Open Access Article
Open Access ArticleInterrogation of drug effects on HeLa cells by exploiting new AFM mechanical biomarkers†
Xiaoling Yun‡
ab,
Mingjie Tang‡b,
Zhongbo Yangb,
Jonathan J. Wilkschc,
Peng Xiud,
Haiyang Gaoae,
Feng Zhang*a and
Huabin Wang *bcf
*bcf
aSchool of Life Science, Inner Mongolia Agricultural University, Hohhot 010018, China. E-mail: fengzhang1978@hotmail.com
bChongqing Key Laboratory of Multi-Scale Manufacturing Technology, Chongqing Institute of Green and Intelligent Technology, Chinese Academy of Sciences, Chongqing 400714, China. E-mail: wanghuabin@cigit.ac.cn
cDepartment of Microbiology and Immunology, University of Melbourne, Parkville, Victoria 3010, Australia
dDepartment of Engineering Mechanics, Soft Matter Research Center, Zhejiang University, Hangzhou 310027, China
eDepartment of Biomedical Engineering, School of Basic Medical Sciences, Guangzhou Medical University, Guangzhou 511436, China
fKey Laboratory of Interfacial Physics and Technology, Chinese Academy of Sciences, Shanghai 201800, China
First published on 12th September 2017
Abstract
New mechanical biomarkers were discovered and used to investigate drug effects on HeLa cells. HeLa cells were measured using advanced atomic force microscopy (AFM) techniques before and after treatment with an anticancer drug (docetaxel, a microtubule polymerizer) and the biomechanical properties of cells were analyzed using theoretical models. The biomechanical results show that the length of the surface brush, the factor of viscosity, and the adhesion work of untreated HeLa cells reduced from 920 nm, 0.72 and 4.12 × 10−16 N m to 673 nm, 0.52 and 5.37 × 10−17 N m for treated cells, respectively. In order to reveal the underlying mechanisms of these changes, transmission electron microscopy (TEM) and western blotting assays were employed to characterize structural changes of the cells. Compared to the untreated cells, TEM images confirmed a thinner brush layer on the surface of treated cells while the western blotting of tubulin extracted from the cells indicated an increased microtubule concentration for the treated cells. These changes are consistent with previous studies on the influence of docetaxel on cells. Based on the above results, we conclude that docetaxel can lead to an increased microtubule network in HeLa cells, a thinner brush layer on HeLa cell surfaces and that the length of surface brush, the factor of viscosity, and the adhesion work can be exploited as effective mechanical biomarkers to evaluate drug effects on HeLa cells. This work demonstrates a new AFM-based technique that can be exploited to assess drug action on cells and is promising for the application of drug screening.
Introduction
The discovery of drugs has made many diseases, particularly acute disorders, treatable or manageable, resulting in improvements in quality of life and increased life expectancy.1,2 However, the rising costs associated with drug discovery and evaluation has meant that innovations in this industry are urgently needed for continual development.1 Many approved drugs target cell membrane proteins and/or cytoskeletons (actin filaments, microtubules and intermediate filaments).3,4 Therefore, evaluation of the influence of drugs on cell properties is of great importance for designing and screening drug candidates, as well as for understanding the biological processes occurring in cells upon drug exposure. Investigating drug–cell interactions from the viewpoint of cell mechanics provides a novel way to evaluate drugs.5 Indeed, in recent years it has been widely recognized that cell mechanics are closely related to cell structures, and determine many biological processes and cellular functions.6,7 Therefore, an evaluation of mechanical biomarkers of cells following drug treatment is an important step for drug discovery and evaluation, because useful information regarding the efficacy and safety of the drugs can be obtained according to the change of biomarkers.5Atomic force microscopy (AFM) is a powerful tool for high-resolution imaging and mechanical measurement of living cells in near-physiological conditions, and enables systematic, in situ characterization of the mechanisms governing drug–cell interactions at the nanometer scale.11 There has been an increasing appreciation that cell mechanics can serve as an effective, label-free biomarker for the indication of cell state.8–10 In particular, AFM-based mechanical biomarkers are thought to be very promising in assisting drug discovery from the perspective of biomechanics, compared to the traditional biochemical approaches such as western blotting, X-ray crystallography and radioimmunoassay.5
The Young's modulus of the cell obtained from the Hertz–Sneddon model has often been used as a mechanical biomarker of cell state in drug assays. The drug action can be assessed by monitoring the cell elastic (Young's modulus) change following drug treatment.4,9,10 Rotsch et al.4 found that both cytochalasin and latrunculin can cause a significant decrease in cell Young's modulus because the two drugs can disaggregate the actin filaments of fibroblasts; and Fang et al.10 revealed that N-methyl-D-aspartate (NMDA) can result in an increased Young's modulus of human neuroblastoma SH-SY5Y cells due to the binding of NMDA to the NMDA receptors on the cell membrane, which eventually triggers the opening of ligand-gated ion channel and facilitates an influx of Ca2+ into the cells. From these studies, it is evident that Young's modulus can act as a mechanical biomarker for cytoskeleton changes. Despite its active role in the assessment of drug actions, the Hertz–Sneddon model is based on a number of assumptions in the analysis of cell Young's modulus. More specifically, the cells have been considered as a kind of homogenetic, isotropic and linearly elastic material with an infinite thickness, a smooth surface and an axisymmetry and infinitesimal deformation during compression.12,13 Consequently, the Young's modulus of cells has its limitations (like depth dependence), which impedes its potential applications as an effective mechanical biomarker to assess cell state.14
