Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Systematic analysis of structural and activity relationships between conventional hierarchical and analog series-based scaffolds
Dagmar Stumpfe,
Dilyana Dimova and
Jürgen Bajorath
and
Jürgen Bajorath *
*
Department of Life Science Informatics, B-IT, LIMES Program Unit Chemical Biology and Medicinal Chemistry, Rheinische Friedrich-Wilhelms-Universität, Dahlmannstr. 2, D-53113 Bonn, Germany. E-mail: bajorath@bit.uni-bonn.de; Fax: +49-228-2699-341; Tel: +49-228-2699-306
First published on 27th March 2017
Abstract
The concept of molecular scaffolds is widely applied in medicinal and computational chemistry to represent core structures of compounds and series. A hierarchical organization of compounds has long dominated scaffold design and generation. Recently, so called ‘analog series-based’ (ASB) scaffolds have been introduced as an alternative category of scaffolds. ASB scaffolds are designed to represent analog series and take reaction information into account, and do not follow a molecular hierarchy. We report a large-scale comparison of ASB scaffolds representing more than 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 analog series with activity against more than 1200 targets and their corresponding hierarchical scaffolds. Most ASB and conventional hierarchical scaffolds were structurally distinct. However, many ASB scaffolds contained conventional scaffolds as substructures or shared smaller substructures with these scaffolds. Although ASB scaffolds and their corresponding hierarchical scaffolds often shared the same target annotations, ASB scaffolds further distinguished between closely related compound series with different activities that yielded the same conventional scaffolds. Taken together, the findings reported herein reveal that ASB scaffolds further extend current core structure representations for the analysis of structure–activity relationships.
000 analog series with activity against more than 1200 targets and their corresponding hierarchical scaffolds. Most ASB and conventional hierarchical scaffolds were structurally distinct. However, many ASB scaffolds contained conventional scaffolds as substructures or shared smaller substructures with these scaffolds. Although ASB scaffolds and their corresponding hierarchical scaffolds often shared the same target annotations, ASB scaffolds further distinguished between closely related compound series with different activities that yielded the same conventional scaffolds. Taken together, the findings reported herein reveal that ASB scaffolds further extend current core structure representations for the analysis of structure–activity relationships.
Introduction
The scaffold concept is of central relevance in medicinal chemistry and chemoinformatics.1–3 Scaffolds are generated to represent core structures of compounds and used, for example, to structurally organize compound data sets, assess chemical diversity, aid in the analysis of structure–activity relationships (SARs), or identify preferred templates for compound design.1–3 In chemoinformatics and computational chemistry, the scaffold concept is also applied to assess the ability of computational methods to identify or construct structurally diverse compounds having similar activity.3,4 In addition to studying structural relationships between scaffolds directly, they are also used to map target annotations of compounds sharing the same scaffold.3 Therefore, annotations of corresponding compounds are assigned to the scaffold, which then represents the activities of a series of compounds as well as its core structure. This makes it possible to explore SARs at the level of scaffolds.Although scaffolds have been described and represented in different ways,1–3 most popular approaches have applied a hierarchical organization of compounds.5–7 The hierarchy distinguishes ring systems as core structures from substituents and aliphatic linker fragments and also involves molecular decomposition steps to reduce compounds to scaffolds and further abstract from scaffolds.5–7 Hierarchical organization of compounds and scaffolds taking activity annotations into account has enabled systematic SAR exploration and the identification and prioritization of molecular core structures for the design of new active compounds.6,7
The original hierarchical definition of scaffolds that has paved the way for systematic computer-aided exploration of scaffolds and become a mainstay in medicinal chemistry was introduced by Bemis and Murcko.5 According to this generally applicable definition, scaffolds are extracted from compounds by removing all substituents while retaining ring systems and linker fragments between rings. It follows that so defined Bemis and Murcko (BM) scaffolds must contain rings structures and that the addition of a ring to a compound generates a new scaffold. During the past two decades, BM scaffolds have become the gold standard for scaffold analysis in medicinal chemistry and chemoinformatics.
Recently, a new category of scaffolds has been introduced designed to complement the hierarchical view of scaffolds by further increasing their medicinal chemistry focus.8 These analog series-based (ASB) scaffolds are derived from series of analogs, i.e. multiple compounds, whereas BM scaffolds are obtained from individual compounds. In addition, ASB scaffolds are non-hierarchical and account for synthetic relationships between analogs.8 Thus, the design of ASB and BM scaffolds fundamentally differs and hence these scaffold categories are conceptually distinct.
