Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Neutral linear supramolecular polymers constructed by three different interactions†
Qi
Wang
*ab,
Peng
Zhang
a,
Yuanyuan
Li
a,
Lu
Tian
a,
Ming
Cheng
b,
Feng
Lu
a,
Xiaomei
Lu
c,
Quli
Fan
*a and
Wei
Huang
c
aKey Laboratory for Organic Electronics and Information Displays (KLOEID), Institute of Advanced Materials (IAM), Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing University of Posts & Telecommunications, 9 Wenyuan Road, Nanjing 210023, China. E-mail: iamqwang@njupt.edu.cn; iamqlfan@njupt.edu.cn
bState Key Laboratory of Coordination Chemistry, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, China
cKey Laboratory of Flexible Electronics (KLOFE), Institute of Advanced Materials (IAM), Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing Tech University (NanjingTech), 30 South Puzhu Road, Nanjing 211816, China
First published on 6th June 2017
Abstract
Neutral linear supramolecular polymers were efficiently constructed by the combination of quadruple hydrogen bonding, pillar[5]arene-based molecular recognition and π–π donor–acceptor interactions. The disassembly of the supramolecular polymers can be effectively controlled by adding the competitive adiponitrile guest.
Supramolecular polymers, in which non-covalent interactions are always employed to hold the monomeric units together, not only exhibit polymer-like properties, but also possess new structures with unique functions such as stimuli-responsiveness, recyclability, self-healing and degradability.1 Therefore, supramolecular polymers are expected to provide a wide range of potential applications as functional adaptive materials.2 As we know, nature commonly utilizes the combination of different non-covalent interactions to create complicated and functionally ordered architectures such as DNA and so on.3 Motivated by this, scientists have been trying to fabricate supramolecular polymers with unprecedented properties and intriguing functions by self-assembly of multiple non-covalent interactions.4 For instance, Huang’s group constructed a homogeneous linear supramolecular copolymer by orthogonal coordination-driven self-assembly, crown-ether-based [2]rotaxane host–guest interactions and quadruple hydrogen bonding of ureidopyrimidinone (UPy) in 2016.5 Recently, based on the unification of quadruple hydrogen bonding and crown-ether-based bistable [c2]daisy chain rotaxane molecular recognition, Giuseppone’s group constructed a molecular-machine-based supramolecular polymer which could exhibit reversible sol–gel transitions, controlled by pH actuation.6
Pillararenes7 have captured more and more attention and provided a useful platform for the construction of supramolecular polymers8 because of their unique symmetrical pillar architecture, facile functionalization and adjustable host–guest properties.9 Until now, lots of pillararene-based neutral supramolecular polymers have been constructed by employing two kinds of non-covalent interactions.10 For example, Li’s group provided a new strategy for the construction of neutral linear supramolecular polymers by the marriage of endo-cavity and exo-wall complexation of a pillar[5]arene in 2015.11 Recently, Shi’s group constructed a novel linear supramolecular polymer by orthogonal pillar[5]arene-based molecular recognition, Eu(III)-coordination and π–π donor–acceptor interactions.12 However, to the best of our knowledge, pillar[5]arene-based neutral supramolecular polymers constructed by combining the themes of three non-covalent interactions are rare. Herein, we report novel neutral linear supramolecular polymers constructed by three kinds of non-covalent recognition motifs including quadruple hydrogen bonding, pillar[5]arene-based molecular recognition and π–π donor–acceptor interactions. Firstly, we designed an unsymmetrical guest 1 possessing a UPy unit and a carbamate moiety, which can dimerize to form an unusual homoditopic AA-type supramonomer. According to Li’s report, pillar[5]arene 2 binds with molecule 3 to form a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 sandwich-type exo-wall complex driven by π–π donor–acceptor interactions, which was seen as a homoditopic BB-type supramonomer.11 As a result, neutral linear supramolecular polymers were successfully constructed in the A2B2 form as suggested in Scheme 1.
1 sandwich-type exo-wall complex driven by π–π donor–acceptor interactions, which was seen as a homoditopic BB-type supramonomer.11 As a result, neutral linear supramolecular polymers were successfully constructed in the A2B2 form as suggested in Scheme 1.