In terms of structures, the majority of cells have a brush layer (various membrane protrusions, corrugations and glycocalyx),14–16 cell membranes, cytoskeleton, cytosol and various internal organelles.17 Because such complicated structures can undoubtedly endow cells a variety of other mechanical-related properties besides the Young's modulus, such as brush properties, surficial adhesion and viscosity of cells, effective mechanical biomarkers other than Young's modulus could also be potentially significant for understanding cell states, especially for in vitro evaluation of drug effects on cells. Recently, we investigated the mechanical properties and biophysical roles of hierarchical structures (such as capsular polysaccharides, fimbriae, cell wall and cytoplasm) of prokaryotic (bacterial) cells using both AFM nanomechanics and physical theoretical models.18–20 In the present work, we aimed to explore new mechanical biomarkers, other than the Young's modulus, of eukaryotic cells for drug assays. The new mechanical biomarkers include the length of surface brush, the factor of viscosity, and the adhesion work.
HeLa cells have often been used as eukaryotic-cell models to investigate cell–drug interactions.21–25 Luo et al.21 found that selenium nanoparticles can influence cell morphology based on AFM observation. Kim et al.22 investigated the effect of paclitaxel on HeLa cells using AFM and found that surface roughness of cells is increased. In addition, Zou et al.25 found the inhibiting effects of both docetaxel and oxaliplatin on the proliferation of HeLa cells occur in a dose-dependent manner. However, until now only limited knowledge is available on the mechanical information regarding HeLa cell–drug interactions.
Docetaxel is a typical anticancer drug and its cytotoxicity on HeLa cells has been investigated by using conventional biological methods.24–26 In order to obtain an in-depth understanding of HeLa cell–drug interactions for the ultimate purpose of drug evaluation, we investigated HeLa cell–docetaxel interactions based on advanced AFM nanomechanics. Very interestingly, we found new mechanical biomarkers that can be used to effectively evaluate the drug effects, which was further confirmed by transmission electron microscopy (TEM) imaging of the cells and western blotting of the β-subunit of tubulin extracted from the cells. The biomarkers discovered through AFM-based nanomechanical measurements include: (i) the length of cell surface brush, (ii) the factor of cell viscosity, and (iii) the cell adhesion work. The results show that these three physical parameters of the docetaxel-treated HeLa cells are significantly smaller than the untreated controls, indicating that such parameters can be used as mechanical biomarkers for the assessment of docetaxel effects on HeLa cells. The technique demonstrated in this work could also be potentially used to evaluate other cell–drug interaction systems.
Experimental section
Cell culture
HeLa cells (CHI Scientific Incorporation, China) were cultured in cell culture media including Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS, 10%), penicillin (100 units per mL), and streptomycin (100 units per mL) in 25 cm2 tissue culture flasks (Corning Inc., New York, USA) in a 5% CO2 humidified atmosphere at 37 °C. All chemicals used in the culture media were purchased from Thermo Fisher Scientific (China branch), Shanghai, China. The culture media was changed every other day until the cells were ∼90% confluent (3–4 days) for subsequent subculture. Afterwards, the media was removed and cells were washed with 2 mL of phosphate buffer solution (PBS, pH 7.4, Gibco), treated with 2 mL 0.25% trypsin–EDTA (Gibco) for 2 minutes, and neutralized with 4 mL cell culture media. The culture solution containing cells was gently mixed with a pipette several times until ∼95% cells were removed from the wall of the flask. The cell solution was transferred to a 15 mL tube (Corning Inc., New York, USA) and centrifuged at 1000 rpm for 3 minutes. The pellets were re-suspended in 1 mL culture media and the prepared cells were ready for use in the following experiments.Cell viability test, optical microscopy observation and cell sample preparation for AFM measurements
Docetaxel was obtained from Southwest Hospital, China, which was first dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 mg mL−1 and further diluted to 10, 20, 30, 40 and 50 μg mL−1 in cell culture media.The viability and proliferation of the cells were analyzed using a commercial cell counting kit (7Sea Cell Counting Kit (CCK-8), 7Sea Biotech, Shanghai, China), according to the manufacturer's instructions. More details can be found in the ESI.† The inhibition rate was calculated based on the measured absorbance value (OD450) of the wells, according to the formula:
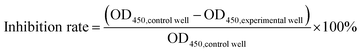 | (1) |
Following the CCK-8 assay, the plates were further examined using an inverted optical microscope (Olympus Corporation, Tokyo, TH4-200, Japan) to observe the cell morphology. To prepare cell samples for AFM measurements, HeLa cell solution prepared as described in “Cell Culture” was diluted in the cell culture media to a density of 1 × 105 cells per mL and 5 mL of the cell solution was seeded in a Petri-dish (60 mm × 15 mm, Corning Inc., New York, USA) coated with poly-L-lysine (P2100, 5 mg mL−1 in Milli-Q water (18.2 MΩ cm−1), Solarbio, Beijing, China) and cultured for 12 h. The media was then removed and the Petri-dish was washed with 2 mL PBS before the addition of 4 mL cell culture media with 0.5% DMSO (control group) or 4 mL docetaxel solution (50 μg mL−1, experimental group). Cells were then incubated for a further 12 h at 37 °C at 5% CO2. Finally, the culture media was removed from the Petri-dish and replaced with 5 mL of PBS, and the Petri-dish was used immediately for AFM experiments. We note that poly-L-lysine is a widely used biocompatible material for facilitating the attachment of cells on the substrate for AFM measurement and did not show an obvious influence on cell viability, as reported by others.27
AFM imaging and force measurement
AFM experiments were performed at room temperature using the Dimension Edge Instrument (Bruker Nano Surfaces, Santa Barbara, USA). Soft AFM cantilevers (MLCT, Bruker Nano Inc., Camarillo, USA) with a nominal spring constant of 0.03 N m−1 were used. The sensitivity and spring constant of the cantilevers were calibrated against the surface of a Petri-dish in PBS buffer using the AFM software.18,19 Contact mode AFM was used to image cells with an optimized scan rate (0.2–0.5 Hz) and minimized load. To provide a statistical analysis of the cell height, ∼30 cells for each type of the untreated and docetaxel-treated HeLa cells were measured from three independent experiments. The data are presented as the averaged value ± standard deviation. The statistical significance (p-value) of the data was analyzed by a one-way ANOVA test using Origin 8.5 software (OriginLab Co., Northampton, USA) and P < 0.05 was considered statistically significant.An optical microscope coupled with the AFM system was used to initially image cell samples. The AFM tip was then accurately positioned on the apex of an individual target cell. To ensure that the data obtained could be analyzed with statistics, at least 10 force curves were collected at a loading/unloading speed of 1 μm s−1 with a threshold loading force of ∼3 nN on each cell, and more than 20 cells from at least three independent experiments were measured. Under this threshold loading force, the cell structure appeared unaffected, since abrupt drops or spikes in the approach force curves were not observed. It has been demonstrated that if a cell was punctured by the AFM tip, abrupt drops or spikes should appear in the approach force curve.28 The collected raw force curves were converted into force–distance curves by correcting cantilever bending through removing the cantilever bending from the piezo displacement using a self-written procedure based on Igor Pro (Version 6.04, Wavemetrics Inc., Lake Oswego, USA).
Force data analysis
To analyze the force profiles, it is essential to determine the point of contact, i.e., zero tip-sample distance. Under the current experimental conditions, the contact is defined as the piezo displacement where the cantilever starts to deflect upward. The force–distance curve includes two curves: the approach curve and retraction curve. Negative distance values in the approach curve indicates that the tip pressed into the cell, while in the retraction curve it indicates the tip is still in contact with the cell. The initial part of the approach curve was fitted to a modified Pincus theory (eqn (2)), from which the length of the cell surface brush can be obtained. The Pincus theory was originally proposed for the compression of a polymer brush29 and later modified to fit the surface polymers of a bacterial cell.18,19,30 The modified Pincus theory can be written as:
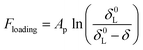 | (2) |
From the retraction curve, the adhesion work of the tip–cell interaction can be calculated by integrating the area between the negative portion of the retraction curve and the zero force line.31 In addition, the hysteresis between the approach and retraction curves indicating energy dissipation during the indentation measurement was analyzed by introducing the factor of viscosity, φ, a parameter characterizing the relative viscous/elastic behavior of materials under external stresses (eqn (3)):32,33
| φ = viscous energy/(viscous energy + elastic energy) | (3) |
Igor Pro (Version 6.04, Wavemetrics Inc., Lake Oswego, USA) and Matlab (Version R2010a, MathWorks Inc., Natick, USA) procedures were written for the analysis of force curves to extract physical parameters regarding the cell surface brush, the adhesion work and the factor of viscosity. Origin 8.5 was used for the statistical analysis of the brush length, the factor of viscosity and the adhesion work, which were fitted to a Gaussian function. The data are presented as the fitted value ± standard deviation. The significance of the data was analyzed by one-sample t-test analysis of hypothesis testing and P < 0.05 was considered statistically significant.