In this work, we have systematically analyzed structural and activity relationships between ASB and BM scaffolds derived from a wide spectrum of bioactive compounds. The analysis uncovered a variety of relationships between these scaffold categories. In addition, ASB scaffolds were capable of distinguishing between different activities of closely related analog series, which was not possible on the basis of BM scaffolds. ASB scaffolds also revealed chemical modifications that rendered analogs active against different targets.
Material and methods
Analog series-based (ASB) scaffold concept
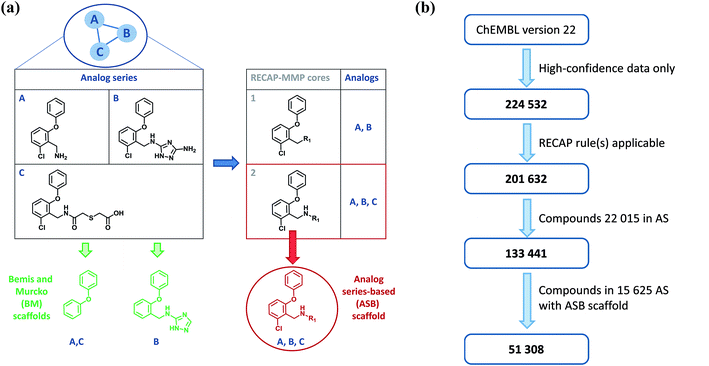
Individual analog series from which ASB scaffolds are extracted are identified by applying a variant of the matched molecular pair (MMP) formalism.9 An MMP is defined as a pair of compounds that are only distinguished by a chemical modification at a single site.9 Therefore, the MMP consists of the shared MMP core and a pair of exchanged substituents. For the identification of analog series, MMPs are generated on the basis of retrosynthetic (RECAP) rules,10 rather than random bond fragmentation, yielding RECAP-MMPs.11 In the next step, a global network representation is generated in which nodes represent compounds and edges pairwise RECAP-MMP relationships.12 In this network, each separate cluster represents a unique series of analogs.12 From systematically identified analog series (AS), ASB scaffolds are isolated. An AS typically contains multiple RECAP-MMP cores and a search is carried out for a core that captures all MMP relationships within the series. If more than one core meets this criterion, the largest one is selected. The qualifying RECAP-MMP core represents the ASB scaffold of the series.8 The generation of ASB scaffolds was reported in detail8 and is illustrated in Fig. 1a. From the ASB scaffold, all analogs comprising the corresponding AS (as well as new analogs) can be generated following chemical reaction rules, which represents one of the key features of this scaffold definition. The ASB scaffold representing an AS also specifies a single substitution site where different R-groups distinguish analogs. Fig. 1b illustrates the workflow and provides compound statistics for ASB scaffold analysis, as further discussed in the following.Compounds and activity data
Bioactive compounds with high-confidence activity data were assembled from ChEMBL version 22.13 Therefore, the following selection criteria were applied: first, only compounds involved in direct interactions (type “D”) with human targets at the highest confidence level (score 9) were selected. Second, only numerically specified equilibrium constants (Ki values) or IC50 values were considered as potency measurements. If multiple Ki or IC50 values for the same target were reported for the same compound, their geometric mean was calculated as the final potency annotation, provided all values were within the same order of magnitude. Otherwise, the values were discarded. Applying these selection criteria, a total of 224![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 532 unique compounds were obtained with activity against a total of 1687 targets.
532 unique compounds were obtained with activity against a total of 1687 targets.
Scaffolds
RECAP-MMPs were systematically generated for the pool of active compounds and organized into AS via the network approach.12 A total of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 015 unique AS comprising 133
015 unique AS comprising 133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 441 compounds were obtained. For 15
441 compounds were obtained. For 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 625 of these AS (71%), a qualifying RECAP-MMP core representing the ASB scaffold was identified, as illustrated in Fig. 1b. Series with ASB scaffolds contained 51
625 of these AS (71%), a qualifying RECAP-MMP core representing the ASB scaffold was identified, as illustrated in Fig. 1b. Series with ASB scaffolds contained 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 308 compounds, with two to 60 compounds per AS. We note that AS were systematically identified exclusively on the basis of structural criteria before target annotations and potency values were mapped to each compound.
308 compounds, with two to 60 compounds per AS. We note that AS were systematically identified exclusively on the basis of structural criteria before target annotations and potency values were mapped to each compound.