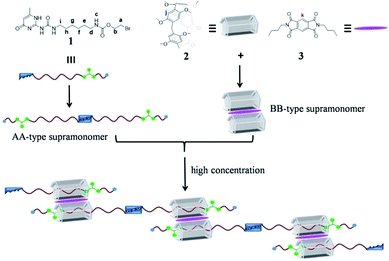 | ||
| Scheme 1 The chemical structures of 1, 2 and 3 and a cartoon representation of the formation of the neutral linear supramolecular polymers. | ||
A well-designed guest molecule 1, which bears a UPy unit and a carbamate moiety, was obtained in a moderate yield using commercially available starting materials according to the reported methods.13 Initially, the host–guest interaction between 1 and 2 was investigated by 1H NMR spectroscopy (Fig. 1, and S5, ESI†). As shown in Fig. 1a, in the spectrum of individual 1, the Upy N–H signals showed large downfield shifts (between 10 and 13.5 ppm), which gave direct evidence for the dimerization of UPy units to form an unusual homoditopic AA-type supramonomer.14 After mixing 1 and 2, an evident upfield chemical shift and a broadening effect of the methylene protons (Hd–g) on 1 were clearly observed, indicating that 1 threaded into the cavity of 2.
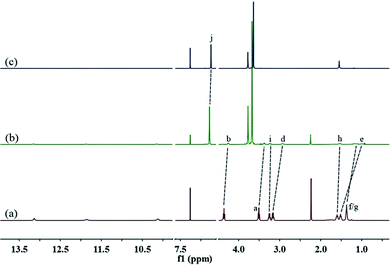 | ||
| Fig. 1 Partial 1H NMR spectra (400 MHz, CDCl3, 298 K) of: (a) 1 (8.00 mM); (b) 1 (8.00 mM) and 2 (12.00 mM); and (c) 2 (12.00 mM). | ||
Subsequently, two-dimensional 1H NOESY was also carried out to study the self-assembly of 1 and 2 in CDCl3 solution. Correlation signals were observed between the methylene protons (Hd–i) of guest 1 and Hj of host 2, indicating the formation of a host–guest inclusion complex between 1 and 2 (Fig. S6, ESI†). In addition, a Job plot based on the 1H NMR data demonstrated that the complex had 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (Fig. S7, ESI†). By using a nonlinear curve-fitting analysis based on the 1H NMR titration experiments, the association constant (Ka) of 2 ⊃ 1 was determined to be 85 ± 4.1 M−1 (Fig. S9, ESI†).
1 stoichiometry (Fig. S7, ESI†). By using a nonlinear curve-fitting analysis based on the 1H NMR titration experiments, the association constant (Ka) of 2 ⊃ 1 was determined to be 85 ± 4.1 M−1 (Fig. S9, ESI†).
As above-mentioned, the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 sandwich-type complex between 2 and 3 formed via π–π donor–acceptor interactions can be seen as a homoditopic BB-type supramonomer. Inspired by this, we investigated the possibility of constructing the neutral linear supramolecular polymers (12·22·3) by three kinds of non-covalent recognition motifs. Initially, when mixing 1, 2, and 3 in a 2
1 sandwich-type complex between 2 and 3 formed via π–π donor–acceptor interactions can be seen as a homoditopic BB-type supramonomer. Inspired by this, we investigated the possibility of constructing the neutral linear supramolecular polymers (12·22·3) by three kinds of non-covalent recognition motifs. Initially, when mixing 1, 2, and 3 in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
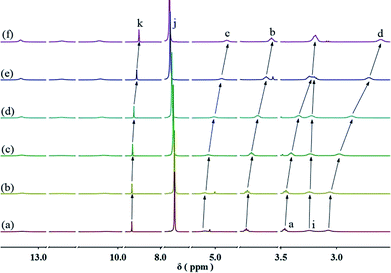
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio in CHCl3 at low concentration, a pale red color from π–π stacking between 2 and 3 appeared immediately. As the concentration increased, the color of the solution gradually became dark red (Fig. S10, ESI†). Thereafter, concentration-dependent 1H NMR spectroscopy was performed to reveal the possibility of generating the expected supramolecular polymers. As shown in Fig. 2, the large downfield chemical shifts and lower intensities observed on UPy N–H indicated the dimerization of the UPy motifs. With increasing concentration, the signals of protons Ha–d of 1 and proton Hk of 3 underwent substantial upfield shifts (Fig. 2, and S11, ESI†). Moreover, all the signals became broad at a high concentration, which confirmed the formation of aggregates with a high molecular