TEM imaging
HeLa cells prepared as described in “Cell Culture” were cultured in 25 cm2 tissue culture flasks with 5 mL of culture media containing 0.5% DMSO (control group) or docetaxel solution (50 μg mL−1, experimental group) for 12 h. Afterwards, the cells were collected by washing, trypsinization, neutralization and centrifugation, as described above in “Cell culture”. The cell precipitates from both the control and docetaxel-treated samples were then fixed with 2.5% glutaraldehyde in 2 mL of 0.1 M PBS buffer for 4 h at 4 °C and post-fixed in 1% osmium tetraoxide at 4 °C. The samples were washed with PBS buffer, dehydrated with a graded series of ethyl alcohol (50%, 70%, and 90%), embedded in resin, sectioned using ultramicrotome and stained with uranyl acetate and lead citrate.23 The stained samples were then viewed under TEM (JEOL-1400, JEOL Ltd., Tokyo, Japan) at a voltage of 80 kV. A magnification of 4000× was used in imaging.Western blot assay
HeLa cells prepared as described in “Cell culture” were cultured in cell culture media containing 0.5% DMSO (control group) or media containing docetaxel (50 μg mL−1, experimental group) for 12 h and then harvested as described above in “Cell culture” and washed three times with PBS buffer. Detailed information regarding the western blot assay can be found in the ESI.†Results and discussion
Cell viability tests
The effect of docetaxel on the viability of HeLa cells was tested using the CCK-8 colorimetric assay, as detailed in “Experimental section”. The inhibition rate of HeLa cells was calculated by comparing the OD450 of cells exposed to 0.5% DMSO (reference control) to cells exposed to varying concentrations of docetaxel (10, 20, 30, 40 and 50 μg mL−1). As shown in Fig. 1, the inhibition rate of HeLa cells increased with increasing concentration of docetaxel, ranging from ∼27% at 10 μg mL−1 to ∼84% at 50 μg mL−1. The docetaxel concentrations used, and the inhibition rate observed in this experiment is very similar to that of Zou et al., who tested the viability of HeLa cells upon treatment with docetaxel using a MTT assay method.25Optical microscopy of HeLa cell morphology
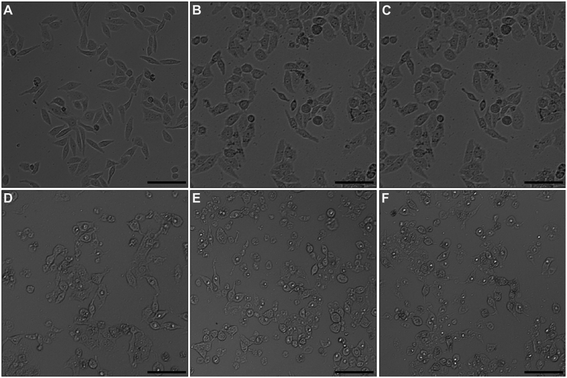
Optical microscopic images of docetaxel-treated and untreated HeLa cells are shown in Fig. 2. Untreated HeLa cells displayed a normal, spindle-like morphology (Fig. 2A). However, docetaxel-treated HeLa cells took on a more rounded shape and the number of adherent cells gradually decreased with increasing docetaxel concentration (Fig. 2B–F). These results indicated that docetaxel can induce changes in cell morphology and reduced cell growth and/or adherence in a dose-dependent manner, which is consistent with other results reported on the effects of docetaxel on HeLa cells,25 and is also similar to that of HeLa cells exposed to, selenium nanoparticles (a potential anticancer drug).21AFM study on single cell morphology
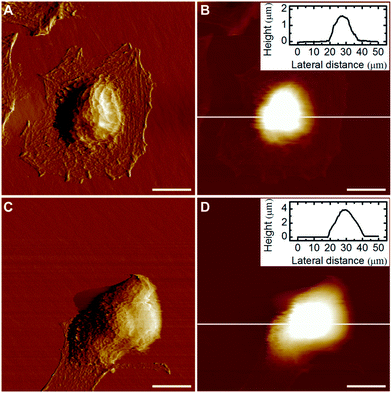
An in-depth study on the morphology of individual HeLa cells was performed using AFM imaging in a liquid environment. The general shape of both the control and drug-treated cells was consistent with their corresponding optical images (Fig. 3). However, AFM images can provide other details on cell morphology compared with the optical microscopy. For example, the lamellipodia can be clearly observed surrounding the untreated cell (Fig. 3A), however is greatly reduced in the docetaxel-treated cell (Fig. 3C). In addition, the apparent height of the docetaxel-treated cell is significantly higher than the untreated HeLa cell, as shown in Fig. 3B and D, respectively. It is possible that the height of cells measured by AFM is smaller than the true height of cells because the AFM tip–cell and substrate–cell interactions can deform the cells, resulting in a lower measured height. Nonetheless, the