From all 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 308 compounds yielding ASB scaffolds, Bemis and Murcko (BM) scaffolds were then extracted. All calculations were carried out using in-house Perl and Python scripts with the aid of KNIME14 protocols and the OpenEye chemistry toolkit.15
308 compounds yielding ASB scaffolds, Bemis and Murcko (BM) scaffolds were then extracted. All calculations were carried out using in-house Perl and Python scripts with the aid of KNIME14 protocols and the OpenEye chemistry toolkit.15
Results and discussion
The scaffold statistics
From 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 308 compounds, which were active against 1251 targets, 15
308 compounds, which were active against 1251 targets, 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 625 analog series-based (ASB) scaffolds were obtained. These compounds yielded 22
625 analog series-based (ASB) scaffolds were obtained. These compounds yielded 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 224 unique Bemis and Murcko (BM) scaffolds, thus enabling a large-scale comparison of corresponding ASB and BM scaffolds.
224 unique Bemis and Murcko (BM) scaffolds, thus enabling a large-scale comparison of corresponding ASB and BM scaffolds.
Different from ASB scaffolds, analog series (AS) can contain one or more BM scaffolds. For 7971 AS producing ASB scaffolds, a single BM scaffold was obtained, resulting in 6771 unique BM scaffolds. Accordingly, in these cases, there was a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 correspondence of ASB and BM scaffolds. Furthermore, for 7654 AS with ASB scaffolds, multiple BM scaffolds were obtained, with two to 34 scaffolds per AS, yielding a total of 17
1 correspondence of ASB and BM scaffolds. Furthermore, for 7654 AS with ASB scaffolds, multiple BM scaffolds were obtained, with two to 34 scaffolds per AS, yielding a total of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 830 unique BM scaffolds. Overall, only 3322 unique BM scaffolds (15.0%) corresponded to more than one ASB scaffold from different AS, indicating that ASB scaffolds captured series-specific chemical information. Otherwise, a higher degree of correspondence between BM and multiple ASB scaffolds would be anticipated.
830 unique BM scaffolds. Overall, only 3322 unique BM scaffolds (15.0%) corresponded to more than one ASB scaffold from different AS, indicating that ASB scaffolds captured series-specific chemical information. Otherwise, a higher degree of correspondence between BM and multiple ASB scaffolds would be anticipated.
Structural relationships
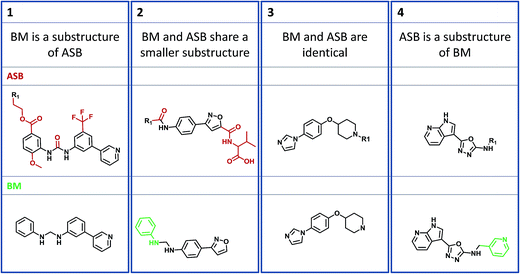
Four structural relationships between ASB and BM scaffolds were investigated, as illustrated in Fig. 2. A BM scaffold might represent a substructure of an ASB scaffold and vice versa (examples 1 and 4 in Fig. 2). In these instances, the larger scaffold provides additional structural information for a given AS. Furthermore, a BM and ASB scaffold might share a smaller substructure (example 2). In such cases, the ASB and BM scaffold capture different structural details. Finally, an ASB and BM scaffold might be identical (example 3).Table 1 reports the results of systematic structural comparisons of ASB and BM scaffolds at the level of individual AS. If an AS produced multiple BM scaffolds, combinations of different relationships were also possible.
| Structural relationship(s) | #ASs | % | |
|---|---|---|---|
| a The distribution of structural relationships between ASB and BM scaffolds according to Fig. 2 is reported. Relationships were detected on the basis of individual analog series (AS). Combinations of different relationships (e.g. 1 + 2) were possible if an AS yielded more than one BM scaffold. Three dominant relationships are shown in bold. | |||
| 1 | BM is a substructure of ASB | 5734 | 36.7 |
| 2 | BM and ASB share a smaller substructure | 5436 | 34.8 |
| 3 | BM and ASB are identical | 155 | 1.0 |
| 4 | ASB is a substructure of BM | 220 | 1.4 |
| 1 + 2 | BM is a substructure of ASB, BM and ASB share a smaller substructure | 3929 | 25.1 |
| Other | 1 + 3, 2 + 3, 2 + 4, and 3 + 4 | 151 | 1.0 |
Identical ASB and BM scaffolds were only detected in 155 cases (1.0% of all ASB scaffolds), confirming that most ASB and BM scaffolds derived from the same AS were structurally distinct. However, in 5734 instances (36.7% of all ASB scaffolds), a BM scaffold was a substructure of the ASB scaffold. By contrast, the alternative scenario that an ASB scaffold was a substructure of a BM scaffold was only rarely observed (1.4%). Thus, more than a third of ASB scaffolds contained invariant substituents from AS that were removed when BM scaffold(s) were generated, as illustrated by example 1 in Fig. 2. If compounds comprising an AS contain conserved substituents, the ASB scaffold takes this information into account and – as a consequence – represents a higher degree of chemical exploration than a corresponding BM scaffold.