weight. Diffusion-ordered 1H NMR spectroscopy (DOSY) was further employed to confirm the formation of a concentration-dependent supramolecular polymer. As the concentration of the mixtures of 3 with 2 equiv. of 1 and 2 increased from 4.00 mM to 50.0 mM, the measured weight average diffusion coefficient decreased significantly from 5.33 × 10−10 m2 s−1 to 2.65 × 10−10 m2 s−1, suggesting the gradual formation of high-molecular-weight polymeric aggregates (Fig. 3). To further study the self-assembly process of 1, 2, and 3 to form the neutral linear supramolecular polymers, viscosity measurements were carried out in CHCl3 using a micro-Ubbelohde viscometer. As presented in Fig. 4, the aggregates assembled from the monomers presented a viscosity transition, which was characterized by a change in the slope of the double logarithmic plots of the specific viscosity versus the concentration. In the low concentration range, the curve slope was 0.71, which indicates the predominance of cyclic oligomers in dilute solutions. When the concentration exceeded the critical polymerization concentration (CPC, 7 mM), an obvious increase in the viscosity was observed (slope = 1.10), suggesting a transition from small oligomers to larger linear supramolecular polymers. Moreover, large spherical aggregates were distinctly observed in the TEM image, which may have originated from further assembly of the linear supramolecular polymers (Fig. S12, ESI†).
1 molar ratio in CHCl3 at low concentration, a pale red color from π–π stacking between 2 and 3 appeared immediately. As the concentration increased, the color of the solution gradually became dark red (Fig. S10, ESI†). Thereafter, concentration-dependent 1H NMR spectroscopy was performed to reveal the possibility of generating the expected supramolecular polymers. As shown in Fig. 2, the large downfield chemical shifts and lower intensities observed on UPy N–H indicated the dimerization of the UPy motifs. With increasing concentration, the signals of protons Ha–d of 1 and proton Hk of 3 underwent substantial upfield shifts (Fig. 2, and S11, ESI†). Moreover, all the signals became broad at a high concentration, which confirmed the formation of aggregates with a high molecular weight. Diffusion-ordered 1H NMR spectroscopy (DOSY) was further employed to confirm the formation of a concentration-dependent supramolecular polymer. As the concentration of the mixtures of 3 with 2 equiv. of 1 and 2 increased from 4.00 mM to 50.0 mM, the measured weight average diffusion coefficient decreased significantly from 5.33 × 10−10 m2 s−1 to 2.65 × 10−10 m2 s−1, suggesting the gradual formation of high-molecular-weight polymeric aggregates (Fig. 3). To further study the self-assembly process of 1, 2, and 3 to form the neutral linear supramolecular polymers, viscosity measurements were carried out in CHCl3 using a micro-Ubbelohde viscometer. As presented in Fig. 4, the aggregates assembled from the monomers presented a viscosity transition, which was characterized by a change in the slope of the double logarithmic plots of the specific viscosity versus the concentration. In the low concentration range, the curve slope was 0.71, which indicates the predominance of cyclic oligomers in dilute solutions. When the concentration exceeded the critical polymerization concentration (CPC, 7 mM), an obvious increase in the viscosity was observed (slope = 1.10), suggesting a transition from small oligomers to larger linear supramolecular polymers. Moreover, large spherical aggregates were distinctly observed in the TEM image, which may have originated from further assembly of the linear supramolecular polymers (Fig. S12, ESI†).
