P. Ortega, B. Klajnert, E. Pedziwiatr-Werbicka, D. Shcharbin, M. Bryszewska, M. Maly, J. Maly, M. J. Serramia, R. Lorente and M. A. Munoz-Fernandez, in Dendrimers in Biomedical Applications, The Royal Society of Chemistry, 2013, pp. 40–55, 10.1039/9781849737296-00040.
- L. Chonco, J. F. Bermejo-Martin, P. Ortega, D. Shcharbin, E. Pedziwiatr, B. Klajnert, F. J. de la Mata, R. Eritja, R. Gomez, M. Bryszewska and M. A. Munoz-Fernandez, Org. Biomol. Chem., 2007, 5, 1886–1893 CAS.
- E. Pedziwiatr, D. Shcharbin, L. Chonco, P. Ortega, F. J. de la Mata, R. Gomez, B. Klajnert, M. Bryszewska and M. A. Munoz-Fernandez, J. Fluoresc., 2009, 19, 267–275 CrossRef CAS PubMed.
- J. F. Bermejo, P. Ortega, L. Chonco, R. Eritja, R. Samaniego, M. Mullner, E. de Jesus, F. J. de la Mata, J. C. Flores, R. Gomez and A. Munoz-Fernandez, Chemistry, 2007, 13, 483–495 CrossRef CAS PubMed.
- M. J. Serramia, S. Alvarez, E. Fuentes-Paniagua, M. I. Clemente, J. Sanchez-Nieves, R. Gomez, J. de la Mata and M. A. Munoz-Fernandez, J. Controlled Release, 2015, 200, 60–70 CrossRef CAS PubMed.
- S. T. Hemp, A. E. Smith, J. M. Bryson, M. H. Allen Jr and T. E. Long, Biomacromolecules, 2012, 13, 2439–2445 CrossRef CAS PubMed.
- C. Ornelas-Megiatto, P. R. Wich and J. M. Frechet, J. Am. Chem. Soc., 2012, 134, 1902–1905 CrossRef CAS PubMed.
- S. T. Hemp, M. H. Allen Jr, M. D. Green and T. E. Long, Biomacromolecules, 2012, 13, 231–238 CrossRef CAS PubMed.
- E. Guénin, A.-C. Hervé, V. Floch, S. Loisel, J.-J. Yaouanc, J.-C. Clément, C. Férec and H. des Abbayes, Angew. Chem., Int. Ed., 2000, 39, 629–631 CrossRef.
- V. Floch, S. Loisel, E. Guenin, A. C. Hervé, J. C. Clément, J. J. Yaouanc, H. des Abbayes and C. Férec, J. Med. Chem., 2000, 43, 4617–4628 CrossRef CAS PubMed.
- E. Picquet, K. Le
Ny, P. Delépine, T. Montier, J.-J. Yaouanc, D. Cartier, H. des Abbayes, C. Férec and J.-C. Clément, Bioconjugate Chem., 2005, 16, 1051–1053 CrossRef CAS PubMed.
- M.-A. Lacour, M. Zablocka, A.-M. Caminade, M. Taillefer and J.-P. Majoral, Tetrahedron Lett., 2009, 50, 4870–4873 CrossRef CAS.
- X. Wang, N. Shao, Q. Zhang and Y. Cheng, J. Mater. Chem. B, 2014, 2, 2546–2553 RSC.
- S. Biswas, N. S. Dodwadkar, A. Piroyan and V. P. Torchilin, Biomaterials, 2012, 33, 4773–4782 CrossRef CAS PubMed.
- A. Krupková and J. Čermák, J. Inorg. Organomet. Polym. Mater., 2011, 22, 470–477 CrossRef.
- T. Strašák, J. Čermák, J. Sýkora, J. Horský, Z. Walterová, F. Jaroschik and D. Harakat, Organometallics, 2012, 31, 6779–6786 CrossRef.
- A. Krupková, J. Čermák, Z. Walterová and J. Horský, Anal. Chem., 2007, 79, 1639–1645 CrossRef PubMed.
- A. Krupková, J. Čermák, Z. Walterová and J. I. Horský, Macromolecules, 2010, 43, 4511–4519 CrossRef.
- E. Arnáiz, L. I. Doucede, S. García-Gallego, K. Urbiola, R. Gómez, C. Tros de Ilarduya and F. J. de la Mata, Mol. Pharm., 2012, 9, 433–447 CrossRef PubMed.
- P. Ortega, B. M. Cobaleda, J. M. Hernandez-Ros, E. Fuentes-Paniagua, J. Sanchez-Nieves, M. a. P. Tarazona, J. L. Copa-Patino, J. Soliveri, F. J. de la Mata and R. Gomez, Org. Biomol. Chem., 2011, 9, 5238–5248 CAS.
- H. K. Haruo Nakayama, N. Yamamoto, Y. Akagi and H. Matsui, Bull. Chem. Soc. Jpn., 1989, 62, 985–992 CrossRef.
- D. Appelhans, B. Klajnert-Maculewicz, A. Janaszewska, J. Lazniewska and B. Voit, Chem. Soc. Rev., 2015, 44, 3968–3996 RSC.
- A. Janaszewska, K. Maczynska, G. Matuszko, D. Appelhans, B. Voit, B. Klajnert and M. Bryszewska, New J. Chem., 2012, 36, 428–437 RSC.
- S. V. Boddapati, P. Tongcharoensirikul, R. N. Hanson, G. G. M. D'Souza, V. P. Torchilin and V. Weissig, J. Liposome Res., 2005, 15, 49–58 CrossRef CAS PubMed.
- L. Sliwka, K. Wiktorska, P. Suchocki, M. Milczarek, S. Mielczarek, K. Lubelska, T. Cierpial, P. Lyzwa, P. Kielbasinski, A. Jaromin, A. Flis and Z. Chilmonczyk, PLoS One, 2016, 11, e0155772 Search PubMed.
- K. Chiba, K. Kawakami and K. Tohyama, Toxicol. in Vitro, 1998, 12, 251–258 CrossRef CAS PubMed.
- J. O. Daiss, S. Duda-Johner, C. Burschka, U. Holzgrabe, K. Mohr and R. Tacke, Organometallics, 2002, 21, 803–811 CrossRef CAS.
- A. W. van der Made, P. W. N. M. van Leeuwen, J. C. de Wilde and R. A. C. Brandes, Adv. Mater., 1993, 5, 466–468 CrossRef CAS.
- C. I. Bayly, P. Cieplak, W. Cornell and P. A. Kollman, J. Phys. Chem., 1993, 97, 10269–10280 CrossRef CAS.
- F.-Y. Dupradeau, A. Pigache, T. Zaffran, C. Savineau, R. Lelong, N. Grivel, D. Lelong, W. Rosanski and P. Cieplak, Phys. Chem. Chem. Phys., 2010, 12, 7821–7839 RSC.
- M. W. Schmidt, K. K. Baldridge, J. A. Boatz, S. T. Elbert, M. S. Gordon, J. H. Jensen, S. Koseki, N. Matsunaga, K. A. Nguyen, S. Su, T. L. Windus, M. Dupuis and J. A. Montgomery Jr, J. Comput. Chem., 1993, 14, 1347–1363 CrossRef CAS.
- M. S. Gordon and M. W. Schmidt, in Theory and Applications of Computational Chemistry: the first forty years, ed. C. E. Dykstra, G. Frenking, K. S. Kim and G. E. Scuseria, Elsevier, 2005, pp. 1167–1189, DOI:citeulike-article-id:3013672.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- D. A. Case, R. M. Betz, W. Botello-Smith, D. S. Cerutti, T. E. Cheatham III, T. A. Darden, R. E. Duke, T. J. Giese, H. Gohlke, A. W. Goetz, N. Homeyer, S. Izadi, P. Janowski, J. Kaus, A. Kovalenko, T. S. Lee, S. LeGrand, P. Li, C. Lin, T. Luchko, R. Luo, B. Madej, D. Mermelstein, K. M. Merz, G. Monard, H. Nguyen, H. T. Nguyen, I. Omelyan, A. Onufriev, D. R. Roe, A. Roitberg, C. Sagui, C. L. Simmerling, J. Swails, R. C. Walker, J. Wang, R. M. Wolf, X. Wu, L. Xiao, D. M. York and P. A. Kollman, AMBER 2016, University of California, San Francisco, 2016 Search PubMed.
- J.-H. Lii and N. L. Allinger, J. Comput. Chem., 1991, 12, 186–199 CrossRef CAS.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- J.-P. Ryckaert, G. Ciccotti and H. J. C. Berendsen, J. Comput. Phys., 1977, 23, 327–341 CrossRef CAS.
- X. Wu and B. R. Brooks, Chem. Phys. Lett., 2003, 381, 512–518 CrossRef CAS.
- A. W. Gotz, M. J. Williamson, D. Xu, D. Poole, S. Le Grand and R. C. Walker, J. Chem. Theory Comput., 2012, 8, 1542–1555 CrossRef CAS PubMed.
- D. R. Roe and T. E. Cheatham, J. Chem. Theory Comput., 2013, 9, 3084–3095 CrossRef CAS PubMed.
- N. A. Baker, D. Sept, S. Joseph, M. J. Holst and J. A. McCammon, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 10037–10041 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: ESI focuses on the following additional information: graphs from 1H, 13C and 29Si NMR analysis (Fig. S1–S69); ESI HRMS of compound 31 and 32 (Tables S1 and S2); details on the solubility of prepared compounds in water (Table S3); calculated values of electrostatic potential on the molecular surface of G3 dendrimers (Table S4); evaluation of G2 and G1 dendrimers cytotoxicity based on IC50 values (Fig. S70 and S71). See DOI: 10.1039/c7ra01845b |
|
| This journal is © The Royal Society of Chemistry 2017 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *b,
Dominika Wróbelb,
Marek Malýc,
Regina Hermab,
Jan Čermákad,
Monika Müllerováa,
Lucie Červenková Št′astnáa and
Petra Cuřínováa
*b,
Dominika Wróbelb,
Marek Malýc,
Regina Hermab,
Jan Čermákad,
Monika Müllerováa,
Lucie Červenková Št′astnáa and
Petra Cuřínováa
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)