As a second dominant relationship, 5436 ASB and BM scaffolds (34.8%) shared a smaller substructure. In these cases, the BM scaffold had to contain at least one additional ring that was not conserved within the AS and therefore not contained in the corresponding ASB scaffold, as illustrated by example 2 in Fig. 2. This frequent relationship reflected a conceptual weakness of BM scaffolds for the representation of core structures: because the additional ring was not invariant, it was not part of the common core but rather a substituent distinguishing different analogs within the AS.
For AS yielding more than one BM scaffold, the combination of these relationships, i.e. a BM scaffold was a substructure of the ASB scaffold and another BM scaffold and the ASB scaffold shared a smaller substructure, was also frequently observed, with 3929 instances (25.1% of all ASB scaffolds). By contrast, combinations of other structural relationships were only rarely detected (Table 1).
Activity relationships
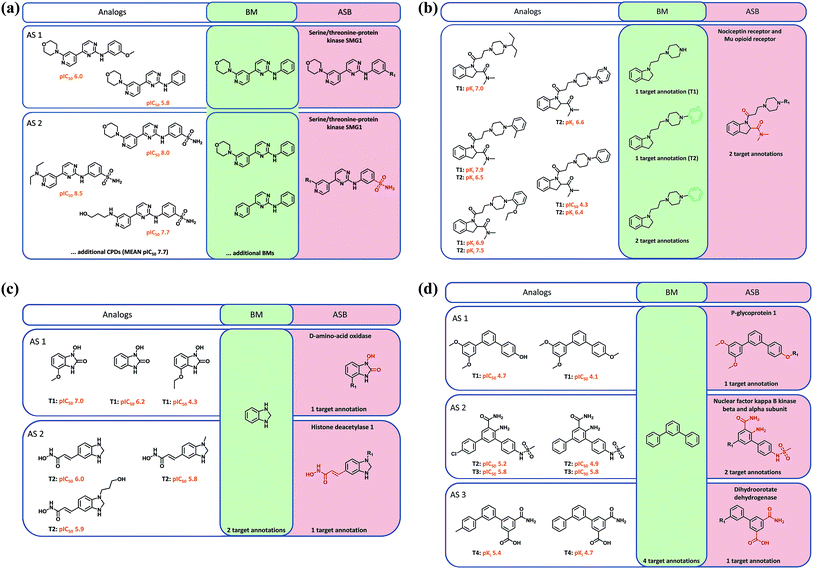
Mapping of target annotations to all AS with ASB scaffolds revealed that 9497 and 6128 AS were associated with single- and multi-target activities, respectively. The activities of all compounds represented by a given (ASB or BM) scaffold were assigned to this scaffold. Accordingly, the scaffold was associated with the union of all target annotations. By definition, an ASB scaffold was associated with target annotations of all compounds comprising the AS. On the other hand, a BM scaffold originating from the same AS might have the same number of target annotations (in the case of single- or multi-target activities) or a smaller number of annotations (multi-target activities). For BM scaffolds corresponding to more than one ASB scaffold from different AS, target annotations of compounds from these AS were combined and assigned to the BM scaffold.Four different activity relationships between annotated ASB and BM scaffolds were examined. First, the ASB and one or more BM scaffolds might have identical target annotations (Fig. 3a). Second, at least one BM scaffold (originating from different AS) might have more target annotations than the ASB scaffold as shown, for example, in Fig. 3c and d. Third, the ASB scaffold might have more target annotations than at least one BM scaffold, as shown in Fig. 3b. Fourth, relationships were considered variable if at least one BM scaffold originating from multiple AS had more target annotations than a corresponding ASB scaffold and one or more other BM scaffolds had fewer annotations than the corresponding ASB scaffold.