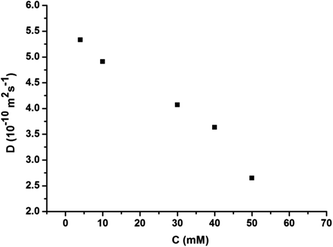 | ||
| Fig. 3 The concentration dependence of the diffusion coefficient D (400 MHz, CDCl3, 298 K) of the mixed solution of 3 and 2 equiv. of 1 and 2. | ||
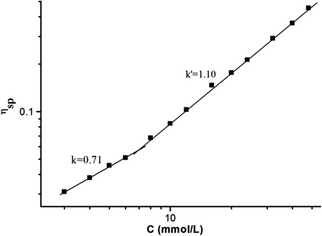 | ||
| Fig. 4 The specific viscosity of the mixed solution of 3 and 2 equiv. of 1 and 2versus the concentration of 3 (CHCl3, 298 K). | ||
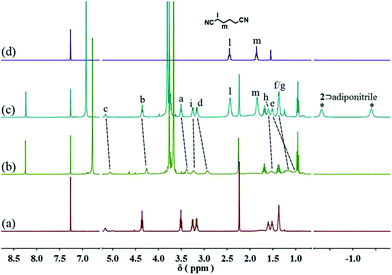
Since pillar[5]arene shows very strong binding affinities with adiponitrile,15 we envisioned that the resulting neutral linear supramolecular polymers could be depolymerized by addition of a competitive guest. When 2.5 equiv. of adiponitrile was added into a solution of 3 (10 mM) with 2 equiv. of 1 and 2 in CDCl3, the 1H NMR signals of the free neutral guest 1 and the newly generated complex 2⊃adiponitrile were observed, which confirmed the disassembly of the supramolecular polymers (Fig. 5 and S13, ESI†).
In summary, we demonstrated that novel neutral linear supramolecular polymers composed of 1, 2, and 3 could be successfully constructed by combining three kinds of non-covalent recognition motifs including quadruple hydrogen bonding, pillar[5]arene-based molecular recognition and π–π donor–acceptor interactions, and this was collectively confirmed by 1H NMR, DOSY, viscosity, and TEM. Furthermore, the resulting supramolecular polymers can be effectively depolymerized by addition of a competitive guest. This work provides a new and convenient way to construct degradable supramolecular polymeric materials with tailored properties. Our future work will focus on developing more functional supramolecular polymers with rationally designed structures, by self-assembly of multiple non-covalent interactions.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 21602112) and the China Postdoctoral Science Foundation (Grant No. 2017M611876), and sponsored by the Scientific Starting Fund from Nanjing University of Posts and Telecommunications (NUPTSF) (Grant No. NY215057) and the Natural Science Foundation of the Jiangsu Higher Education Institutions (Grant No. 16KJB150031).Notes and references
- (a) C. Fouquey, J.-M. Lehn and A.-M. Levelut, Adv. Mater., 1990, 2, 254 CrossRef CAS; (b) C. Schmuck and W. Wienand, Angew. Chem., Int. Ed., 2001, 40, 4363 CrossRef CAS; (c) T. F. A. D. Greef, M. M. J. Smulders, M. Wolffs, A. P. H. J. Schenning, R. P. Sijbesma and E. W. Meijer, Chem. Rev., 2009, 109, 5687 CrossRef PubMed; (d) M. Burnworth, L. Tang, J. R. Kumpfer, A. J. Duncan, F. L. Beyer, G. L. Fiore, S. J. Rowan and C. Weder, Nature, 2011, 472, 334 CrossRef CAS PubMed; (e) E. A. Appel, J. del Barrio, X. J. Loh and O. A. Scherman, Chem. Soc. Rev., 2012, 41, 6195 RSC; (f) E. Elacqua, D. S. Lye and M. Weck, Acc. Chem. Res., 2014, 47, 2405 CrossRef CAS PubMed; (g) L. Yang, X. Tan, Z. Wang and X. Zhang, Chem. Rev., 2015, 115, 7196 CrossRef CAS; (h) A. Winter and U. S. Schubert, Chem. Soc. Rev., 2016, 45, 5311 RSC.