apparent height measured by AFM imaging is still a very useful parameter to evaluate the influence of docetaxel on the morphology of HeLa cells, given that imaging conditions used in our experiments, including the substrate, tip and buffer and imaging parameters, were highly consistent. By measuring ∼30 cells for each type of the treated and untreated HeLa cells from three independent experiments, it was found that the average height of docetaxel-treated HeLa cells was 3.73 ± 0.53 μm, significantly higher (at the 0.05 level) than the untreated HeLa cells with an average height of 2.43 ± 0.59 μm. These results indicate that docetaxel has effects on HeLa cells and dramatically changed the cell structure, as evidenced by the morphological changes of increased height and reduced lamellipodia. The disappearance of lamellipoda of HeLa cells has also been observed by AFM for cells treated with selenium nanoparticles.21Biomechanical study of HeLa cells
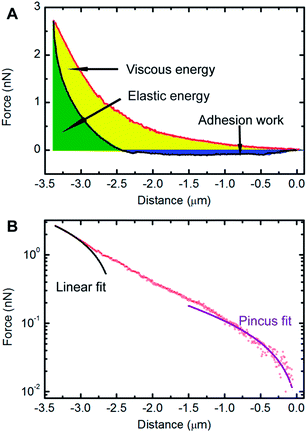
The benchmark test concentration (BTC) of a drug is the concentration beyond which there is no further significant response, and can be used as an appropriate way to evaluate drug efficacy.25 In our experimental conditions, the BTC of docetaxel for HeLa cells was 50 μg mL−1 according to a pre-test. Therefore, a concentration of 50 μg mL−1 was chosen to treat HeLa cells for subsequent AFM studies. These experiments involved generating and analyzing force–distance curves to extract biomechanical parameters of cells. A typical force–distance curve collected on a HeLa cell is shown in Fig. 4A, from which a hysteresis between the approach and retraction curves is observed, which is common for soft materials due to energy dissipation.34 The yellow area of the curve (the region between the approach curve, retraction curve and zero-force line) represents the viscous energy, while the green area of the curve (the region between the positive force portion of the retraction force curve and the zero-force line) represents the elastic energy.32 It is clear that the cell cannot be modeled as an ideal elastic body due to the viscous deformation observed. The factor of viscosity, φ, as described in “Experimental section” was used to evaluate the deformation properties of the cell. In addition, the adhesion between the AFM tip and cell surface existed during the withdrawing process of the tip, which contributes to the measured adhesion work. The adhesion work was calculated by the integration of the area between the zero-force line and the negative force section of the retraction force curve. As shown in Fig. 4B, the approach curve was fitted to the modified Pincus theory (see “Force data analysis” in “Experimental section”), from which the properties of the cell brush can be extracted. The brush layer was estimated as the distance in the force–distance curve where the modified Pincus theory no longer fits the initial part of the approach curve. In this example, the region between 0 to −1.01 μm was well-fitted to the modified Pincus theory, which estimated that the length of cell brush was 1.01 μm. Detailed information on the data fitting can be found in the ESI.†The results of the extracted physical parameters from more than 200 force curves analyzed for both the untreated (control) or docetaxel-treated cells are summarized in Table 1. It was observed that the values of surface brush length, factor of viscosity and adhesion work of untreated cells were significantly higher than docetaxel-treated cells. The brush length, factor of viscosity and adhesion work of untreated cells reduced from 920 nm, 0.72 and 4.12 × 10−16 N m to 673 nm, 0.52 and 5.37 × 10−17 N m for treated cells, respectively. Based on the above results, it is evident that the properties of HeLa cells have been substantially changed following 50 μg mL−1 docetaxel treatment.