Table 2 reports the distribution of these activity relationships between ASB and BM scaffolds. The majority of ASB scaffolds (70.0%) and corresponding BM scaffolds had identical target annotations including 7737 and 3202 ASB scaffolds associated with single- and multi-target activities, respectively. In addition, for 18.7% of all ASB scaffolds (1760 with single- and 1160 with multi-target activities), there was at least one corresponding BM scaffold with additional target annotations from different AS. Furthermore, 8.6% of ASB scaffolds had more target annotations than at least one corresponding BM scaffolds. Variable activity relationships with multiple BM scaffolds were only detected for 2.7% of the ASB scaffolds.
| a The distribution of activity relationships between ASB and BM scaffolds is reported. Relationships were detected on the basis of individual analog series (AS). For each of four possible relationships, the number of ASB scaffolds and corresponding percentage of all ASB scaffolds are given. Relationships were considered variable if at least one BM scaffold originating from multiple AS had more target annotations than a corresponding ASB scaffold, whereas at least one other BM scaffold had fewer annotations than the corresponding ASB scaffold. | |
|---|---|
| 1 | BM and ASB have identical target annotations |
• 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 939 (7737 single-target AS and 3202 multi-target AS) 939 (7737 single-target AS and 3202 multi-target AS) |
|
| • 70.0% | |
| 2 | BM with more target annotations than ASB |
| • 2920 (1760 single-target AS and 1160 multi-target AS) | |
| • 18.7% | |
| 3 | ASB with more target annotations than BM(s) |
| • 1350 (multi-target AS only) | |
| • 8.6% | |
| 4 | Variable |
| • 416 (multi-target AS only) | |
| • 2.7% | |
Thus, despite abundant structural differences between ASB and corresponding BM scaffolds, differences in target annotations were overall only limited at the level of individual AS, even when annotations for BM scaffolds originating from more than one AS were combined.
Structure–activity relationships
A major motivation for the ‘meta level’ assignment of compound activities to corresponding scaffolds is the dissection of multifaceted SARs within AS and across related AS. Annotated scaffolds are useful tools for SAR analysis, especially when AS are large and/or exhibit multi-target activities. Structural differences between ASB and BM scaffolds, as discussed above, are relevant for SAR exploration. Fig. 3 provides representative examples. Fig. 3a shows compounds from two structurally related AS of serine/threonine kinase inhibitors. One series had consistently higher, the other consistently lower potency. These two AS shared one BM scaffold representing a subset of analogs that differed at the given substitution site. Analogs from the first AS had a hydrogen or methoxy group at this position, whereas the analogs from the more potent second AS contained a sulfonamide group. This sulfonamide was a hallmark of all potent compounds comprising the second AS and therefore a part of its ASB scaffold. Fig. 3b depicts an AS consisting of compounds active against the nociceptin and/or mu opioid receptor that contained three BM scaffolds, which were distinguished by different rings and represented analogs with varying target annotations. The ASB scaffold representing all compounds including those with dual–receptor activity defined the invariant core of the AS and identified a substitution site that distinguished analogs with different target annotations.In Fig. 3c, two AS with activity against distinct enzymes are displayed that yielded the same benzimidazolidine BM scaffold. The ASB scaffold of each series contained the benzimidazolidine and specific substituents that were characteristic of each AS. These chemically more differentiated ASB scaffolds exclusively represented analogs that were either active against D-amino-acid oxidase or histone deacetylase 1. Similarly, in Fig. 3d, compounds from three AS are shown that were active against distinct targets but yielded the same BM scaffold. The ASB scaffold of each AS contained invariant substituents at different phenyl ring positions and distinguished between compounds of each AS and their specific activities. Hence, in these cases, multi-target SARs were resolved at the level of ASB scaffolds, which represented distinct core structures characteristic of related AS with different activities.
Concluding remarks
Bemis and Murcko (BM) scaffolds have long been the gold standard for representing core structures of compounds and series. Analog series-based (ASB) scaffolds have recently been introduced as an alternative category of scaffolds. By design, ASB scaffolds are distinct from BM scaffolds because they are non-hierarchical, no central role is assigned to ring structures, and they are generated from multiple analogs, rather than individual compounds. In addition, reaction information is considered in deriving ASB scaffolds. As we have reported, ASB scaffolds are obtained from many but not all AS, depending on structural relationships between analogs.Herein we have presented a large-scale comparison of ASB and BM scaffolds to investigate relationships between scaffolds of different design and their utility for SAR exploration. To enable a direct comparison, corresponding ASB and BM scaffolds were extracted from more than 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 analog series (AS) with activity against more than 1200 targets. The vast majority of corresponding ASB and BM scaffolds were structurally distinct but formed systematic structural relationships. However, about a third of all ASB scaffolds contained corresponding BM scaffolds as substructures and another third shared smaller substructures with BM scaffolds. These relationships and their combination involved nearly all ASB scaffolds (97%). We also found that the majority of ASB and BM scaffolds shared the same target annotations. However, ASB scaffolds typically provided a higher-resolution view of SARs than BM scaffolds and further differentiated between related AS with different activities sharing the same BM scaffold(s). Distinct ASB scaffolds of related AS exclusively represented compounds having the same activity.