- (a) S. Burattini, B. W. Greenland, D. Chappell, H. M. Colquhoun and W. Hayes, Chem. Soc. Rev., 2010, 39, 1973 RSC; (b) T. Aida, E. W. Meijer and S. I. Stupp, Science, 2012, 335, 813 CrossRef CAS; (c) N. L. Strutt, D. Fairen-Jimenez, J. Iehl, M. B. Lalonde, R. Q. Snurr, O. K. Farha, J. T. Hupp and J. F. Stoddart, J. Am. Chem. Soc., 2012, 134, 17436 CrossRef CAS; (d) J. Tian, T.-Y. Zhou, S.-C. Zhang, S. Aloni, M. V. Altoe, S.-H. Xie, H. Wang, D.-W. Zhang, X. Zhao, Y. Liu and Z.-T. Li, Nat. Commun., 2014, 5, 5574 CrossRef CAS; (e) R. Dong, Y. Zhou, X. Huang, X. Zhu, Y. Lu and J. Shen, Adv. Mater., 2015, 27, 498 CrossRef CAS.
- (a) E. Winfree, F. Liu, L. A. Wenzler and N. C. Seeman, Nature, 1998, 394, 539 CrossRef CAS; (b) J. D. Hartgerink, E. Beniash and S. I. Stupp, Science, 2001, 294, 1684 CrossRef CAS; (c) X. Yan, F. Wang, B. Zheng and F. Huang, Chem. Soc. Rev., 2012, 41, 6042 RSC; (d) S.-L. Li, T. Xiao, C. Lin and L. Wang, Chem. Soc. Rev., 2012, 41, 5950 RSC; (e) X. Yan, D. Xu, X. Chi, J. Chen, S. Dong, X. Ding, Y. Yu and F. Huang, Adv. Mater., 2012, 24, 362 CrossRef CAS.
- (a) R. J. Wojtecki, M. A. Meador and S. J. Rowan, Nat. Mater., 2011, 10, 14 CrossRef CAS; (b) C.-H. Wong and S. C. Zimmerman, Chem. Commun., 2013, 49, 1679 RSC; (c) X. Ma and H. Tian, Acc. Chem. Res., 2014, 47, 1971 CrossRef CAS; (d) X.-Y. Hu, T. Xiao, C. Lin, F. Huang and L. Wang, Acc. Chem. Res., 2014, 47, 2041 CrossRef CAS; (e) P. Wei, X. Yan and F. Huang, Chem. Soc. Rev., 2015, 44, 815 RSC; (f) D. S. Kim, J. Chang, S. Leem, J. S. Park, P. Thordarson and J. L. Sessler, J. Am. Chem. Soc., 2015, 137, 16038 CrossRef CAS; (g) Z. Zhou, X. Yan, T. R. Cook, M. L. Saha and P. J. Stang, J. Am. Chem. Soc., 2016, 138, 806 CrossRef CAS.
- P. Wei, X. Yan, T. R. Cook, X. Ji, P. J. Stang and F. Huang, ACS Macro Lett., 2016, 5, 671 CrossRef CAS.
- A. Goujon, G. Mariani, T. Lang, E. Moulin, M. Rawiso, E. Buhler and N. Giuseppone, J. Am. Chem. Soc., 2017, 139, 4923 CrossRef CAS PubMed.
- (a) T. Ogoshi, S. Kanai, S. Fujinami, T.-A. Yamagishi and Y. Nakamoto, J. Am. Chem. Soc., 2008, 130, 5022 CrossRef CAS; (b) D. Cao, Y. Kou, J. Liang, Z. Chen, L. Wang and H. Meier, Angew. Chem., Int. Ed., 2009, 48, 9721 CrossRef CAS; (c) C. Li, Chem. Commun., 2014, 50, 12420 RSC; (d) T. Ogoshi, T.-A. Yamagishi and Y. Nakamoto, Chem. Rev., 2016, 116, 7937 CrossRef CAS; (e) S. Dasgupta and P. S. Mukherjee, Org. Biomol. Chem., 2017, 15, 762 RSC.
- (a) M. Xue, Y. Yang, X. Chi, Z. Zhang and F. Huang, Acc. Chem. Res., 2012, 45, 1294 CrossRef CAS; (b) H. Zhang and Y. Zhao, Chem.–Eur. J., 2013, 19, 16862 CrossRef CAS; (c) N. L. Strutt, H. Zhang, S. T. Schneebeli and J. F. Stoddart, Acc. Chem. Res., 2014, 47, 2631 CrossRef CAS; (d) Q. Wang, M. Cheng, Y. Zhao, L. Wu, J. Jiang, L. Wang and Y. Pan, Chem. Commun., 2015, 51, 3623 RSC; (e) Q. Wang, M. Cheng, Y. Cao, J. Jiang and L. Wang, Acta Chim. Sin., 2016, 74, 9 CrossRef CAS; (f) Y. Wang, G. Ping and C. Li, Chem. Commun., 2016, 52, 9858 RSC.