| Parameter | Control | Treated | *p value |
|---|---|---|---|
| Brush length (nm) | 920 ± 116 | 673 ± 56 | <0.05 |
| Factor of viscosity | 0.72 ± 0.02 | 0.52 ± 0.01 | <0.05 |
| Adhesion (N m) | (4.12 ± 0.13) × 10−16 | (5.37 ± 0.43) × 10−17 | <0.05 |
Cell surface brush changes
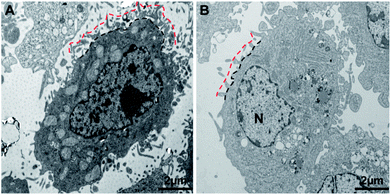
From the nanomechanical measurements, we observed that the cell surface brush length was significantly reduced by ∼250 nm following docetaxel treatment. These results were further confirmed by TEM images of the cells with or without docetaxel treatment (Fig. 5). The TEM images showed that the treated cells (Fig. 5B) displayed shorter and less abundant brush projections on the cell surface and also showed a smaller nucleus (N) and an absent/fragmented nucleolus (NL), compared to the untreated cells (Fig. 5A). The estimated thicknesses for the untreated (n = 15) and treated cells (n = 15) from three independent experiments are 980 ± 170 nm and 650 ± 130 nm, respectively. The TEM observations are highly consistent with the report by Xiao et al., who observed that in comparison with the control, HeLa cells treated with soyasaponin (an antitumor agent) have smaller nuclei, fragmented nucleoli and microvilli with substantially decreased length and abundance.23 The high consistency between the TEM imaging and AFM mechanical measurement strongly suggests that the brush length obtained from the modified Pincus theory fitting to the approach force curve can be used as an effective mechanical biomarker to detect the effect of docetaxel on HeLa cells. Since HeLa cells treated by docetaxel have reduced brush length and abundance compared to untreated cells (Fig. 5), it is reasonable to observe that the measured adhesion work is also reduced (Table 1), since less brush–tip interactions will occur in the process of withdrawing the AFM tip from the treated cell.Cytoskeletal changes
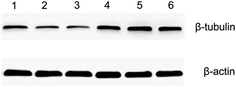
Both the optical microscopy and AFM images showed that the morphology of HeLa cells changed from a spindle-like shape to a rounded structure following exposure to docetaxel. AFM images also showed that the height of HeLa cells increased and lacked almost all lamellipodia after docetaxel treatment (Fig. 3). These changes could suggest that the internal structure of HeLa cells had changed, since cell morphology is influenced by the cytoskeleton, while the lamellipodia are determined by intracellular actin molecules.6,35It has been reported that docetaxel can bind to the β-subunit of tubulin in tumor cells and enhance microtubule polymerization, resulting in cell mitosis inhibition and ultimately cell apoptosis.26 Our western blot results show that the amount of β-subunit of tubulin is increased (the bands marked by 4, 5 and 6 in Fig. 6) after the cells were treated with docetaxel, consistent with immunofluorescent microscopy studies of microtubule changes upon the addition of docetaxel to HeLa cells.36 The results suggest that the amount of microtubules increased in HeLa cells treated with docetaxel. This could explain the reduced factor of viscosity obtained from AFM nanomechanics (Table 1), in that an increased amount of microtubules could increase the elasticity of the HeLa cells and decrease the viscous deformation, relatively, resulting in a lower factor of viscosity for docetaxel-treated HeLa cells. The immediate implication of this result is that the factor of viscosity could be used to predict internal structural changes of HeLa cells treated by docetaxel.
Spedden et al. have reported that the elasticity of living cells increased 35% after the addition of Taxol which is a well studied drug with known microtubule stabilizing effects in cells.37 By combining with fluorescence microscopy, they confirmed that compared to the untreated cells, the concentration/network of microtubules in the treated cells increased and was positively related to the local elasticity of the cells. In our experiments, we also confirmed that the addition of docetaxel increased the concentration of microtubules using the western blot assay. In addition, from the definition of the factor of viscosity, it is not difficult to understand that the higher the cellular elasticity (indicating higher elastic energy) the lower the factor of viscosity. As can be seen from Table 1, compared to the untreated cells, the factor of viscosity of the docetaxel-treated cells decreased, which means the elastic energy was more predominant. Therefore, our results are consistent with the work by Spedden et al. However, it is necessary to point out that although our evidence shows that the increase of microtubule concentration contributed to the decrease of the factor of viscosity, other factors such as nucleus change might also involve more or less in this change, which needs to be further explored.
Nanomechanical biomarkers
Based on above results and discussions, we observed that the brush length, the factor of viscosity and the adhesion work of HeLa cells exposed to docetaxel are significantly different from the cells without exposure. As confirmed by the TEM and western blot experiments, these physical parameters obtained via AFM mechanical measurements are related to the cytoskeleton and cell surface brush, which could be used as effective nanomechanical biomarkers to detect the efficacy of docetaxel on HeLa cells. Although the modified Pincus theory has in the past been used to investigate prokaryotic (bacterial) cells, it has not been used to study the surface brush of eukaryotic cells.18,19 The surface brush of normal and cancerous eukaryotic cell lines has been studied using another polymer brush theory, but is effective on eukaryotic cells only by employing a colloid probe, which is inherently less capable of the high spatial and force resolution that sharp AFM tips can offer,14,15 as demonstrated in the present study. The factor of viscosity has often been used to evaluate inorganic or organic materials, but has not been widely used to study the mechanical properties of eukaryotic cells. We found that the factor of viscosity is a very useful physical parameter for cell nanomechanics. In addition, although the adhesion work has been used to differentiate normal and cancerous cells, it has not been exploited to evaluate the effects of anticancer drugs on cells to our knowledge.38 Consequently, we have demonstrated a new and meaningful technique with promising applications in drug screening. It needs to keep in mind that cells are viscoelastic materials the operational parameters in the force measurement such as the loading/unloading rate of the tip and the dwell time for the tip on the cell surface could influence the factor of viscosity and probably the adhesion work.39In this work, we mainly focused on the establishment a label-free biomechanical markers-based technique to assess the action of docetaxel on HeLa cells, so we only compared the mechanical properties of the untreated cells with the cells treated by the drug for 12 h and did not investigate the biomechanical changes with varying exposure times. However, from previous studies we know that for a docetaxel dosage ≤ 50 μg mL−1, the inhibitory action of docetaxel on HeLa cells increased with the exposure time for at least up to 72 hours.40 Therefore, it is very likely that within a docetaxel dosage ≤ 50 μg mL−1 and a time period ≤ 72 h, the brush length, the factor of viscosity and the adhesion work of the drug treated cells will decrease with the exposure time. In addition, it has been proved that docetaxel could be used as an effective anticancer agent for various cancerous cell lines by inducing cell death through impairing the dynamic of microtubules and promoting the polymerization of microtubules.41 Therefore, the technique demonstrated in this work could be exploited to assess the action of docetaxel on cancerous cells from patients, as far as structural changes occurred in the cells upon drug treatment.