000 analog series (AS) with activity against more than 1200 targets. The vast majority of corresponding ASB and BM scaffolds were structurally distinct but formed systematic structural relationships. However, about a third of all ASB scaffolds contained corresponding BM scaffolds as substructures and another third shared smaller substructures with BM scaffolds. These relationships and their combination involved nearly all ASB scaffolds (97%). We also found that the majority of ASB and BM scaffolds shared the same target annotations. However, ASB scaffolds typically provided a higher-resolution view of SARs than BM scaffolds and further differentiated between related AS with different activities sharing the same BM scaffold(s). Distinct ASB scaffolds of related AS exclusively represented compounds having the same activity.
Taken together, the results of our analysis suggest that ASB scaffolds represent an attractive extension of current core structure representations and further increase the utility of scaffolds for SAR exploration.
Acknowledgements
We also thank OpenEye Scientific Software for a free academic license. D. S. is supported by Sonderforschungsbereich 704 of the Deutsche Forschungsgemeinschaft.References
- Y. Hu, D. Stumpfe and J. Bajorath, J. Chem. Inf. Model., 2011, 51, 1742–1753 CrossRef CAS PubMed.
- C. M. Marson, Chem. Soc. Rev., 2011, 40, 5514–5533 RSC.
- Y. Hu, D. Stumpfe and J. Bajorath, J. Med. Chem., 2016, 59, 4062–4076 CrossRef CAS PubMed.
- A. Schuffenhauer, WIREs. Comput. Mol. Sci., 2012, 2, 842–867 CrossRef CAS.
- G. W. Bemis and M. A. Murcko, J. Med. Chem., 1996, 39, 2887–2893 CrossRef CAS PubMed.
- S. J. Wilkens, J. Janes and A. I. Su, J. Med. Chem., 2005, 48, 182–193 CrossRef PubMed.
- A. Schuffenhauer, P. Ertl, S. Roggo, S. Wetzel, M. A. Koch and H. Waldmann, J. Chem. Inf. Model., 2007, 47, 47–58 CrossRef CAS PubMed.
- D. Dimova, D. Stumpfe, Y. Hu and J. Bajorath, Future Sci. OA, 2016, 2, FSO149 CrossRef PubMed.
- P. W. Kenny and J. Sadowski, in Chemoinformatics in Drug Discovery, ed. T. I. Oprea, Wiley-VCH, Weinheim, Germany, 2004, pp. 271–285 Search PubMed.
- X. Q. Lewell, D. B. Judd, S. P. Watson and M. M. Hann, J. Chem. Inf. Comput. Sci., 1998, 38, 511–522 CrossRef CAS PubMed.
- A. de la Vega de León and J. Bajorath, MedChemComm, 2014, 5, 64–67 RSC.
- D. Stumpfe, D. Dimova and J. Bajorath, J. Med. Chem., 2016, 59, 7667–7676 CrossRef CAS PubMed.
- A. P. Bento, A. Gaulton, A. Hersey, L. J. Bellis, J. Chambers, M. Davies, F. A. Krüger, Y. Light, L. Mak, S. McGlinchey, M. Nowotka, G. Papadatos, R. Santos and J. P. Overington, Nucleic Acids Res., 2014, 42, D1083–D1090 CrossRef CAS PubMed.
- M. R. Berthold, N. Cebron, F. Dill, T. R. Gabriel, T. Kötter, T. Meinl, P. Ohl, C. Sieb, K. Thiel and B. Wiswedel, in Studies in Classification, Data Analysis, and Knowledge Organization, ed. C. Preisach, H. Burkhardt, L. Schmidt-Thieme and R. Decker, Springer, Berlin, Germany, 2008, pp. 319–326 Search PubMed.
- OEChem TK, OpenEye Scientific Software, Inc., Sante Fe, NM, USA, 2012 Search PubMed.
| This journal is © The Royal Society of Chemistry 2017 |