- (a) Z.-Y. Li, Y. Zhang, C.-W. Zhang, L.-J. Chen, C. Wang, H. Tan, Y. Yu, X. Li and H.-B. Yang, J. Am. Chem. Soc., 2014, 136, 8577 CrossRef CAS; (b) C.-L. Sun, J.-F. Xu, Y.-Z. Chen, L.-Y. Niu, L.-Z. Wu, C.-H. Tung and Q.-Z. Yang, Chin. Chem. Lett., 2015, 26, 843 CrossRef CAS; (c) K. Yang, Y. Pei, J. Wen and Z. Pei, Chem. Commun., 2016, 52, 9316 RSC; (d) C. Sathiyajith, R. R. Shaikh, Q. Han, Y. Zhang, K. Meguellati and Y.-W. Yang, Chem. Commun., 2017, 53, 677 RSC; (e) J. Murray, K. Kim, T. Ogoshi, W. Yao and B. C. Gibb, Chem. Soc. Rev., 2017, 46, 2479 RSC.
- (a) X.-Y. Hu, P. Zhang, X. Wu, W. Xia, T. Xiao, J. Jiang, C. Lin and L. Wang, Polym. Chem., 2012, 3, 3060 RSC; (b) X.-Y. Hu, X. Wu, Q. Duan, T. Xiao, C. Lin and L. Wang, Org. Lett., 2012, 14, 4826 CrossRef CAS; (c) X. Wang, K. Han, J. Li, X. Jia and C. Li, Polym. Chem., 2013, 4, 3998 RSC; (d) P. Chen, J. H. Mondal, Y. Zhou, H. Zhu and B. Shi, Polym. Chem., 2016, 7, 5221 RSC.
- S. Wang, Y. Wang, Z. Chen, Y. Lin, L. Weng, K. Han, J. Li, X. Jia and C. Li, Chem. Commun., 2015, 51, 3434 RSC.
- L. Shangguan, H. Xing, J. H. Mondal and B. Shi, Chem. Commun., 2017, 53, 889 RSC.
- H. Hofmeier, A. El-ghayoury, A. P. H. J. Schenning and U. S. Schubert, Chem. Commun., 2004, 318 RSC.
- (a) A. T. ten Cate and R. P. Sijbesma, Macromol. Rapid Commun., 2002, 23, 1094 CrossRef CAS; (b) S.-L. Li, T. Xiao, Y. Wu, J. Jiang and L. Wang, Chem. Commun., 2011, 47, 6903 RSC; (c) X. Yan, T. R. Cook, J. B. Pollock, P. Wei, Y. Zhang, Y. Yu, F. Huang and P. J. Stang, J. Am. Chem. Soc., 2014, 136, 4460 CrossRef CAS; (d) L. Wang, Z.-L. Gong, S.-Y. Li, W. Hong, Y. W. Zhong, D. Wang and L.-J. Wan, Angew. Chem., Int. Ed., 2016, 55, 12393 CrossRef CAS; (e) Q. Wang, M. Cheng, J.-L. Jiang and L.-Y. Wang, Chin. Chem. Lett., 2017, 28, 793 CrossRef CAS.
- (a) X. Shu, S. Chen, J. Li, Z. Chen, L. Weng, X. Jia and C. Li, Chem. Commun., 2012, 48, 2967 RSC; (b) J. Wu, S. Sun, X. Feng, J. Shi, X.-Y. Hu and L. Wang, Chem. Commun., 2014, 50, 9122 RSC; (c) X. Wu, M. Ni, W. Xia, X.-Y. Hu and L. Wang, Org. Chem. Front., 2015, 2, 1013 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic procedures, characterizations, NMR spectra, and the calculation of binding constants. See DOI: 10.1039/c7ra05351g |
| This journal is © The Royal Society of Chemistry 2017 |