Conclusions
In summary, an AFM force measurement technique was used to investigate the biomechanical properties of HeLa cells with or without treatment by an anticancer drug, docetaxel. It was observed that the brush length, the factor of viscosity and the adhesion work of HeLa cells subjected to docetaxel treatment were reduced, compared to untreated HeLa cells. These changes can be attributed to the influence of docetaxel on the cell skeleton (mainly microtubules) and the cell surface brush, which were shown to be altered by TEM imaging and western blot analysis of tubulin production. The above results indicated that the brush length, the factor of viscosity and the adhesion work measured by AFM could be used as effective mechanical biomarkers for the evaluation of the effects of docetaxel on HeLa cells. To the best of our knowledge, these mechanical biomarkers have not yet been used for drug assays. The technique demonstrated in this work could also be used in other drug–cell interaction systems if the drug causes cytoskeletal changes or cell brush alterations, which is true for many anticancer drugs. For example, we have also interrogated the interactions between curcumin (an anticancer drug) and SH-SY5Y neuronal cells and found that these newly reported mechanical biomarkers can be used to evaluate the cell state (unpublished data). We hope these new nanomechanical biomarkers demonstrated in this study could be exploited to help improve the efficiency of drug screens and discovery of new drugs.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (2016YFC0101002 and 2016YFC0101300), Central Government Supported Key Instrument Program of China (YXGYQ201700136), National Natural Science Foundation of China (11504372, 21407145, 11574268, and 11604332), Chongqing Science & Technology Commission (cstc2013yykfC00007, cstc2014jcyjA10002, cstc2015jcyjA10057 and YJ500061LH1), Chinese Academy of Sciences (R52A500Z10), and Natural Science Foundation of Inner Mongolia Autonomous Region of China (2013MS1121, 2015MS0806, and 2016MS0211).Notes and references
- S. M. Paul, D. S. Mytelka, C. T. Dunwiddie, C. C. Persinger, B. H. Munos, S. R. Lindborg and A. L. Schacht, Nat. Rev. Drug Discovery, 2010, 9, 203–214 CAS
.
- I. Khanna, Drug Discovery Today, 2012, 17, 1088–1102 CrossRef PubMed
.
- C. Zheng, L. Han, C. Yap, Z. Ji, Z. Cao and Y. Chen, Pharmacol. Rev., 2006, 58, 259–279 CrossRef CAS PubMed
.
- C. Rotsch and M. Radmacher, Biophys. J., 2000, 78, 520–535 CrossRef CAS PubMed
.
- M. Li, L. Liu, N. Xi and Y. Wang, Acta Pharmacol. Sin., 2015, 36, 769–782 CrossRef CAS PubMed
.
- D. A. Fletcher and R. D. Mullins, Nature, 2010, 463, 485–492 CrossRef CAS PubMed
.
- D. Di Carlo, J. Lab. Autom., 2012, 17, 32–42 CrossRef CAS PubMed
.
- X. Mao and T. J. Huang, Lab Chip, 2012, 12, 4006–4009 RSC
.
- W. A. Lam, M. J. Rosenbluth and D. A. Fletcher, Blood, 2007, 109, 3505–3508 CrossRef CAS PubMed
.
- Y. Fang, C. Y. Iu, C. N. Lui, Y. Zou, C. K. Fung, H. W. Li, N. Xi, K. K. Yung and K. W. Lai, Sci. Rep., 2014, 4, 7074 CrossRef PubMed
.
- K. J. Van Vliet and P. Hinterdorfer, Nano Today, 2006, 1, 18–25 CrossRef
.
- M. Li, L. Liu, N. Xi and Y. Wang, Chin. Sci. Bull., 2014, 59, 4020–4029 CrossRef
.
- K. D. Costa, Dis. Markers, 2004, 19, 139–154 CrossRef
.
- N. Guz, M. Dokukin, V. Kalaparthi and I. Sokolov, Biophys. J., 2014, 107, 564–575 CrossRef CAS PubMed
.
- S. Iyer, R. Gaikwad, V. Subba-Rao, C. Woodworth and I. Sokolov, Nat. Nanotechnol., 2009, 4, 389–393 CrossRef CAS PubMed
.
- E. Kipps, D. S. Tan and S. B. Kaye, Nat. Rev. Cancer, 2013, 13, 273–282 CrossRef CAS PubMed
.
- D. Zink, A. H. Fischer and J. A. Nickerson, Nat. Rev. Cancer, 2004, 4, 677–687 CrossRef CAS PubMed
.
- H. Wang, J. J. Wilksch and T. Lithgow, Soft Matter, 2013, 9, 7560–7567 RSC
.
- H. Wang, J. J. Wilksch, R. A. Strugnell and M. L. Gee, ACS Appl. Mater. Interfaces, 2015, 7, 13007–13013 CAS
.
- A. Mularski, J. J. Wilksch, H. Wang, M. A. Hossain, J. D. Wade, F. Separovic, R. A. Strugnell and M. L. Gee, Langmuir, 2015, 31, 6164–6171 CrossRef CAS PubMed
.
- H. Luo, F. Wang, Y. Bai, T. Chen and W. Zheng, Colloids Surf., B, 2012, 94, 304–308 CrossRef CAS PubMed
.
- K. S. Kim, C. H. Cho, E. K. Park, M.-H. Jung, K.-S. Yoon and H.-K. Park, PLoS One, 2012, 7, e30066 CAS
.
- J. X. Xiao, G. Q. Huang, C. P. Zhu, D. D. Ren and S. H. Zhang, Toxicol. In Vitro, 2007, 21, 820–826 CrossRef CAS PubMed
.
- N. Tang, Q. Wang, D. Wu, S. Zhang, Y. Zhang and L. Tao, Mol. Med. Rep., 2013, 8, 638–644 CrossRef PubMed
.
- Y. Zou, W. Gou, M. Pei, J. Xiao and X. Liu, J. Med. Coll. PLA, 2010, 25, 204–211 CrossRef CAS
.
- K. Gelmon, Lancet, 1994, 344, 1267–1272 CrossRef CAS
.
- T. Ando and Y. Yonamoto, Appl. Magn. Reson., 2015, 46, 977–986 CrossRef CAS
.
- I. U. Vakarelski, S. C. Brown, K. Higashitani and B. M. Moudgil, Langmuir, 2007, 23, 10893–10896 CrossRef CAS PubMed
.
- G. Subramanian, D. R. M. Williams and P. A. Pincus, Macromolecules, 1996, 29, 4045–4050 CrossRef CAS
.
- F. Gaboriaud, M. L. Gee, R. Strugnell and J. F. L. Duval, Langmuir, 2008, 24, 10988–10995 CrossRef CAS PubMed
.
- F. T. Arce, R. Avci, I. B. Beech, K. E. Cooksey and B. Wigglesworth-Cooksey, Biophys. J., 2004, 87, 4284–4297 CrossRef CAS PubMed
.
- H. Wang, J. J. Wilksch, L. Chen, J. W. H. Tan, R. A. Strugnell and M. L. Gee, Langmuir, 2017, 33, 100–106 CrossRef CAS PubMed
.
- K. E. Bremmell, A. Evans and C. A. Prestidge, Colloids Surf., B, 2006, 50, 43–48 CrossRef CAS PubMed
.
- F. Gaboriaud, B. S. Parcha, M. L. Gee, J. A. Holden and R. A. Strugnell, Colloids Surf., B, 2008, 62, 206–213 CrossRef CAS PubMed
.
- T. J. Mitchison and L. P. Cramer, Cell, 1996, 84, 371–379 CrossRef CAS PubMed
.
- T. Wang, D. Zhu, G. Liu, W. Tao, W. Cao, L. Zhang, L. Wang, H. Chen, L. Mei, L. Huang and X. Zeng, RSC Adv., 2015, 5, 50617–50627 RSC
.
- E. Spedden, J. D. White, E. N. Naumova, D. L. Kaplan and C. Staii, Biophys. J., 2012, 103, 868–877 CrossRef CAS PubMed
.
- S. E. Cross, Y.-S. Jin, J. Tondre, R. Wong, J. Rao and J. K. Gimzewski, Nanotechnology, 2008, 19, 384003 CrossRef PubMed
.
- M. Radmacher, M. Fritz, C. M. Kacher, J. P. Cleveland and P. K. Hansma, Biophys. J., 1996, 70, 556–576 CrossRef CAS PubMed
.
- Y.-L. Zou, J. Feng, W.-L. Gou, M.-L. Pei, F.-L. Fan and Z.-M. Zhang, Acad. J. Xi'an Jiaotong Univ., 2010, 22, 260–264 CAS
.
- H.-V. Héctor, P. José and M.-B. Gema, Cell Cycle, 2007, 6, 780–783 CrossRef PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available: CCK-8 assay, western blot assay and the Pincus theory and data fitting. See DOI: 10.1039/c7ra06233h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |